Abstract
Nanotechnology is playing an increasing role in targeted drug delivery into pathological tissues. Drug-loaded pharmaceutical nanocarriers can be delivered into diseased sites by passive targeting (spontaneous accumulation of nanocarriers in the areas with affected vasculature) or by active targeting (via site-specific ligands attached to the surface of drug-loaded nanocarriers). Subsequent level of targeting requires cellular internalization of nanocarriers and their specific association with certain individual cell organelles. The control over intracellular distribution of pharmaceutical nanocarriers requires effective and non-invasive methods of their visualization inside cells. In an attempt to enhance cellular internalization of pharmaceutical nanocarriers and their association with mitochondria specifically, we have prepared three types of cationic liposomes and investigated their intracellular distribution. The analysis was performed using Raman microspectroscopy in combination with optical microscopy, in order to provide morphological information as well as biochemical signatures of the sample. It was demonstrated that the Raman microscopy allows to evaluate the extent of mitochondrial association depending on the liposome composition.
Keywords: Raman microscopy, cationic liposomes, cellular uptake, intracellular localization
INTRODUCTION
Pharmaceutical nanocarriers are increasingly used in basic medical research as well as in clinical medicine as delivery systems for drugs and imaging agents.1–3 Several nanoparticulates, such as nanospheres, nanocapsules, micelles, liposomes, and dendrimers have been applied to serve as drug carriers, allowing for an enhanced delivery of loaded drugs compared to free drugs. Nano-encapsulation of drugs also prevents premature degradation and excretion as well as circumvents non-specific toxicity.4–7
A biocompatible drug-carrier system is required for in vivo administration, in order to circumvent toxicity due to the delivery system itself. In our research, we have used liposomes, which are artificial phospholipid vesicles with size ranging from 50 to 1000 nm and can be loaded with a variety of drugs.6,8
Efficacy and specificity of drug delivery systems, including liposomes, can be further enhanced with various liposomal modifications with cationic compounds, which augment their internalization and may even increase affinity for specific organelles, depending on the modification type. Among those organelles of interest, lysosomes may be targeted for the treatment of lysosomal storage diseases,9 nuclei - for gene delivery applications,10,11 mitochondria - for initiating apoptosis in target cells.12,13 Major advantages of cationic liposomes are being explored in areas such as genetic therapy, since encapsulation of the genetic material is vastly enhanced. However, there are some drawbacks with the use of cationic liposomes such as non-specificity on a sub-cellular level as well as some inherent cytotoxicity.10,14,15 In lieu of these events, research has been underway to develop novel and enhance current cationic lipid derivatives, which may circumvent these drawbacks.
We have chosen to test three distinct cationic components to modify the composition of the liposomes, in order to annotate, characterize, and distinguish their individual uptake and intracellular dynamics patterns. The three cationic modifications utilized greatly enhance cellular internalization of the nano-carriers. The systems analyzed include 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP)-, stearyl amine (SA)-, and stearyltriphenylphosphonium (STPP)-modified liposomes. Out of the three cationic modification units, STPP was shown to accumulate specifically within the mitochondria-rich regions inside cells.16,17
Visualization of the nano-assemblies and their fate in situ however, commonly require diverse labels or tags to be used for their detection in vitro and in vivo. However, imaging modalities, which rely on extensive labeling procedures, such as fluorescence microscopy, may not be the optimal tool in non-invasive imaging. Thus, to follow the fate of liposomes inside cells, we utilize a biochemical imaging technique, which is based on the vibrational spectroscopy coupled to optical microscopy, and renders a non-invasive chemical information of any system probed within the biological environment, via the identification and annotation of molecular signatures of all components involved.
Vibrational (infrared and Raman) micro-spectroscopy allows for the identification of molecular fingerprints of the functional groups within any biological species. For cells, such fingerprints arise from the functional groups contained within proteins, carbohydrates, nucleic acids, lipids and phospholipids, among others, which constitute basic building blocks of the cell. Vibrational spectra may be obtained by illuminating the specimen either with broadband infrared radiation and monitoring its absorption, or with narrowband visible light from a laser and detecting scattered photons. The latter approach detects inelastically scattered photons and is known as the Raman effect. Here, we have used Raman micro-spectroscopic imaging technique in order to achieve biochemical functional maps of the three cationic liposomal drug-delivery systems in vitro.
Cellular spectra exhibit features originating from peptides, DNA/RNA, lipids, etc., within the lower wavenumber spectral region, termed “fingerprint region,” (750 – 1800 cm−1) as well as the C-H stretching region (2800 – 3100 cm−1) of the spectrum.18 Any biocompatible nanoparticle employed – such as liposomes – would also exhibit vibrational peaks overlapping with signal originating from various intracellular compartments. Hence, in order to increase the sensitivity to the nanoparticle system within the intracellular environment, we exploit the properties of isotopic (deuterium) labeling to produce vibrational signals that do not occur in the un-deuterated compounds. Also, subsequent biochemical contrast may be further achieved with employment of statistical methods of analysis of hyper-spectral datasets, such as spectral un-mixing method of analysis - Vertex Component Analysis (VCA), see discussion below.
EXPERIMENTAL METHODS
Liposome Preparation
Liposomes were prepared from 1,2-Dipalmitoyl-d62-sn-glycero-3-phosphocholine DPPC-d62 and cholesterol (85:15 mol%) by the lipid film hydration method. (DPPC-d62), 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) were obtained from Avanti Polar Lipids, Inc (Alabaster, AL). The dried lipid film prepared from the chloroform solution of DPPC-d62 and cholesterol was hydrated and vortexed with HEPES buffered saline (HBS), pH 7.4, at final lipid concentration of 5 mg/ml. The resulting liposomes were sized through double stacked 200 nm pore size polycarbonate membranes (Nucleopore) using a hand-held extruder (Avanti). Liposomal sizes were on average 175 nm, enumerated in Table 1. When required, 1.5mol % of stearyl amine (SA), or stearyltriphenylphosphonium (STPP) (kindly provided by Dr Hanson, Northeastern University) or 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) (Sigma) was added during liposome preparation.
Table 1.
Characteristics of liposome preparations.
| Formulation | Size (nm), SD | Zeta potential (mV), SD |
|---|---|---|
| STPP-modified Liposomes | 175 ± 12 | 14 ± 1 |
| DOTAP-modified Liposomes | 178 ± 33 | 12 ± 2 |
| SA-modified Liposomes | 172 ± 21 | 13 ± 2 |
Liposome size and size distribution were determined by Coulter N4 MD Submicron Particle Size Analyzer (Beckman-Coulter, Fullerton, CA); the results are given in Table 1. The zeta-potential values of the various liposomal formulations were measured at 25°C in water using the Zeta Plus Particle Analyzer (Brookhaven Instruments Corp, Santa Barbara, CA).
Cell culture
Human HeLa cells (cell line CCL-2, ATCC, Manassas, VA) were grown in 75 cm2 culture flasks (Fisher Scientific) with 7 mL of Dulbecco’s Modified Eagle’s medium (DMEM, ATCC) and 10% fetal bovine serum (FBS, ATCC). Cells were incubated at 37 °C and 5% CO2. Cells were grown on CaF2 windows to avoid background signals from the regular glass slides.
Prior to Raman biochemical analysis, cells were fixed in 10% formalin (Sigma-Aldrich, St. Louis, MO) and subsequently submerged into the phosphate-buffered saline.
Raman Data Acquisition
The cellular images presented in this paper were recorded using a CRM200 confocal Raman microscope (WITec GmbH, Ulm, Germany) employing 488 nm (ca. 40 mW out of fiber, with ~5 mW on sample) excitation wavelength from a solid state laser (Spectral Physics model “Cyan” laser) and a water immersion objective (60x/NA 1.00, WD 2.0 mm), with resolution or step size of 500 nm at 250 ms per pixel. The exciting laser radiation was coupled into a Zeiss microscope through a wavelength-specific single-mode optical fiber. The incident laser beam was collimated via an achromatic lens and passed a holographic band-pass filter before being focused onto the sample through the microscope objective.
The sample was located on a piezo-electrically driven microscope scanning stage with an x,y resolution of ca. 3 nm and a repeatability of ±5 nm and z resolution of ca. 0.3 nm and ±2 nm repeatability. The sample was scanned through the laser focus in a raster pattern at a constant stage speed of fractions of a micrometer per second. The continuous motion prevents sample degradation at the focal point of the laser beam.
Raman backscattered radiation was collected through the microscope objective and passed through a holographic edge filter to block Rayleigh scattering and reflected laser light before being focused into a multimode optical fiber. The 50 µm diameter multimode output fiber provided the optical apertures for the confocal measurement. The light emerging from the output optical fiber was dispersed by a 30 cm focal length, f/4 CzernyTurner monochromator, incorporating interchangeable gratings (1800/mm, blazed at 500 nm, and 600/mm, blazed at 500 nm). The light was finally detected by a back-illuminated deep-depletion, 1024 × 128 pixel charge-coupled device camera operating at −82 °C.
Multivariate Data Analysis: Vertex Component Analysis (VCA)
All cellular Raman images were analyzed using a multivariate color decomposition - VCA.19 It decomposes the spectrum of each pixel into contributions from the most dissimilar spectra of the dataset, which are considered the “pure component” or “endmember” spectra. Each endmember spectrum has a corresponding abundance or intensity variation associated with a region within the cellular architecture. In other words, each Raman spectrum can be represented as a vector defined by intensity measurements of channels or frequencies. The Raman-collected spectra discussed here are represented by VCA in a 1024-dimensional space, since there are 1024 frequency points, which comprise the spectrum. VCA then proceeds with a step-by-step reduction of the dimensionality of the system via a simplex-method, until we get a predetermined geometrical shape, such as a tetrahedron, which would yield us 4 spectral endmembers. Thus, from an n-simplex, VCA reduces the problem to (n - 1)-simplex after a specific amount of iterations.
If more than one mixed spectrum is considered and the spectra are identical, they will have identical vectors with the same orientation. Spectra, which have same composition but different total intensities, will have different length superimposed vectors. If we plot a multitude of data points collected at two specified frequencies, we get a two-dimensional plane with discrete spectra as endpoints of a line. If three component spectra are needed to describe the material, they will be the corners of a triangle in a two-dimensional space, etc. These endpoint spectra represent the closest to being pure component spectra and are termed “endmembers”.
The rendered endmembers and their abundance vectors are scaled and converted to monochrome intensity images of the specimen of interest. Ultimately three such abundance maps are overlaid and converted to a red, green, and blue (RGB) map, showing the intensity of each of the rendered endmembers. The intensities depict the extent of spectral contribution of the endmember spectrum to each of the pixels within the dataset. An overlay of these RGB color maps yields a pseudo-color map with mixed colors indicating the intensity values of each of three endmembers.
RESULTS AND DISCUSSION
Deuteration of the phospholipids enhances the specificity and sensitivity of the nanoparticulate detection due to presence of C-D stretching intensities within the biological spectrum. Sensitivity of biochemical imaging to the liposomal systems has thus been increased with the use of isotopic labeling, namely via deuteration of the aliphatic side chains of 1,2-Dipalmitoyl-d62-sn-glycero-3-phosphocholine (DPPC-d62). The deuterated liposomes were prepared by the liquid film hydration method, as discussed in the experimental section and subsequently modified with three distinct cationic moieties, namely DOTAP, SA, and STPP. Sizes and zeta potentials of the three cationic liposomes are shown in Table 1.
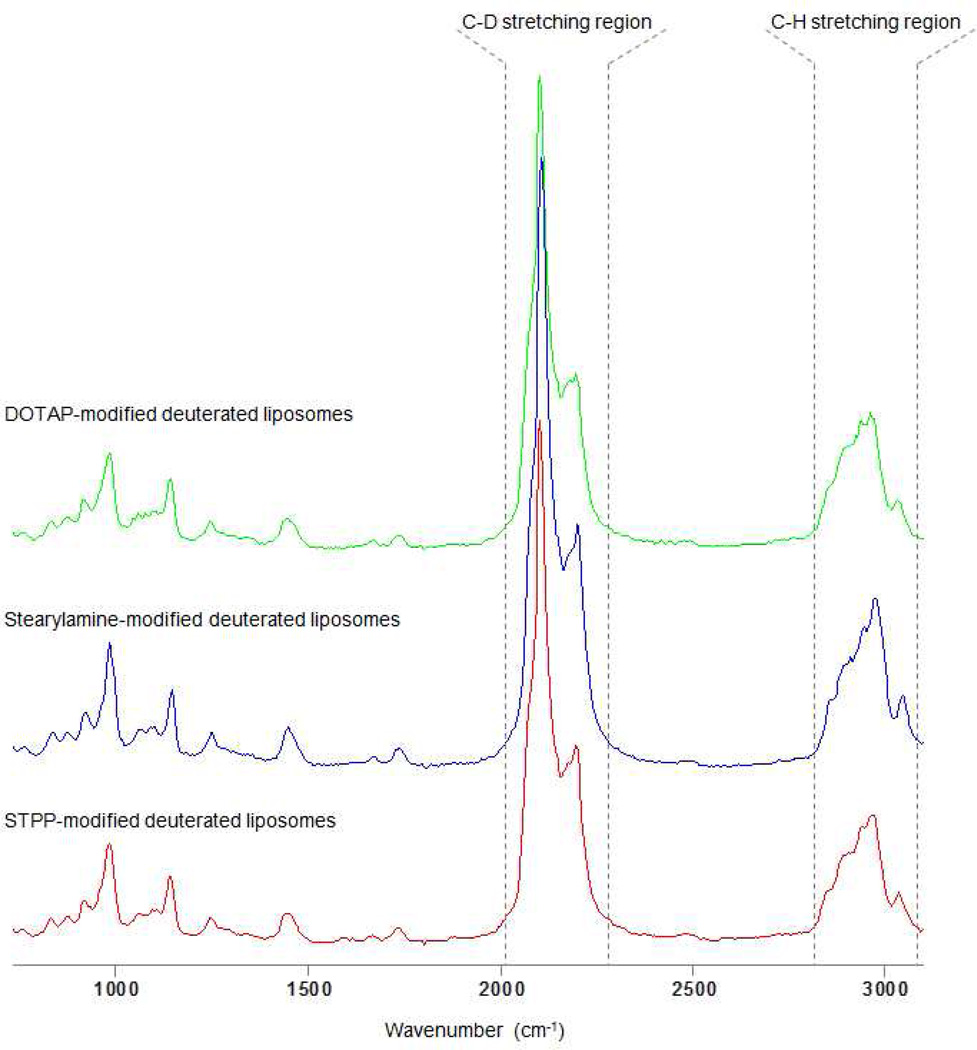
Biochemical analysis of the prepared nanoparticles reveals their characteristic chemical signatures, which fall into two distinct spectral regions, namely the fingerprint region (750 – 1800 cm−1) as well as the C-H stretching region (2800 – 3100 cm−1). Their corresponding spectra are shown in Figure 1. Carbon – deuterium (C-D) stretching vibrations fall within a spectral region devoid of any other spectral signatures of bio-organic materials, 2050 – 2300 cm−1. Sensitivity of detection of the nanoparticle-characteristic signal within the intra-cellular milieu is thus greatly enhanced due to the isolated vibrational intensities of the C-D bond, allowing for single-particle tracking capabilities. Deuterium alters the physicochemical properties of the labeled molecule only minimally, due to deuterium atoms being covalently bonded to the carbon atoms on the aliphatic side chains of DPPC-d62 and do not exchange with other protons of the cell.
Figure 1.
Raman spectra of the liposome preparations modified with three distinct cationic moieties: DOTAP, SA, and STPP. Spectra were collected from dried liposomal solutions onto a CaF2 slides.
The highest peak intensities seen in the spectra in Figure 1 are at 2100 cm−1 and 2175 cm−1, originating from the C-D stretching vibrations of the aliphatic side chains within liposomes. C-H stretching vibrations of the remaining C-H bonds are less intense and are observed in the region of 2800 – 3020 cm−1, while other liposome-characteristic peaks are seen within the fingerprint region (750 – 1800 cm−1).
Incubations with liposomes were carried out at 37 °C, at a lipid concentration of 1 mg/ml. Human HeLa cell cultures were incubated for 1, 3, and 6 hrs with a liposome-containing medium, after which they were fixed in 10% formalin and submerged into phosphate-buffered saline for biochemical analysis. Fixation of the cellular samples was carried out primarily due to required data acquisition times for hyperspectral datasets (with a 0.25 sec/pixel dwell time and upward of 10,000 pixels/cell, yields an estimated 30 min to 1hr data accumulation time/cell).
Formalin was chosen due to ease of handling and extensive employment in the biological field as an optimal fixing medium.20 Along with biological data, spectroscopic analysis has been performed in order to verify whether chemical information from a fixed specimen could be compromised due to fixation. Various groups all over the world have tested, annotated, and validated the various fixatives utilized for preparation of biological specimen for optical or histochemical analysis. The results underscored the need for consistent fixation methods employed for a particular experiment. Fixation of cultured or exfoliated cellular samples demonstrated main differences due to the incubation times in various fixatives.21 This finding shows that any one specific method of fixation employed would yield similar results as long as this method is performed throughout the experiment. Also, the authors found that relevant information regarding diseased (i.e. cancerous) cells versus normal cell is still much greater then the variation between the minute intensity changes due to fixatives.
The time points for the time-course experiment were chosen based on previous studies with cationic modifications of liposomes.4,14,22,23 It was seen that within minutes cationic liposomes, micelles, and various other polymeric nanostructures were seen within the cellular periphery. Likewise, incubation for just a few hours resulted in subsequent translocation of the nanoassemblies through the cytosol, where upon cellular sorting destinations were established depending on the composition of the nanostructures. Hence, after a 1 hr incubation of the HeLa cell cultures with our liposomal nanoparticles, consistent presence of the liposomes were observed within every cell analyzed. Shorter incubation times resulted in nanoparticle aggregations in fewer cells. There have been previous reports regarding saturation behavior of various nanoparticles, showing linear dependancies at low concentrations and reaching a plateau at high dosages.24 As we have tried to avoid a saturation of the cell cultures with the liposomes in order to detect their uptake and translocation kinetics, the 1 hr incubation was our starting point.
The vast amount of data recorded from each cell analyzed (ca. 10,000 individual pixel spectra/cell recorded and referred to as a hyper-spectral dataset) bears information regarding each of many biochemical components within each of the sub-micron pixels. This implies that the single spectrum collected from such a volume exhibits a superposition of information of various cellular compartments, which are nested within the cell. Thus a univariate representation (intensity image based on any one distinct spectral region, i.e. The C-H stretching region) of the cellular architecture would yield a topographic rendering of the cellular morphology, delineating mainly density variations. Hence, statistical methods of analysis, such as Vertex Component Analysis (VCA), are utilized in order to achieve superior biochemical contrast, de-convolute and un-mix the abundant information collected, sorting it into distinct and manageable classes based on their chemical composition.19
The VCA algorithm decomposes all spectra within any one dataset in terms of the most dissimilar spectra, termed endmembers, based on principle that biological samples are comprised of “pure” component spectra (i.e. DNA, lipids, or proteins).25 Abundances of each such distinct endmember within the dataset can be represented by a monochrome intensity or abundance plot. When overlaying any three of such monochrome intensity maps, each one is assigned a red, green or blue intensity scale to produce a composite false color RGB image, or a functional biochemical map, of the cell. The principle sub-cellular regions detected by this method of analysis are those related to internalized liposomes, which are assigned a red channel, the nuclei and cell body proteins (blue channel), and various membrane-rich organelles such as the mitochondria, ER and Golgi, or vesicular compartments (green channel).
Cells were incubated for 1 hr, 3 hr, and 6 hr in liposome-containing medium to establish their uptake patterns and intracellular dynamics. The kinetics of internalization patterns of cationic liposomes are thus seen to drastically differ from earlier described results with non-modified, neutral liposomes, which demonstrated significantly slower sub-cellular influx.22 The initial penetration of the non–modified neutral liposomes was detected after a 12 hr incubation of the HeLa cell culture with the liposomes (not shown here).22 However, the initial uptake of cationic liposomes is accelerated, with detectable inclusions observed within an hour of incubation, as is seen in Figure 2, when vast amounts of partially aggregated deuterated liposomes are seen mostly at the cell periphery (red aggregates). Depth profiles, or axial scans, of the cells (data not shown) were also taken and ascertain a full penetration of the liposomes into the cytosol, not their precipitation on the cell surface. The setup of the Raman micro-spectral imaging system allows for confocal imaging, ensuring for a precise slice of a cell to be imaged at any one point in time. Since optical fibers are utilized and act as pinholes to eliminate background information and out-of-focus light, allowing signal to be collected from a specific focal plane within a sample. Hence a 3-dimensional reconstruction may be feasible with this setup by co-addition of confocal lateral slices taken from a sample.26,27
Figure 2.
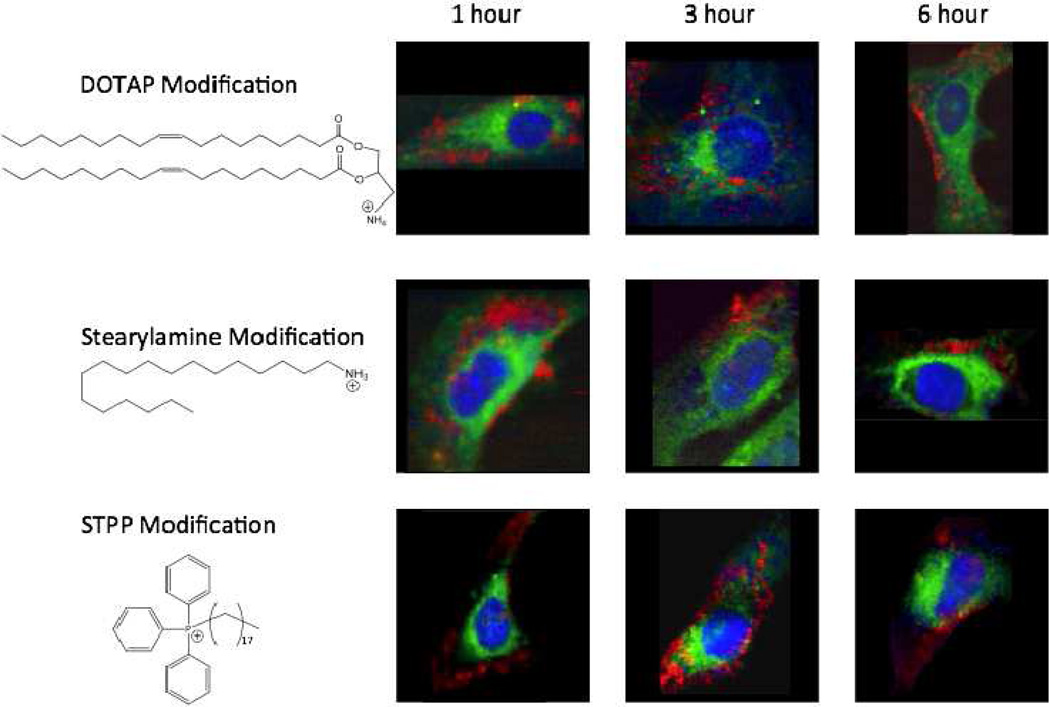
Raman images of HeLa cells incubated with DOTAP-modified liposomes (top row), SA-modified liposomes (middle row), and STPP-modified liposomes (bottom row), structures of which are shown on the left. Cell cultures were incubated for 1, 3, and 6 hrs, with DOTAP-, SA-, or STPP-modified liposomes. Generated VCA-abundance images. The colors are representative of nuclei and cell body (blue), mitochondria-rich regions (green), and internalized liposomes (red).
Apart from different kinetics of the non-modified and the cationic liposomes, the dynamics of the two systems display observable differences in the amounts of liposomes internalized. This is easily detected by analyzing the sample at a 500 nm spatial resolution. While non-modified liposomes are sequestered within the cytosol in small quantities, inclusions of which range from 1–3 µm, the mass of intracellular aggregates of cationic liposomes is vastly greater. It is interesting to note that the liposome internalization proceeds rather fast and its outcome does not seem to vary drastically between different incubation times. Favorable cytosolic areas for nanoparticle segregation are not very evident, since the nanoparticles seem to be simply sequestered within the periphery of the cell, with some speckling of the perinuclear region. The cells thus seem to retain large quantities of the modified liposomes within the cell body, without being functionally compromised.28
Visual observation of the three incubation periods of DOTAP- and SA-modified cationic liposomes did not demonstrate distinct variability of liposomal internalizations. Three and six hours of incubation of the HeLa cell cultures with the aforementioned liposomal modifications did not result in their increased penetration or any variability within translocation patterns of the two cationic systems (Figure 2, top and middle rows). Also, the majority of the internalized DOTAP- and SA-modified liposomes did not seem to follow any specific pattern of translocation or compartmentalization within the cell body. Most of the signal detected for these two cationic systems was seen to aggregate at the periphery of the cell, with a few distinct pixels of liposome-characteristic signal within the perinuclear region.
The alteration of the composition of the cationic liposomes with a moiety known to have an affinity for mitochondria (STPP) renders an entirely different biochemical image. Visual analysis of initial stages (1 hr incubation) of internalization of STPP-containing cationic liposomes does not show drastic differences from the behaviors of the DOTAP- and SA-modified systems. However, after a 3 hr incubation of the HeLa cell culture with STPP-modified liposomes, sub-cellular preferences are evident after the spectral analysis.
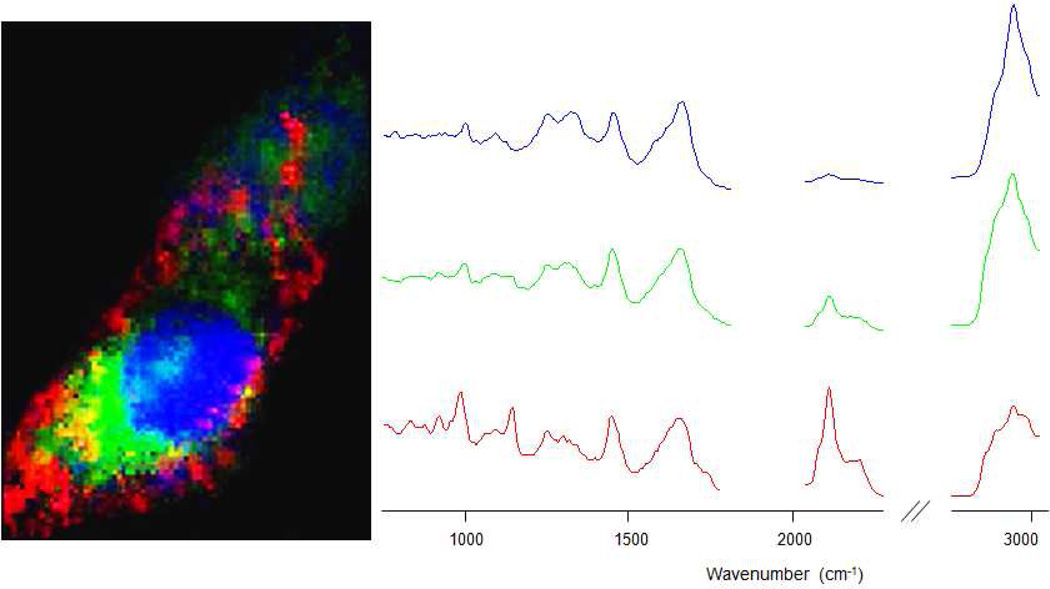
A distinct variance in sub-cellular localization behaviors of the DOTAP-modified and SA-modified cationic liposomes from those of STPP-modified cationic liposomes is seen solely after application of multivariate statistical methods of analysis. Figure 3 demonstrates the biochemical information gained upon application of multivariate analyses on the spectral dataset. The three color channels show a rendition of a VCA-analyzed HeLa cell incubated with STPP-modified liposomes for 3 hrs. The three most distinct biochemical environments within the cell are pseudo-colored blue (nucleus/cell body), green (mitochondria-rich regions) and red (internalized liposomes). These intensity maps of each of the sub-cellular environments were generated from the three most distinct endmember spectra within the hyperspectral dataset, discerned by the VCA, and are shown in Figure 3. Nucleus and the cell body, pseudo-colored blue, exhibit chemical features pertaining to DNA/RNA and various proteins, which is seen from the corresponding color-coded endmember spectrum. The highest peak intensity is centered at 2920 cm−1 within the C-H stretching region, other DNA/RNA/protein scattering intensities of the biological samples are seen in the fingerprint region (750–1800 cm−1), while the 2050–2300 cm−1 (C-D stretching) spectral region, characteristic of nanoparticle accumulations, remains bare. The bright red inclusions within the cytosol and their corresponding endmember spectrum express high intensity peaks within the C-D stretching region, due to a superposition of an intense signal pertaining to STPP-modified liposomes onto the biochemical compartment encapsulating them. The green abundances within the image along with their corresponding spectrum, on the other hand, possesses chemical properties pertaining to mitochondrial aggregations, while also showing a distinct C-D stretching vibrational peak. This is due to the STPP moiety on the deuterated liposomes, which demonstrates not only an enhanced internalization effect but also a clear affinity for the mitochonrdia-rich perinuclear regions.
Figure 3.
Raman image of a HeLa cell incubated with STPP-modified liposomes for 3hrs. Nucleus and cell body (blue), mitochondria-rich regions (green), and internalized liposomes aggregations (red). Color-coded corresponding spectra are shown on the right.
This result demonstrates the sensitivity of the micro-spectroscopy coupled with statistical means of hyperspectral analysis. While univariate imaging would be sensitive to intense C-D stretching vibrations of the cationic liposomes, affinity of STPP-liposomes to the mitochondria may be missed entirely. This is due to smaller intensities of the C-D stretching vibrations as well as variation of the spectral signatures, which exhibit chemical patterns of the mitochondria-rich regions (green spectrum in Figure 3). VCA is thus capable of distinguishing the liposomes co-localized with distinct sub-cellular organelles. Whether liposomes are fully or partially intact is still under investigation, however previous studies have reported seeing a fusion of the liposomal monomers with the sub-cellular organelles upon their degradation.22,29
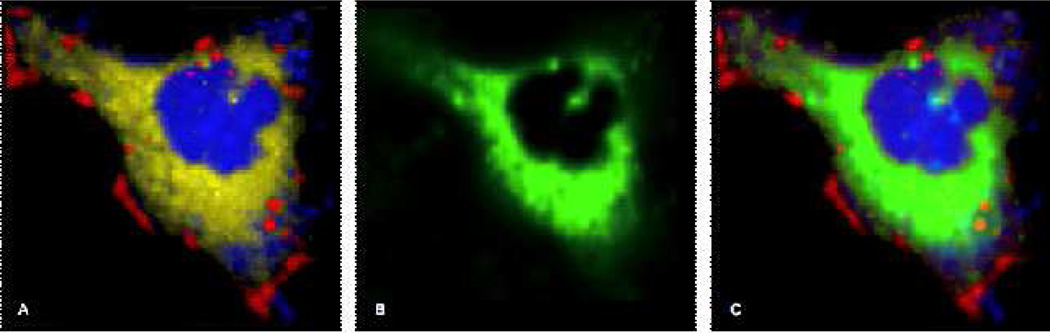
The analysis of trafficking patterns of mitochondria-specific liposomes and their ultimate destination is corroborated with use of Mitotracker® dye, which labels mitochondrial aggregations (shown in Figure 4). Both Raman biochemical mapping and fluorescence imaging have been performed on the same cell for demonstration purposes. The dark red regions visible in the periphery of the cell, as well as speckling the cell body, reflect the intracellular presence of the STPP-modified liposomes, throughout the incubation period. In Figure 4B, green intensity reflects the fluorescence of the Mitotracker® dye, depicting the mitochondria. Figure 4C shows an overlay of the VCA intensity map with the fluorescence of mitochondria. It is interesting to note that the VCA analysis, in Figure 4A, has delineated a super-imposable image of mitochondrial aggregations, which are shown in yellow intensities for reference. Furthermore, the VCA analysis of the dataset revealed an appearance of peaks in the C-D spectral region (spectra not shown) originating from the yellow intensities, which detail the mitochondria-rich regions within the cell in Figure 4A. Hence, apart from information gained regarding the sorting of internalized liposomes into distinct vesicular compartments (red intensities in Figure 4A), VCA analysis extracts and classifies biochemical information from the STPP-modified liposomes and co-localizes it with mitochondria (yellow intensities in Figure 4A). This demonstrates favorability or affinity of the STPP-modified cationic liposomes to specific cellular organelles.
Figure 4.
Raman images of HeLa cell incubated for 3 hours STPP-modified liposomes. (A) blue region denotes nuclear and cytoplasmic protein content; red—internalized liposomes; yellow—membrane-rich organelles. (B) is the fluorescence image of the Mitotracker of the cell. (C) is the analyzed image in A overlayed with B fluorescence stain Mitotracker®, pseudo-colored green.
Raman microspectroscopy in synergy with multivariate methods of analysis thus offer an advantage over diverse staining techniques such as fluorescence microscopy. In combination with optical microscopy, Raman spectroscopic images display a biochemical snapshot of a cell, along with any exogenous systems therein. The use of the multivariate statistics in conjunction with Raman micro-spectral methods reveals sub-cellular organelles (such as mitochondria, nuclei, nucleoli, and vesicles) contrasted from the exogenous nanocarrier systems, without the implementation of organelle-specific stains. Synergistic approach to biochemical imaging using Raman micro-spectral mapping with VCA has proven superior to tracking single fluorosphores, as follows from Figure 4. Apart from the ability of the statistical analysis to distinguish sub-cellular compartments without fluorescing labels, the chemical features of the mitochondria reveal C-D stretching vibrations, which can be utilized to determine the dynamics and physicochemical behaviors of administered system, i.e., the exchange of C-D rich lipids from the liposomes with undeuterated lipids of naturally lipid-rich organelles. Distinct patterns of translocation of the cationic liposomes are also revealed and annotated without the use fluorophores.
The general pattern of uptake of liposomes with cationic modifications show enhanced internalization patterns from the earliest incubation point analyzed. However, a mere charge modification to the surface of liposomes does not dictate their intracellular localization patterns. We have thus shown that with the employment of cationic compounds with distinct affinities for the sub-cellular compartments, such as the mitochondria, the liposomal nanoparticles are no longer haphazardly distributed within the cytosol, but concentrate within the mitochondria-rich sub-cellular regions.
ACKNOWLEDGEMENTS
Partial support for this research was provided by NIH grants, CA 090346 (to Max Diem) and CA128486-02S1 (to Vladimir Torchilin) and are gratefully acknowledged.
REFERENCES
- 1.Bawa R. Nanoparticle-based therapeutics in humans – a survey. Nanotech. Law Business. 2008;5(2):1–21. [Google Scholar]
- 2.Torchilin V. Structure and design of polymeric surfactant-based drug delivery systems. J. Control Release. 2001;73(2–3):1–36. doi: 10.1016/s0168-3659(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 3.Alexis F, Rhee J, Richieb J, Radovic-Moreno A, Langer R, Farokhzad O. New frontiers in nanotechnology for cancer treatment. Urologic Oncology: Seminars and Original Investigations. 2008;26(1):74–85. doi: 10.1016/j.urolonc.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Torchilin V. Recent approaches to intracellular delivery of drugs and DNA and organelle targeting. Annu. Rev. Biomed. Eng. 2006;8:343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 5.Torchilin V. Drug Targeting. European J. Pharm. Sci. 2000:S81–S91. doi: 10.1016/s0928-0987(00)00166-4. [DOI] [PubMed] [Google Scholar]
- 6.Torchilin V. Micellar nanocarriers: pharmaceutical perspectives. Pharm. Res. 2006;24(1):1–16. doi: 10.1007/s11095-006-9132-0. [DOI] [PubMed] [Google Scholar]
- 7.Couvreur P, Vauthier C. Nanotechnology: intelligent design to treat complex disease. Pharm. Res. 2006;23(12):1417–1450. doi: 10.1007/s11095-006-0284-8. [DOI] [PubMed] [Google Scholar]
- 8.Torchilin V. Multifunctioanl Nanocarriers. Adv. Drug Deliv. Rev. 2006;58(14):1532–1555. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Giannotti M, Esteban O, Oliva M, García-Parajo M, Sanz F. pH-responsive polysaccharide-based polyelectrolyte complexes as nanocarriers for lysosomal delivery of therapeutic proteins. Biomacromol. 2011;12(7):2524–2533. doi: 10.1021/bm2003384. [DOI] [PubMed] [Google Scholar]
- 10.Canine B, Hatefi A. Development of recombinant cationic polymers for gene therapy research. Ad.v Drug Deliv. Rev. 2010;62(15):1524–1529. doi: 10.1016/j.addr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pack D, Hoffman A, Pun S, Stayton P. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 12.Weissig V, Cheng S, D'Souza G. Mitochondrial pharmaceutics. Mitochondrion. 2004;3(4):229–244. doi: 10.1016/j.mito.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Boddapati S, Tongcharoensirikul P, Hanson R, D'Souza G, Torchilin V, Weissig V. Mitochondriotropic liposomes. J. Liposome Res. 2005;15(1–2):49–58. doi: 10.1081/lpr-64958. [DOI] [PubMed] [Google Scholar]
- 14.Lonez C, Vandenbranden M, Ruysschaert J. Cationic liposomal lipids: From gene carriers to cell signaling. Progress in Lipid Res. 2008;47(5):340–347. doi: 10.1016/j.plipres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Wasungu L, Hoekstra D. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Control. Release. 2006;116(2):255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 16.Smith R, Porteous C, Gane A, Murphy M. Delivery of bioactive molecules to mitochondria in vivo. Proc. Nat. Ac. Sci. 2003;100(9):5407–5412. doi: 10.1073/pnas.0931245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ojovan S, Knorre D, Markova O, Smirnova E, Bakeeva L, Severin F. Accumulation of dodecyltriphenylphosphonium in mitochondria induces their swelling and ROS-dependent growth inhibition in yeast. J. Bioenerg. Biomembr. 2011;43(2):175–180. doi: 10.1007/s10863-011-9345-8. [DOI] [PubMed] [Google Scholar]
- 18.Naumann D. Mid-Infrared and Raman spectroscopy for medical diagnosis. Applied Spectroscopy Reviews. 2001;36(2–3):239–298. [Google Scholar]
- 19.Nascimento J, Bioucas Dias J. Vertex Component Analysis: A Fast Algorithm to Unmix Hyperspectral Data. IEEE Transactions on Geoscience and Remote Sensing. 2005;43(4):898–910. [Google Scholar]
- 20.Werner M, Chott A, Fabiano A, Battifora H. Effect of formalin tissue fixation and processing on immunohistochemistry. American J. Surgical Path. 2000;24(7):1016–1019. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Mazur A, Marcsisin E, Bird B, Miljković M, Diem M. Evaluating different fixation protocols for spectral cytopathology, Part 1. Anal. Chem. 2012;84(3):1259–1266. doi: 10.1021/ac202046d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthäus C, Kale A, Chernenko T, Torchilin V, Diem M. New ways of imaging uptake and intracellular fate of liposomal drug carrier systems inside individual cells, based on Raman microscopy. Mol. Pharmaceutics. 2008;5(2):287–293. doi: 10.1021/mp7001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aramaki Y. Cationic liposomes induce macrophage apoptosis through mitochondrial pathway. Arch. Biochem. Biophys. 2001;392(2):245–250. doi: 10.1006/abbi.2001.2458. [DOI] [PubMed] [Google Scholar]
- 24.Nativo P, Prior I, Brust M. Uptake and intracellular fate of surface-modified gold nanoparticles. ACS Nano. 2008;2(8):1639–1644. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 25.Miljković M, Chernenko T, Romeo M, Bird B, Matthäus C, Diem M. Label-free imaging of human cells: algorithms for image reconstruction of Raman hyperspectral datasets. Analyst. 2010;135(8):2002–2013. doi: 10.1039/c0an00042f. [DOI] [PubMed] [Google Scholar]
- 26.Wood B, Bambery K, Evans C, Quinn M, McNaughton D. A three-dimensional multivariate image processing technique for the analysis of FTIR spectroscopic images of multiple tissue sections. BMC Med. Imaging. 2006;6:12. doi: 10.1186/1471-2342-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asghari-Khiavi M, Wood B, Mechler A, Bambery K, Buckingham D, Cookec B, McNaughton D. Correlation of atomic force microscopy and Raman micro-spectroscopy to study the effects of ex vivo treatment procedures on human red blood cells. Analyst. 2010;135(3):525. doi: 10.1039/b919245j. [DOI] [PubMed] [Google Scholar]
- 28.Campbell R, Balasubramanian S, Straubinger R. Phospholipid-cationic lipid interactions: influences on membrane and vesicle properties. Biochim. Biophys. Acta. 2001;1512(1):27–39. doi: 10.1016/s0005-2736(01)00290-5. [DOI] [PubMed] [Google Scholar]
- 29.Krafft C, Knetschke T, Funk R, Salzer R. Studies on Stress-Induced Changes at the Subcellular Level by Raman Microspectroscopic Mapping. Anal. Chem. 2006;78(13):4424–4429. doi: 10.1021/ac060205b. [DOI] [PubMed] [Google Scholar]