Abstract
In response to virus infection, cells can alter protein expression to modify cellular functions and limit viral replication. To examine host protein expression during infection with human cytomegalovirus (HCMV), an enveloped DNA virus, we performed a semi-quantitative, temporal analysis of the cell surface proteome in infected fibroblasts. We determined that resident low density lipoprotein related receptor 1 (LRP1), a plasma membrane receptor that regulates lipid metabolism, is elevated early after HCMV infection, resulting in decreased intracellular cholesterol. siRNA knockdown or antibody-mediated inhibition of LRP1 increased intracellular cholesterol, and concomitantly increased the infectious virus yield. Virions produced under these conditions contained elevated cholesterol, resulting in increased infectivity. Depleting cholesterol from virions reduced their infectivity by blocking fusion of the virion envelope with the cell membrane. Thus, LRP1 restricts HCMV infectivity by controlling the availability of cholesterol for the virion envelope and increased LRP1 expression is likely a defense response to infection.
INTRODUCTION
Human cytomegalovirus (HCMV) is a β-herpesvirus that infects more than 60% of adults, resulting in establishment of life-long viral latency (Mocarski et al., 2007). It is a major cause of birth defects, where congenital infection causes disabilities such as hearing loss and retardation (Pereira and Maidji, 2008). HCMV is also an opportunistic agent in immunosuppressed individuals (Britt, 2008), and it might contribute to cardiovascular disease (Streblow et al., 2008), cancer (Soroceanu and Cobbs, 2011) and immune senescence (Moss, 2010).
Like all viruses, HCMV relies on numerous host cell functions for its replication. As a consequence, it has evolved mechanisms to hijack aspects of cell machinery in order to replicate efficiently. HCMV modulates diverse classes of cell surface proteins, including immune surveillance (Park et al., 2010; Wilkinson et al., 2008), signaling (Gredmark et al., 2007; Montag et al., 2011; Popovic et al., 2010), transporter (Yu et al., 2011) and adhesion proteins (Leis et al., 2004); and RNA array studies predict that many more surface proteins might be altered (Browne et al., 2001). However, a comprehensive characterization of HCMV-induced alterations to the cell surface proteome has not been reported. Cell surface proteins comprise more than a third of the human proteome (Josic and Clifton, 2007), they are often altered on diseased cells and they serve as targets for more than two-thirds of existing drugs (Overington et al., 2006). Therefore, it is likely that enhanced understanding of the infected cell surface proteome will yield insights to viral pathogenesis and identify previously unexplored targets for treatment of HCMV- related diseases.
In this study, we compared the cell surface proteome of mock-infected to HCMV-infected fibroblasts. Mass spectrometry (MS)-based proteomics identified 505 affinity-enriched proteins at each time assayed. Using spectral counting analysis, 114 of these proteins were classified as candidates for differential cell surface expression after HCMV infection. Two proteins predicted to increase after infection were the GLUT4 glucose transporter and the LDL receptor-related protein 1 (LRP1). Inhibition of GLUT4 reduces the yield of virus (Yu et al., 2011), presumably by blocking HCMV-mediated induction of glycolysis (Munger et al., 2006; Munger et al., 2008). GLUT4 and LRP1 are delivered to the plasma membrane together as constituents of insulin- responsive vesicles, and LRP1 can influence GLUT4 expression (Jedrychowski et al., 2010).
LRP1 (also known as α2-macroglobulin receptor or CD91) is a ubiquitously expressed, type I transmembrane receptor that is a member of the low density lipoprotein (LDL)-receptor family (Herz et al., 1988). It is implicated in numerous physiologic processes, including the regulation of lipid metabolism, cell migration, the proliferation of vascular smooth muscle cells, and neurodevelopment (Franchini and Montagnana, 2011; Lillis et al., 2008). LRP1 surface expression has been implicated in the development of atherosclerosis (Boucher and Herz, 2010), and knockdown of LRP1 results in an increase in intracellular cholesterol levels in fibroblasts (Terrand et al., 2009; Zhou et al., 2009).
Given the potential for LRP1 to profoundly affect the physiology of HCMV-infected cells, and the possibility that the receptor and HCMV (Soderberg-Naucler, 2008; Streblow et al., 2008) might both influence atherogenic lesion development, we explored the consequences of LRP1 modulation during HCMV infection. HCMV increased its expression both at the RNA and protein level. Knockdown of LRP1 increased cellular and virion cholesterol content, which correlated with increased virion infectivity due to more efficient fusion of the virion envelope with the plasma membrane.
RESULTS
Alterations in cell surface protein abundance after HCMV infection
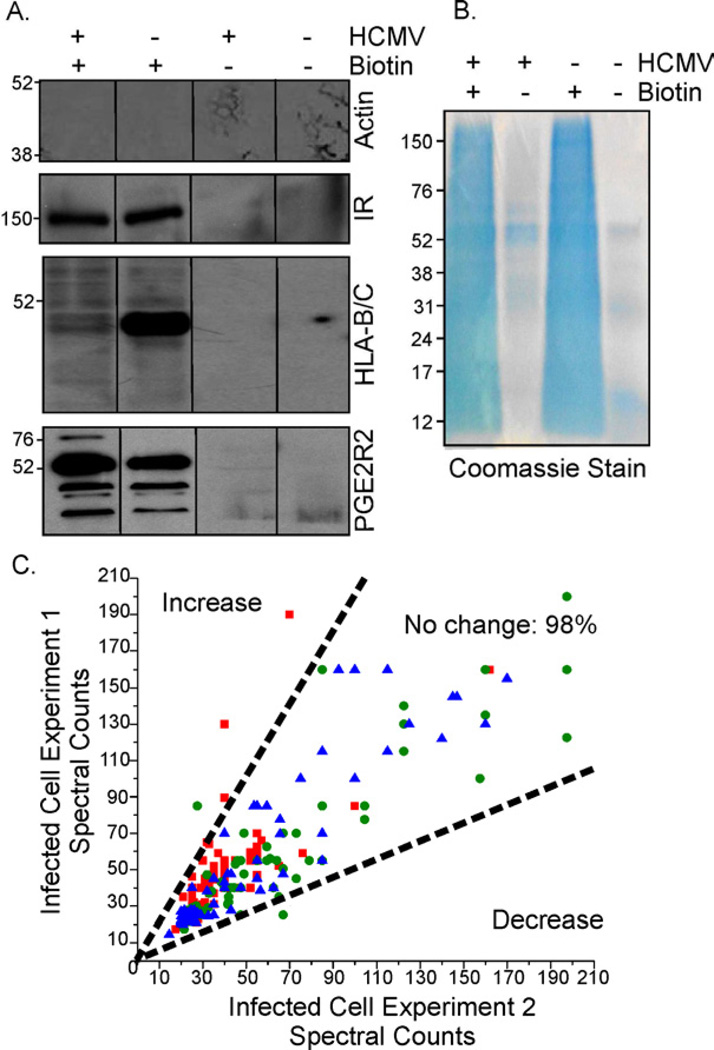
Cell surface proteins regulate many essential cellular processes, ranging from growth to glucose metabolism, and they also serve critical roles in cellular defense against invading pathogens. To monitor alterations to surface proteins during the HCMV replication cycle, fibroblasts were treated during the immediate-early (6 h post-infection, hpi), early (24 hpi), and late (72 hpi) phase of infection with an amine-reactive, thiol-cleavable biotin probe, tagging proteins at the outer surface of the plasma membrane. Cells were subsequently lysed, and biotin-tagged proteins were affinity purified using a NeutrAvidin matrix (Fig. 1A, left panel). Samples that were not biotinylated were purified to monitor non-specific binding to the matrix. Western blot assays of tagged and purified proteins (Fig. 1A, right panel) verified the presence of plasma membrane-localized insulin receptor, MHC-class I receptor, and prostaglandin receptor 2 (PGE2R2), while an abundant intracellular marker, β-actin, was largely absent. In addition, the analysis confirmed that MHC class I molecules are downregulated (del et al., 1992) and PGE2R2 is slightly upregulated (Browne et al., 2001) upon infection, verifying that multiple expression phenotypes were reliably detected. Coomassie staining of avidin-purified proteins in both HCMV-infected and mock-infected cells demonstrated the efficient isolation of biotin-tagged proteins with limited purification of proteins in controls that were not biotinylated (Fig. 1B).
Fig. 1. Identification of cell surface protein alterations in HCMV-infected cells.
Fibroblasts were mock-infected or infected at a multiplicity of 5 IU/cell.
(A) Specificity of biotin labeling: Avidin-purified cell surface protein preparations were assayed by Western blot for β-actin, insulin receptor (IR), MHC-class I receptor (HLA-B/C), and prostaglandin receptor 2 (PGE2R2).
(B) Purification of biotinylated proteins: Avidin-purified samples (~50 µg protein in +biotin samples) were subjected to electrophoresis and Coomassie stained.
(C) Reproducibility of MS data across independent experiments: Normalized spectral counts of infected cell proteins isolated at 6 (■), 24 (●) and 72 hpi (▲) were plotted from two independent experiments. Dotted-lines mark a 2-fold difference in counts.
Cell surface-enriched protein preparations from two independent experiments were subjected to MS analysis. To retain protein identifications that resulted from specific enrichment via the biotin tag, we utilized label-free spectral counting to evaluate protein enrichment in the biotin-labeled versus non-labeled condition. This method uses the total number of MS/MS spectra as a measure of relative protein abundance. In total, 505 proteins were considered enriched (≥ 5-fold) in biotin-labeled samples across all conditions, of which ~62% have been previously annotated as cell surface proteins based on Gene Ontology classification. Since ~20% of the genome is predicted to encode plasma membrane proteins, the MS results also support significant enrichment of cell surface proteins by this approach. Further, the portion of avidin-enriched proteins classified as cell surface residents was similar to that reported for cancer cell lines (Lund et al., 2009).
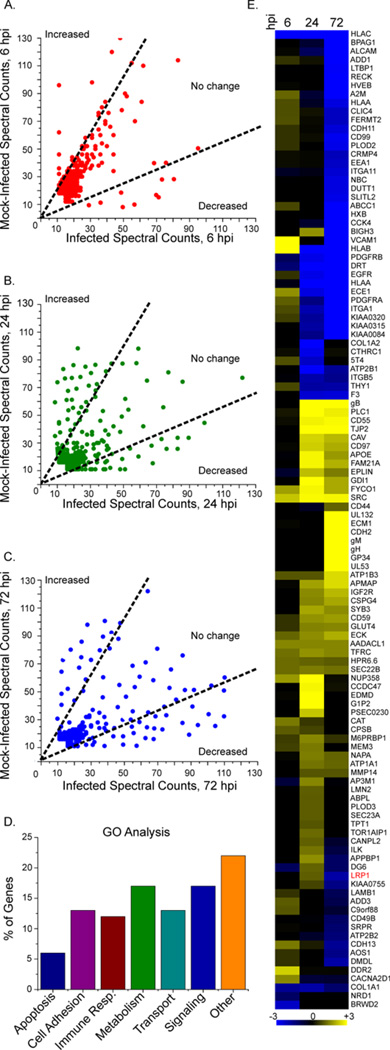
To assess temporal, HCMV-dependent regulation of cell surface proteins, label-free spectral counting was again employed. Appropriate thresholds for distinguishing significant HCMV-induced protein abundance changes were selected by evaluating the spectral count reproducibility between biological replicates. As shown in figure 1C, a threshold of ≥ 2-fold change (minimum 10 spectral counts) resulted in only ~ 2% false positives. Using these criteria, comparison of HCMV- versus mock-infected samples revealed that ~8% of proteins were altered at 6 hpi, ~19% at 24 hpi, and ~24% at 72 hpi (Fig. 2A-C). Gene ontology analysis (Mi et al., 2010) grouped the majority of plasma membrane proteins altered upon infection into six functional classes: apoptosis (e.g. CD99), cell adhesion (CSPG4, VCAM1), immune response (CD55, CD59), metabolism (LRP1, GLUT4), transport (Na+2/K-transporting ATPase-subunits), and signaling (ephrins) (Fig. 2D). As illustrated by hierarchical clustering of altered proteins (Fig. 2E), differential regulation favored upregulation (yellow) at 6 and 24 hpi, while at 72 hpi more proteins were down-regulated (blue). In some cases, proteins were both up and down modulated as infection progressed (Fig. 2E, Table S1), such as vascular cell adhesion protein 1 (VCAM1) and LRP1. VCAM1 was upregulated at 6 hpi, but downregulated at 72 hpi. This is consistent with a previous study that showed HCMV infection of endothelial cells increased cell surface VCAM1, which resulted in an increase in monocyte and T-lymphocyte adhesion within 24 hpi (Shahgasempour et al., 1997). Together, these results indicate that the composition of the cell surface proteome is diverse and dynamic, undergoing marked time-dependent changes during infection.
Fig. 2. Alterations to the cell surface proteome after HCMV infection.
(A-C) Spectral counts of cell surface proteins: Fibroblasts were mock infected or infected (5 IU/cell) and analyzed 6 (A), 24 (B) or 72 h (C) later. Dotted-lines mark a 2-fold difference in counts.
(D) Differentially expressed proteins grouped by gene ontology analysis.
(E) Heat map showing changes in cell surface protein expression after infection as compared to mock infection: Proteins upregulated after infection are yellow, while downregulated proteins are blue. The intensity of the color reflects the spectral count fold-change. Gene names are listed to the right. The average change from two independent experiments is displayed for each time point. Table S1 contains the primary data set underlying this figure.
Experimental knockdown of LRP1 generates virions with enhanced infectivity
MS analysis indicated that LRP1 was transiently elevated at the cell surface (Fig. 2E and Table S1). Given its potential to profoundly affect the physiology of HCMV-infected cells, LRP1 was chosen for detailed analysis.
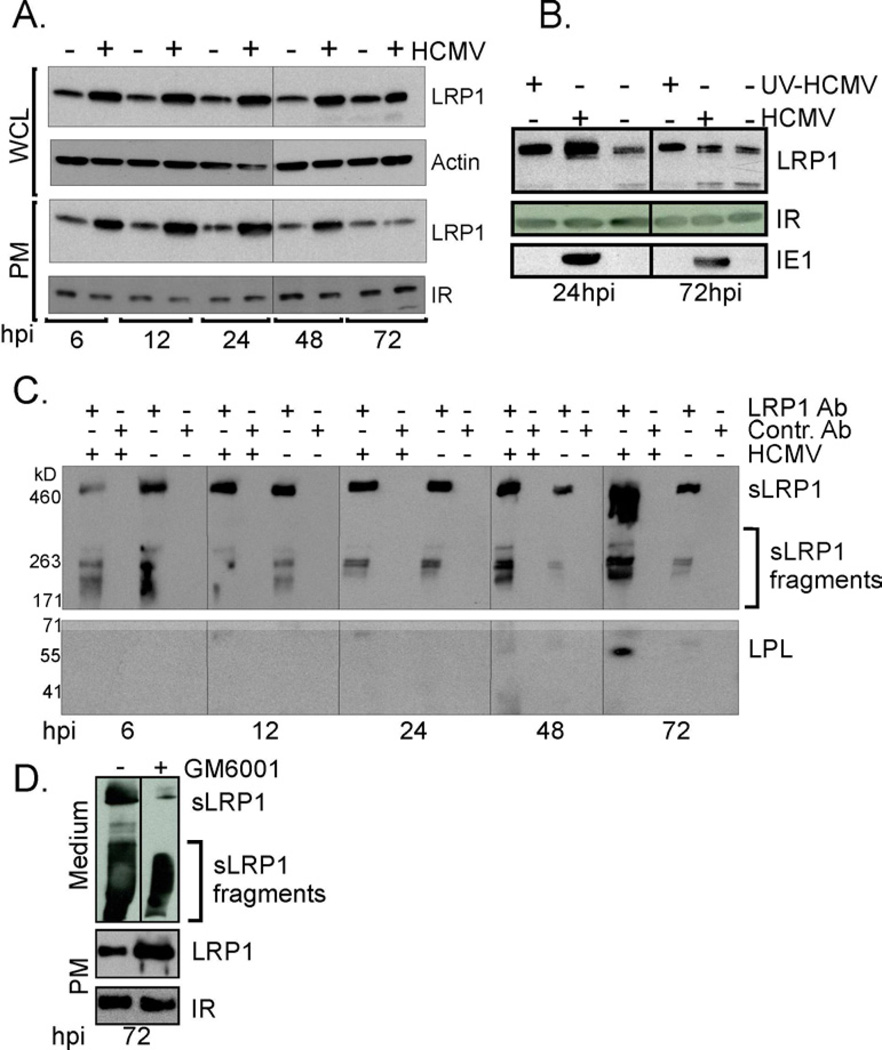
Consistent with earlier transcriptome analysis (Browne et al., 2001), quantification of LRP1 RNA by qRT-PCR showed that it increased by a factor of 5–10 after infection (Fig. 4A, left), and Western blot assays demonstrated that LRP1 protein was upregulated throughout infection in whole cell lysates (Fig. 3A). In contrast, LRP1 expression on the cell surface was elevated from 6 to 48 hpi, but not at 72 hpi (Fig. 3A and 3B). β-actin (whole cell lysate) and insulin receptor (plasma membrane) were assayed as loading controls. UV-inactivated virus also upregulated LRP1 on the cell surface at 24 hpi (Fig. 3B), arguing that virion binding or entry, or virion constituents are at least partially responsible for LRP1 upregulation. The IE1 protein was assayed as a control to confirm that the UV-treated virus was unable to express viral proteins.
Fig. 3. Cell surface LRP1 is elevated by HCMV and released to the medium as infection progresses.
(A) LRP1 is transiently elevated after infection: Whole cell lysates (WCL) and isolated plasma membrane proteins (PM) were assayed by Western blot for LRP1, β-actin and insulin receptor (IR) at the indicated times after mock infection or infection (5 IU/cell).
(B) LRP1 is elevated after infection with UV-inactivated virus: Whole cell lysates were assayed by western blot for the viral IE1 protein and isolated plasma membrane proteins were assayed at the indicated times for LRP1 and IR or at the indicated times after mock infection or infection (5 IU/cell) with HCMV or UV-inactivated HCMV (UV HCMV).
(C) Accumulation of soluble LRP1 (sLRP1): At the indicated times after infection, soluble LRP1 (sLRP1) released into the medium was immunoprecipitated with LRP1 antibody, captured with protein A beads, then assayed by Western blot for LRP1 (top panel) or lipoprotein lipase (LPL) (bottom panel).
(D) sLRP1 levels in the presence of a metalloproteinase-inhibitor: HCMV infected cells (5 IU/cell) were treated with GM6001 (10 µM) at 48 hpi, and LRP1 levels in the medium and at the PM were determined 24 h later by western blot.
Membrane metalloproteinases are activated to cleave LRP1 from the membrane as cellular cholesterol is depleted (Selvais et al., 2011), generating soluble LRP1 (sLRP1, comprised of full length LRP1 α-chain plus fragments of the α-chain). sLRP1 could be immunoprecipitated from the medium at all times after infection, and its amount increased substantially at 72 hpi (Fig. 3C, top panel). sLRP1 in the medium of infected cells was able to interact with its ligands (Fig. 3C, bottom panel) because it co-immunoprecipitated with lipoprotein lipase, a known LRP1 ligand (Boucher and Herz, 2010). GM6001, a broad-spectrum metalloproteinase inhibitor, reduced the level of sLRP1 at 72 hpi (Fig. 3D), accompanied with a corresponding increase in the level of plasma membrane LRP1. We interpret this result to indicate that the reduced level of LRP1 on the cell surface during the late phase of infection results from its cleavage and the release of sLRP1 and sLRP1 fragments into the medium.
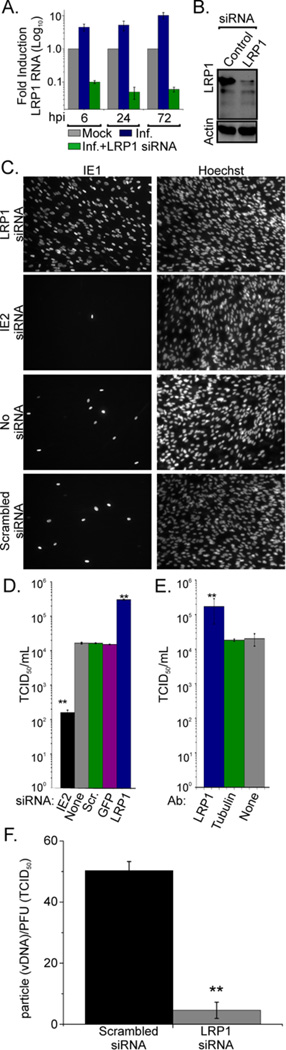
To determine the effect of altering LRP1 expression on virus production, an LRP1-specific siRNA was used to knock down the protein beginning 24 h before infection. LRP1 mRNA and protein were substantially reduced by siRNA treatment at all times tested after infection (Fig. 4A and 4B). Reduced LRP1 expression increased the production of HCMV infectivity by a factor of ~15 (Fig. 4C and D). Experiments included a mock (no siRNA) and scrambled siRNA as negative controls, and an IE2 siRNA (an essential viral protein) as a positive control for protein knockdown resulting in reduced virus replication. A similar increase in infectivity was obtained by treating infected cells with a neutralizing antibody for LRP1 (Fig. 4E), whereas no effect was observed using an isotype-specific control. LRP1 knockdown did not significantly alter the level of viral DNA or a selected subset of viral mRNAs and proteins compared to controls (Fig. S1), suggesting that increased virus release is not due to increased viral protein or nucleic acid accumulation. Importantly, however, virion DNA-normalized virus particles produced in LRP1 knockdown cells were ~15-fold more infectious than particles from cells treated with scrambled siRNA (Fig. 4F). Cells receiving the control siRNA produced virions with a particle to infectious unit ratio of ~50, within the range of infectivity routinely documented for lab-adapted strains of HCMV (Mocarski et al., 2007). In contrast, cells receiving LRP1- specific siRNA generated particles exhibiting a ratio of ~3.5. Thus, reduced LRP1 function does not lead to the production of more virus particles, but rather generates virions with enhanced infectivity.
Fig. 4. Inhibition of LRP1 yields virions with enhanced infectivity.
(A) Efficient LRP1 knockdown at the RNA level: Fibroblasts were transfected with an LRP1-specific siRNA or left untreated. 24 h later, cells were infected (5 IU/cell); and then, after the indicated time intervals, RNA was isolated and assayed by real-time qPCR to determine the levels of LRP1 transcripts. Samples were normalized to GAPDH RNA. Infected cell RNA levels are presented relative to mock.
(B) Efficient LRP1 knockdown at the protein level: Fibroblasts were transfected with an LRP1-specific or scrambled control siRNA. 24 h later, cells were infected, and, after an additional 24 h, whole cell lysates were prepared and assayed by Western blot for LRP1.
(C-D) siRNA-mediated LRP1 knockdown produces enhanced yields of infectious HCMV: Fibroblasts were infected (0.1 IU/cell) at 24 h post transfection with the indicated siRNAs or no siRNA. 96 h later, virus in the medium was assayed by IE1 fluorescence in cultures where nuclei were identified by Hoechst staining (C) or TCID50 assay (D).
(E) Neutralizing antibody to LRP1 produces enhanced yields of infectious HCMV. Fibroblasts were infected (0.1 IU/cell) at 1h after treatment with the indicated antibodies (Ab), and cell-free virus was quantified by a TCID50 assay 96 h later.
(F) LRP1 knockdown generates virions with enhanced infectivity. Particle to IU ratios were calculated by dividing the amount of DNA in virus particles by the virus titer.
Error bars report the standard errors of the means from three experiments, each performed in triplicate. **P<0.001 (t-test, compared to control condition). Fig. S1 contains additional data relevant to this experiment.
LRP1 expression impacts cellular and virion cholesterol levels
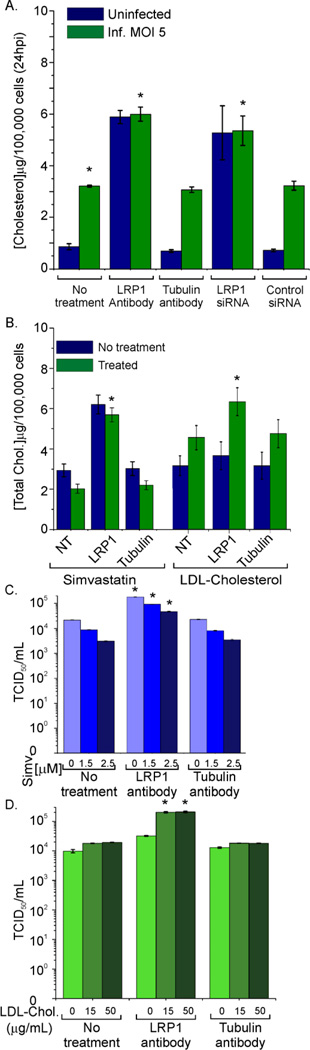
Since knockdown of LRP1 was shown previously to increase intracellular cholesterol levels (Zhou et al., 2009), cholesterol is important for replication of a variety of enveloped viruses (Pollock et al., 2010), and HCMV infection alters cholesterol levels (Abrahamsen et al., 1996), the cholesterol content of LRP1 knockdown cells was examined. As anticipated, when LRP1 activity was reduced by antibody or siRNA treatment, intracellular cholesterol levels assayed in extracts of cells lysed in buffer containing Triton X-100, increased at 24 h after mock or HCMV infection (Fig. 5A). Similar results were obtained using an organic solvent to extract cholesterol (Fig. S2A). Virus infection alone resulted in a significant increase in intracellular cholesterol at 24 hpi. The increase is consistent with earlier work (Abrahamsen et al., 1996) and occurs in spite of the elevation of cell surface LRP1, which would favor its decrease.
Fig. 5. Cholesterol content is increased in LRP1 knockdown cells via increased cholesterol import.
(A) Reduced LRP1 activity increases intracellular cholesterol: Fibroblasts were untreated, transfected with LRP1-specific or scrambled control siRNA for 24 h, or treated with LRP1 or tubulin antibody for 1 h; and then cells were mock infected or infected (5 IU/cell). At 24 hpi, cell cholesterol content was quantified.
(B) Inhibition of cholesterol synthesis had a modest effect, whereas extracellular cholesterol was needed to increase cellular cholesterol after LRP1 inhibition: Cholesterol was quantified after treatment with simvastatin (2.5 µM) or LDL-cholesterol (50 µg/ml) at 24 hpi.
(C) Inhibition of cholesterol synthesis does not block the enhanced yield of infectious virus produced by inhibition of LRP1: Fibroblasts were untreated, treated with simvastatin (2.5 µM), or mock treated; and 30 min later, cells were treated with antibody to LRP1 or tubulin or left untreated for 1 h. Cells were subsequently infected (0.1 IU/cell). At 96 hpi, released virus was quantified by TCID50 assay. (D) Extracellular cholesterol is required for the enhanced yield of infectious virus produced by inhibition of LRP1: Cells were serum starved for 48 h, treated first with the indicated antibodies for 1 h and then with various concentrations of LDL-cholesterol for 1 h. Next, cells were infected (0.1 PFU/cell) and released virus was assayed by TCID50 assay at 96 hpi.
Error bars report the standard error from three experiments with three replicates each. *P<0.001 (t-test, compared to equivalent control condition). Fig. S2 contains additional data relevant to this experiment.
To characterize the molecular mechanisms triggering the increase in cholesterol content of LRP1 knockdown cells, we examined the contribution of de novo cholesterol synthesis to altered cellular levels. Cells were treated with simvastatin to block HMG-CoA reductase, the rate limiting step in cholesterol biosynthesis, and examined its effect on virus release in LRP1- deficient cells. Simvastatin reduced intracellular cholesterol (Fig. 5B, left) and decreased the release of infectious virus into the medium in a dose-dependent manner (Fig. 5C); however, it did not reverse the increase in virus release observed after inhibition of LRP1 with neutralizing antibody (Fig. 5C). In contrast, when serum starved cells were treated with Simvastatin, the increase in cholesterol content and infectivity due to LRP1 inhibition was lost (Fig. S2B). Increases in total cellular cholesterol due to LRP1 inhibition (Fig. 5A) could also be negated in cholesterol-free medium (Fig. 5B, right, blue bars), which was restored by supplementing the medium with LDL-cholesterol (Fig. 5B, right, green bars). Additionally, LDL-cholesterol supplementation also restored the increase in virus release due to LRP1 inhibition (Fig. 5D). LDL-cholesterol and simvastatin treatments were not toxic to the cells (Fig. S2C). Taken together, these data demonstrate that increased cholesterol import, and not de novo synthesis, is largely responsible for the increase in infectious virus released from LRP1 knockdown cells.
HCMV virion cholesterol content affects infectivity
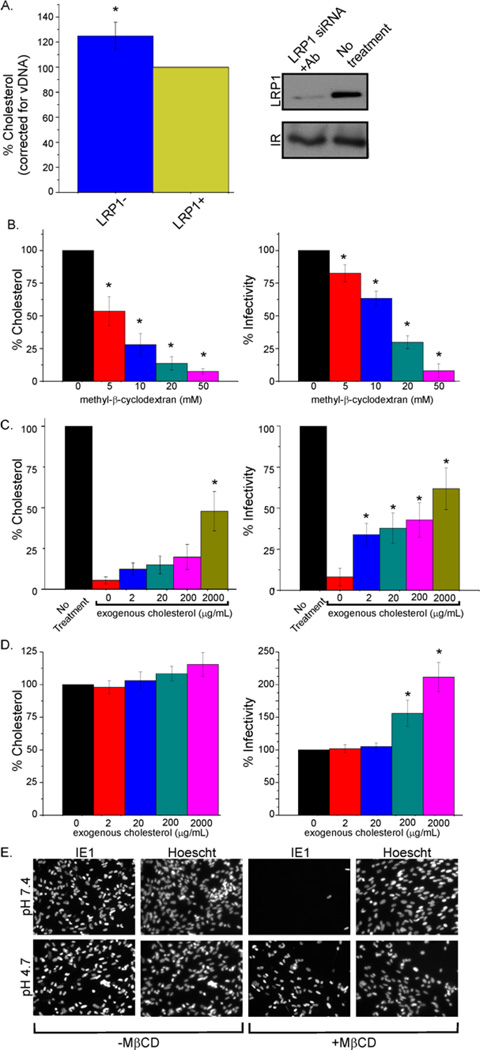
Increased intracellular cholesterol correlated with elevated virion infectivity in LRP1 knockdown cells, raising the possibility that virions released from these cells also have increased cholesterol content. To investigate this, we quantified the envelope cholesterol of gradient-purified virus particles from cells with normal LRP1 function and cells in which LRP1 was inhibited with neutralizing antibody. As predicted, virus released from cells deficient in LRP1 activity (Fig. 6A, right panel) exhibited an increase in virion-associated cholesterol (Fig. 6A, left panel).
Fig. 6. Cholesterol content of virions regulates fusion with the plasma membrane.
(A) LRP1 activity correlates with virion cholesterol level: Fibroblasts were left untreated or transfected with LRP1 siRNA (24 h before infection) and then treated with LRP1 neutralizing antibody 24 h later. After an additional 1 h, cells were infected (0.1 IU/cell). Antibody was reapplied every 48 h. 10 days later, virus was collected, gradient purified, normalized for virion DNA, and assayed for cholesterol content. LRP1 plasma membrane levels were assayed by Western blot to confirm its knockdown.
(B) Depletion of virion cholesterol reduces infectivity: Gradient purified virions were either mock treated or treated with increasing concentrations of MβCD for 30 min at 37°C. Treated virus was pelleted through a sorbitol cushion to remove MβCD and infectivity was assayed by IE1 immunofluorescence.
(C) Treatment of cholesterol-depleted virions with exogenous cholesterol substantially restores infectivity: MβCD-treated (50 mM) virus was incubated with varying concentrations of cholesterol for 30 min at 37°C, pelleted through a sorbitol cushion, and infectivity was assayed.
(D) Exogenous cholesterol treatment of normal virions enhances infectivity: Purified virions were incubated in varying concentrations of cholesterol for 30 min at 37°C, pelleted through a sorbitol cushion, and infectivity was assayed.
(E) Cholesterol-depleted virions enter cells efficiently when normal membrane fusion is bypassed: Fibroblasts were infected (3 IU/cell) with MβCD-treated (50 mM) or mock-treated virus, allowed to sit at 4°C for 30 min to allow virus binding, then treated with buffer at neutral (7.4) or acidic (4.7) pH for 3 min. Cells were assayed for IE1 expression at 24 hpi.
Error bars report the standard error from three experiments with three replicates each. *P<0.001 (t-test, compared to control condition). Fig S3 contains additional data relevant to this experiment.
To more directly explore the role of envelope cholesterol in HCMV infection, we treated purified virions with methyl-β-cyclodextrin (MβCD), which selectively extracts cholesterol from membranes (Ohtani et al., 1989). When virus particles released from LRP1 knockdown cells were treated with 10 mM MβCD (Fig. S3) their cholesterol content and infectivity were reduced to nearly wild-type levels, arguing that the increase in infectivity is in fact due to increased cholesterol content. Furthermore, wild type virions treated with the compound showed a dose- dependent decrease in cholesterol content, with ~90% depletion obtained with 50 mM MβCD (Fig 6B, left). This treatment resulted in a strikingly similar dose-dependent inhibition of HCMV infectivity (Fig. 6B, right). To assess whether the effect of MβCD could be rescued and show that the loss of infectivity was due mainly to cholesterol depletion, exogenous cholesterol was used to replenish the envelopes of MβCD-treated virions. This treatment partially restored envelope cholesterol (Fig. 6C, left) and HCMV infectivity (Fig. 6C, right), again in a dose-dependent manner. The highest dose of exogenous cholesterol tested (2 mg/ml) increased the cholesterol content and infectivity of MβCD-depleted virions by a factor of about 8. Higher cholesterol doses might completely reverse the depletion, but solubility issues limited the concentration used in the experiments. Alternatively, it is possible that MβCD-treatment also depleted the virus of another lipid, which would not be replenished by exogenous cholesterol. The infectivity of purified virions containing a normal level of cholesterol could also be augmented by treating with exogenous cholesterol. Treatment with 2 mg/ml cholesterol increased virion cholesterol content by 10% (Fig. 6D, left) and the resulting infectivity by a factor of 2 (Fig. 6D, right). In sum, these results show that depletion of cholesterol from the virion envelope reduces infectivity and are consistent with the view that LRP1 modulates the infectivity of virions by controlling their cholesterol content.
Envelope cholesterol content regulates HCMV fusion
Infection of host cells by enveloped viruses relies on the fusion of the viral envelope with either the endosomal or plasma membrane of the cell. As such, cholesterol might influence the infectivity of virions by modulating membrane fusion. We tested this notion by asking if the infectivity of MβCD-treated virions could be restored by circumventing the normal membrane fusion process during virus entry. This was done by using low pH to induce fusion of the virus envelope with the plasma membrane (Chang et al., 2010; Mercer and Helenius, 2008; Sun and Whittaker, 2003). Mock-depleted or cholesterol-depleted particles were bound to cells at 4°C then incubated in a low pH buffer (pH 4.7) to allow fusion-deficient virus to enter the cell and replicate. A neutral pH buffer was used as a control. Acid bypass of the fusion event caused a dramatic difference in the ability of cholesterol-depleted virus to enter the cell and express the IE1 immediate-early protein (Fig. 6E). IE1 expression was virtually undetectable when the cholesterol-depleted particles exposed to the cell surface in pH 7.4 buffer. In contrast, cholesterol-depleted particles entered cells and expressed IE1 as efficiently as mock-depleted particles. We conclude that envelope cholesterol is a critical determinant of HCMV fusion with the host cell plasma membrane.
DISCUSSION
The cell surface proteome includes proteins that are physically embedded in the lipid bilayer, proteins anchored to the outer leaflet of the plasma membrane (e.g., glycosylphosphatidylinositol (GPI)-anchored proteins) and proteins that interact with these two classes of membrane proteins. We utilized a proteomic approach, based on the purification membrane-associated protein ectodomains and MS-based label-free spectral counting analysis to identify 114 HCMV-induced changes (Fig. 2E and Table S1) in proteins predicted to be constituents of the cell surface proteome (Mi et al., 2010). We identified 28 altered proteins during the immediate-early phase of the replication cycle (6 hpi); 63 alterations during the early phase (24 hpi) and 83 changes during the late phase (72 hpi). Most of the changes were not verified by additional methods and thus must be considered tentative; however, at least 12 of the predicted alterations were reported previously. Platelet derived growth factor receptors α and β (Gredmark et al., 2007), integrin α-1 (Warren et al., 1994), epidermal growth factor receptor (Fairley et al., 2002), and four HLA class I molecules (van der Wal et al., 2002) were reduced, while CD44 and CD59 adhesion molecules (Ito et al., 1995), complement decay- accelerating factor (Spiller et al., 1996), and GLUT4 (Yu et al., 2011) were elevated.
The analysis identified six viral proteins at the cell surface. Five are known membrane glycoproteins, pUL55 (gB), pUL75 (gH), pUL100 (gM), pUL132 and pRL11, and one, pUL53, forms a complex with a membrane protein. They were detected predominately late after infection, so they are likely de novo synthesized rather than delivered by infecting virions.
Cell-coded LRP1 was upregulated at 6 and 24 hpi and downregulated at 72 hpi. It regulates cholesterol levels in fibroblasts (Terrand et al., 2009; Zhou et al., 2009), and changes in intracellular cholesterol flux can influence numerous cell regulatory processes (Fessler and Parks, 2011). LRP1 upregulates (Zhou et al., 2009) and HCMV downregulates (Sanchez and Dong, 2010) the ABCA1 transporter beginning at 24 hpi, which promotes the efflux of cholesterol from the cell, and both upregulate CD36 (Carlquist et al., 2004; Gaultier et al., 2010), which sponsors cholesterol import.
Western blot analysis confirmed the MS data showing that cell surface LRP1 is transiently elevated during infection (Fig. 3A), showing that the changes occur independently of viral gene expression (Fig. 3B), and the decrease in LRP1 late during infection was shown to result from release of the protein’s ectodomain (sLRP1) into the medium (Fig. 3C). LRP1 was shown to function as an HCMV restriction factor because abrogation of its function by siRNA knockdown (Fig. 4 C and D) or antibody neutralization (Fig. 4E) enhanced the yield of HCMV infectivity by a factor of about 15. The enhanced infectivity was not due to the production of increased numbers of virions, but rather it resulted from increased specific infectivity (IU/particle) of virons (Fig. 4F). Cellular (Fig. 5A) and virion (Fig. 6A) cholesterol content were increased when LRP1 function was inhibited. The increases depended on the availability of extracellular cholesterol (Fig. 5B), and depletion of cholesterol from virions reduced their specific infectivity (Fig. 6B) and inhibited their ability to fuse with the plasma membrane at the start of infection (Fig. 6E). The mechanistic role of cholesterol in HCMV-envelope fusion at the plasma membrane during entry remains unclear, although it is possible that cholesterol is necessary for the organization of HCMV-virus glycoproteins into fusion-competent domains within the envelope. Depletion of cholesterol from HIV-1 (Campbell et al., 2002; Guyader et al., 2002) and influenza virions (Sun and Whittaker, 2003) has also been shown to reduce infectivity by inhibiting internalization at the start of infection.
It is noteworthy that HCMV infection elevated cellular cholesterol by a factor of about three at 24 hpi (Fig. 5A), in spite of the fact that LRP1 was increased at the plasma membrane from 6–48 hpi (Fig. 2E and 3A; Table S1). The increase in total cell cholesterol following infection could result from HCMV-mediated downregulation of the ABCA1 transporter (Sanchez and Dong, 2010). Importantly, when LRP1 function was inhibited by treatment with siRNA or neutralizing antibody, the level of infected cell cholesterol increased at 24 hpi (Fig. 5A), demonstrating that LRP1 is limiting the virus-induced increase in cellular cholesterol.
As the infection enters the late stage (72 hpi and later, Fig. 3A), the level of LRP1 is reduced. The reduction results from the cleavage and release of sLRP1 from the cell surface late during infection with a cellular membrane metalloproteinase (Fig. 3C); however, we do not yet know which metalloproteinase triggers the cleavage. The release might be induced by a late viral gene product that activates a metalloproteinase responsible for the cleavage. Cholesterol has been shown to control LRP1 levels at the plasma membrane by modulating membrane-type 1 matrix metalloproteinase and metalloproteinase-12 activity (Selvais et al., 2011). Whatever the mechanism, reduced LRP1 levels would be expected to increase intracellular cholesterol levels and promote the production of virions with enhanced infectivity. It is possible that sLRP1 competes for ligand binding within the microenvironment of the residual cell-associated LRP1, further reducing its function at the plasma membrane. The soluble protein also might compete with and reduce LRP1 function in nearby uninfected cells, preparing them for infection.
In summary, our results demonstrate that the HCMV-infected cell surface proteome is markedly dynamic in its composition, with a substantial portion of the proteins that we have monitored increasing or decreasing at various times during the virus replication cycle. LRP1 provides an example of this dynamic behavior, and our analysis demonstrates that it functions as an HCMV restriction factor during the early stages of infection (until at least 48 hpi, Fig. 3A). LRP1 limits the availability of cholesterol and, as a consequence, reduces the infectivity of newly produced virions. LRP1 also has been shown to be elevated in HIV-1-infected T cells (Rasheed et al., 2008) and in monocytes of HIV-1-infected individuals whose disease doesn’t progress (Stebbing et al., 2003), so it might exhibit broad spectrum antiviral activity.
EXPERIMENTAL PROCEDURES
Cells and viruses
Human MRC5 fibroblasts (ATCC) were cultured in Dulbecco-modified Eagle medium (DMEM) containing 10% fetal bovine serum. Cell viability was assessed by a trypan blue exclusion assay (Moorman et al., 2010). HCMV was prepared from a bacterial artificial chromosome (BAC) clone of the AD169 strain expressing GFP (BADinGFP) (Wang et al., 2004). Virus stocks were concentrated and partially purified by centrifugation through 20% sorbitol cushions, and in some cases further purified in sodium tartrate gradients (Irmiere and Gibson, 1983).
Virus titers (infectious units, IUs) were determined by tissue culture infectious dose (TCID50) assay or by quantifying infected fibroblasts using IE1-specific antibody (clone 1B-12) (Zhu et al., 1995). Fluorescent images were processed with Q-Capture Pro software (Luo et al., 2007). Error bars represent the standard errors of the means from two or three independent experiments, each performed in triplicate. Virion infectivity was determined by measuring IUs and viral DNA in virions (Womack and Shenk, 2010).
Isolation of cell surface proteins
Confluent fibroblasts were mock infected or infected with HCMV (5 IU/cell). After 6, 24, or 72 h, plasma membrane proteins were isolated using the Pierce cell surface protein isolation kit. In brief, surface proteins were tagged with a thiol-cleavable biotin moiety, sulfo-NHS-SS-biotin [sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate], which contains a 24.3 angstrom spacer arm and reacts with the epsilon-amine of lysine residues, lysed in detergents (0.1% SDS, 1.0% Triton-X, 1.0% sodium deoxycholate) and sonicated. Biotinylated proteins were bound to a NeutrAvidin matrix (Pierce), and labeled proteins were eluted in buffer containing dithiothreitol [50 mM]. MS studies used one 15 cm plate and western blot assays one 3.5 cm plate of fibroblasts per sample.
Mass spectrometry
Filter-aided sample preparation (FASP) (Wisniewski et al., 2009) was used to generate tryspinized proteins for MS. About 1/4 of the 250 µg total protein recovered from the NeutrAvidin matrix was mixed with 400 µl of 0.1M Tris-HCl, pH 8.0, containing 8 M urea (UB) in a YM-10 Microcon device (Millipore). Samples were concentrated at 14,000 × g, then washed with 400 µl of UB. Following centrifugation, 200 µl of UB containing 0.05 M iodoacetamide was added. Samples were mixed on a TOMY shaker for 1 min then incubated at room temperature in the dark for 30 min. Following centrifugation, 200 µl of 0.05 M ammonium bicarbonate (ABC) was added to the filter unit and concentrated. This step was repeated twice. The concentrate was subjected to proteolytic digestion overnight at 37°C in 100 µl of 0.05 M ABC containing 10 ng/µl trypsin (Promega). The peptide digests were collected by centrifugation, and the filter was rinsed with 3 × 50 µl of HPLC grade water and centrifuged. Samples were acidified to a final concentration of 1% trifluoroacetic acid. Vacuum centrifugation was used to concentrate the sample to ~0.5 µg/µl prior to MS analysis.
Tryptic peptides (~2.5 µg) were analyzed (Tsai et al., 2012) by nLC-MS/MS on a Dionex Ultimate 3000 RSLC coupled directly to an LTQ-Orbitrap Velos ETD mass spectrometer (ThermoFisher Scientific) operated in data-dependent acquisition mode with FT preview scan disabled and predictive AGC and dynamic exclusion enabled (repeat count: 1, exclusion duration: 90 sec). A single acquisition cycle comprised a single full-scan mass spectrum (m/z = 350 – 1700) in the orbitrap (R = 30,000 @ m/z = 400), followed by CID fragmentation in the linear ion trap on the top 20 most intense precursor ions (min. signal = 1000) determined from the full-scan mass spectrum.
MS/MS spectra from raw data files were extracted by Proteome Discover (ver. 1.3, ThermoFisher Scientific) and submitted to SEQUEST (ver. 1.20) to search against the UniProt SwissProt sequence database (downloaded 11/2010) containing the subset of human, herpesvirus, and common contaminant sequences (21,570 entries). Peptide spectrum matches (PSMs) were generated by searching MS/MS spectra against indexed peptide databases, generated from the forward and reverse protein sequence entries (Tsai et al., 2012). Q-values for SEQUEST PSMs were calculated using the Percolator algorithm (Spivak et al., 2009) and filtered to achieve a global peptide false discovery rate of ≤ 1 %. Filtered peptide assignments were assembled into protein groups with a minimum of two unique peptides per protein using strict rules of parsimony.
For spectral counting analyses, total assigned spectra for each protein group (including unique and shared sequences) were used. After normalizing each sample set for total spectral counts, proteins with a count below 10 were removed. Specificity of enrichment was calculated for each protein as the spectral count ratio of biotinylated/non-biotinylated. Proteins with less than 5-fold enrichment were removed. HCMV-induced protein alterations were similarly calculated for each protein as the spectral count ratio of infected/uninfected. A change of ≥ 2.0- fold in spectral count ratio was considered differentially regulated since comparison of spectral counts between biological replicates resulted in ~98% of proteins that were changed ≤ 2-fold (Fig. 1C). Subcellular protein localization was obtained using Proteome Discoverer (ver. 1.3, ThermoFisher Scientific).
Inhibition of LRP1 function
To reduce the level of LRP1 protein, siRNA (5’-CAUCGAUCUUCACAAAGGA-3’) was transfected into fibroblasts at about 80% confluency using opti-mem and oligofectamine (Invitrogen). To inhibit LRP1 with neutralizing antibody, fibroblasts were grown to ~100% confluency and incubated at 37°C with 15 µg/ml LRP1 neutralizing antibody (Progen) (Orr et al., 2003). To assay the effect of inhibition, fibroblasts were infected (0.1 IU/cell) at 24 h post siRNA transfection or 60 min after antibody treatment, and infectious progeny was assayed 96 h later.
Analysis of nucleic acids and proteins
Viral DNA and RNA was assayed by qPCR or qRT-PCR, respectively, and proteins were monitored by western blot assay (Womack and Shenk, 2010). Antibodies to the following antigens were used: LRP1 and α-tubulin (Sigma); LDLR, ABCA1, LPL, Insulin Receptor, PGE2R2 and β-actin-HRP (Abcam); HLA-B/C (Santa Cruz); (ICP36)UL144 (Virusys); (pUL123)IE1 (clone 1B-12) (Zhu et al., 1995), (pp28)UL99 (clone 10B4-29) (Silva et al., 2003). To assay cell-free sLRP1, medium was concentrated, sLRP1 was immunoprecipitated using specific antibody or isotype control (R&D systems), and proteins were analyzed by western blot.
Manipulation and quantification of cholesterol levels
To inhibit cholesterol synthesis, fibroblasts were incubated for 90 min prior to infection or mock infection with simvastatin (1.5 or 2.5 µM; Sigma). To inhibit LDLR-mediated cholesterol import, cells were transfected with siRNA (5’-GUCUUUGAGGACAAAGUAU-3’) 24 h prior to infection or mock infection.
For treatment of cells with cholesterol, serum-starved (48 h), confluent fibroblasts received various concentrations of cholesterol + fatty acid-free (>96%) BSA (Sigma) for 1 h before mock infection or infection.
For treatment of virions with cholesterol, purified particles (~107 IU in 50 µl) were incubated in various concentrations of exogenous cholesterol (Sigma) for 30 min at 37°C. Immediately after treatment, virions were centrifuged through a sorbitol cushion to remove exogenous cholesterol, and used to infect fibroblasts (0.5 IU/cell). Virus cholesterol content was determined using the Amplex Red Cholesterol Assay (Invitrogen).
To deplete cholesterol from virions, purified particles (~107 IU in 50 µl) were mock treated (PBS only) or treated with increasing concentrations of methyl-β-cyclodextrin (MβCD) (Sigma) for 30 min at 37°C (Ohtani et al., 1989; Sun and Whittaker, 2003). Immediately after treatment, virions were centrifuged through a sorbitol cushion, and used to infect fibroblasts. For rescue experiments, exogenous cholesterol (Sigma) was added to the virion cyclodextrin mixture at 37°C for 30 min, before centrifugation through a sorbitol cushion.
To quantify cholesterol levels, cells or virions were lysed in PBS containing 1% Triton X-100 and protease inhibitor cocktail (Sigma). Cell lysates were clarified by centrifugation, and cholesterol content was determined using the Amplex Red Cholesterol Assay (Invitrogen). When cells were extracted with an organic solvent [chloroform:isopropanol:NP-40 (7:11:0.1)], the Abcam cholesterol quantification kit was used. Cellular and viral cholesterol content were normalized to cell number and viral DNA, respectively.
Acid Bypass assay
After addition of virus, cells were maintained at 4°C for 30 min to allow virus binding, then treated with buffer at a neutral (Tris, pH 7.4) or acidic (MES, pH 4.7) pH for 3 min (Chang et al., 2010), washed with PBS, and examined for IE1 expression at 24 hpi.
Supplementary Material
HIGHLIGHTS.
HCMV infection of fibroblasts induced changes in 114 cell surface proteins.
LDL receptor-related protein 1 (LRP1) was transiently elevated after infection.
Elevated LRP1 reduced intracellular cholesterol in infected cells.
Inhibition of LRP1 increased intracellular cholesterol and infectious virus yield.
ACKNOWLEDGEMENTS
We thank members of our labs for helpful discussions. The work was supported by grants from U.S. National Institutes of Health to I.M.C. (DP1DA026192), and T.S. (AI87672 and CA82396), HFSPO award RGY0079/2009-C to I.M.C, and an American Heart Association Postdoctoral Fellowship (12POST8850003) to N.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abrahamsen LH, Clay MJ, Lyle JM, Zink JM, Fredrikson LJ, DeSiervo AJ, Jerkofsky MA. The effects of cytomegalovirus infection on polar lipids and neutral lipids in cultured human cells. Intervirology. 1996;39:223–229. doi: 10.1159/000150521. [DOI] [PubMed] [Google Scholar]
- Boucher P, Herz J. Signaling through LRP1: Protection from atherosclerosis and beyond. BiochemPharmacol. 2010 doi: 10.1016/j.bcp.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. JVirol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SM, Crowe SM, Mak J. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS. 2002;16:2253–2261. doi: 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- Carlquist JF, Muhlestein JB, Horne BD, Hart NI, Lim T, Habashi J, Anderson JG, Anderson JL. Cytomegalovirus stimulated mRNA accumulation and cell surface expression of the oxidized LDL scavenger receptor, CD36. Atherosclerosis. 2004;177:53–59. doi: 10.1016/j.atherosclerosis.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Chang SJ, Chang YX, Izmailyan R, Tang YL, Chang W. Vaccinia virus A25 and A26 proteins are fusion suppressors for mature virions and determine strain-specific virus entry pathways into HeLa, CHO-K1, and L cells. JVirol. 2010;84:8422–8432. doi: 10.1128/JVI.00599-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del VM, Hengel H, Hacker H, Hartlaub U, Ruppert T, Lucin P, Koszinowski UH. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide- loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. JExpMed. 1992;176:729–738. doi: 10.1084/jem.176.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairley JA, Baillie J, Bain M, Sinclair JH. Human cytomegalovirus infection inhibits epidermal growth factor (EGF) signalling by targeting EGF receptors. J Gen Virol. 2002;83:2803–2810. doi: 10.1099/0022-1317-83-11-2803. [DOI] [PubMed] [Google Scholar]
- Fessler MB, Parks JS. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J Immunol. 2011;187:1529–1535. doi: 10.4049/jimmunol.1100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini M, Montagnana M. Low-density lipoprotein receptor-related protein 1: new functions for an old molecule. Clin Chem Lab Med. 2011;49:967–970. doi: 10.1515/CCLM.2011.154. [DOI] [PubMed] [Google Scholar]
- Gaultier A, Simon G, Niessen S, Dix M, Takimoto S, Cravatt BF, 3rd, Gonias SL. LDL receptor-related protein 1 regulates the abundance of diverse cell-signaling proteins in the plasma membrane proteome. J Proteome Res. 2010;9:6689–6695. doi: 10.1021/pr1008288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredmark S, Straat K, Homman-Loudiyi M, Kannisto K, Soderberg-Naucler C. Human cytomegalovirus downregulates expression of receptors for platelet-derived growth factor by smooth muscle cells. Journal of virology. 2007;81:5112–5120. doi: 10.1128/JVI.02197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyader M, Kiyokawa E, Abrami L, Turelli P, Trono D. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. Journal of virology. 2002;76:10356–10364. doi: 10.1128/JVI.76.20.10356-10364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- Ito M, Watanabe M, Ihara T, Kamiya H, Sakurai M. Increased expression of adhesion molecules (CD54, CD29 and CD44) on fibroblasts infected with cytomegalovirus. MicrobiolImmunol. 1995;39:129–133. doi: 10.1111/j.1348-0421.1995.tb02179.x. [DOI] [PubMed] [Google Scholar]
- Jedrychowski MP, Gartner CA, Gygi SP, Zhou L, Herz J, Kandror KV, Pilch PF. Proteomic analysis of GLUT4 storage vesicles reveals LRP1 to be an important vesicle component and target of insulin signaling. The Journal of biological chemistry. 2010;285:104–114. doi: 10.1074/jbc.M109.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josic D, Clifton JG. Mammalian plasma membrane proteomics. Proteomics. 2007;7:3010–3029. doi: 10.1002/pmic.200700139. [DOI] [PubMed] [Google Scholar]
- Leis M, Marschall M, Stamminger T. Downregulation of the cellular adhesion molecule Thy-1 (CD90) by cytomegalovirus infection of human fibroblasts. J Gen Virol. 2004;85:1995–2000. doi: 10.1099/vir.0.79818-0. [DOI] [PubMed] [Google Scholar]
- Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor- related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88:887–918. doi: 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund R, Leth-Larsen R, Jensen ON, Ditzel HJ. Efficient isolation and quantitative proteomic analysis of cancer cell plasma membrane proteins for identification of metastasis-associated cell surface markers. JProteomeRes. 2009;8:3078–3090. doi: 10.1021/pr801091k. [DOI] [PubMed] [Google Scholar]
- Luo MH, Rosenke K, Czornak K, Fortunato EA. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase- mediated DNA damage responses during lytic infection. J Virol. 2007;81:1934–1950. doi: 10.1128/JVI.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Shenk T, Pass RF. In: Cytomegaloviruses. Virology Fields, Knipe DM, Howley PM., editors. Philadelphia, PA: Lippincott, Williams and Wilkins; 2007. pp. 2702–2772. [Google Scholar]
- Montag C, Wagner JA, Gruska I, Vetter B, Wiebusch L, Hagemeier C. The latency-associated UL138 gene product of human cytomegalovirus sensitizes cells to tumor necrosis factor alpha (TNF-alpha) signaling by upregulating TNF-alpha receptor 1 cell surface expression. Journal of virology. 2011;85:11409–11421. doi: 10.1128/JVI.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. A targeted spatial- temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. MolCell Proteomics. 2010;9:851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss P. The emerging role of cytomegalovirus in driving immune senescence: a novel therapeutic opportunity for improving health in the elderly. Curr Opin Immunol. 2010;22:529–534. doi: 10.1016/j.coi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS pathogens. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng X-J, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani Y, Irie T, Uekama K, Fukunaga K, Pitha J. Differential effects of alpha- , beta- and gamma-cyclodextrins on human erythrocytes. Eur J Biochem. 1989;186:17–22. doi: 10.1111/j.1432-1033.1989.tb15171.x. [DOI] [PubMed] [Google Scholar]
- Orr AW, Pedraza CE, Pallero MA, Elzie CA, Goicoechea S, Strickland DK, Murphy-Ullrich JE. Low density lipoprotein receptor-related protein is a calreticulin coreceptor that signals focal adhesion disassembly. J Cell Biol. 2003;161:1179–1189. doi: 10.1083/jcb.200302069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Park B, Spooner E, Houser BL, Strominger JL, Ploegh HL. The HCMV membrane glycoprotein US10 selectively targets HLA-G for degradation. J Exp Med. 2010;207:2033–2041. doi: 10.1084/jem.20091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Maidji E. Cytomegalovirus infection in the human placenta: maternal immunity and developmentally regulated receptors on trophoblasts converge. Curr Top Microbiol Immunol. 2008;325:383–395. doi: 10.1007/978-3-540-77349-8_21. [DOI] [PubMed] [Google Scholar]
- Pollock S, Nichita NB, Bohmer A, Radulescu C, Dwek RA, Zitzmann N. Polyunsaturated liposomes are antiviral against hepatitis B and C viruses and HIV by decreasing cholesterol levels in infected cells. ProcNatlAcadSciUSA. 2010;107:17176–17181. doi: 10.1073/pnas.1009445107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Paskas S, Zivkovic M, Burysek L, Laumonnier Y. Human cytomegalovirus increases HUVEC sensitivity to thrombin and modulates expression of thrombin receptors. J Thromb Thrombolysis. 2010;30:164–171. doi: 10.1007/s11239-010-0447-7. [DOI] [PubMed] [Google Scholar]
- Rasheed S, Yan JS, Lau A, Chan AS. HIV replication enhances production of free fatty acids, low density lipoproteins and many key proteins involved in lipid metabolism: a proteomics study. PLoS One. 2008;3:e3003. doi: 10.1371/journal.pone.0003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Dong JJ. Alteration of lipid metabolism in cells infected with human cytomegalovirus. Virology. 2010;404:71–77. doi: 10.1016/j.virol.2010.04.026. [DOI] [PubMed] [Google Scholar]
- Selvais C, D'Auria L, Tyteca D, Perrot G, Lemoine P, Troeberg L, Dedieu S, Noel A, Nagase H, Henriet P, et al. Cell cholesterol modulates metalloproteinase-dependent shedding of low-density lipoprotein receptor-related protein-1 (LRP-1) and clearance function. FASEB J. 2011 doi: 10.1096/fj.10-169508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahgasempour S, Woodroffe SB, Garnett HM. Alterations in the expression of ELAM-1, ICAM-1 and VCAM-1 after in vitro infection of endothelial cells with a clinical isolate of human cytomegalovirus. Microbiol Immunol. 1997;41:121–129. doi: 10.1111/j.1348-0421.1997.tb01177.x. [DOI] [PubMed] [Google Scholar]
- Silva MC, Yu QC, Enquist L, Shenk T. Human cytomegalovirus UL99- encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. Journal of virology. 2003;77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg-Naucler C. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol. 2008;41:218–223. doi: 10.1016/j.jcv.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller OB, Morgan BP, Tufaro F, Devine DV. Altered expression of host- encoded complement regulators on human cytomegalovirus-infected cells. Eur J Immunol. 1996;26:1532–1538. doi: 10.1002/eji.1830260719. [DOI] [PubMed] [Google Scholar]
- Spivak M, Weston J, Bottou L, Kall L, Noble WS. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J Proteome Res. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J, Gazzard B, Kim L, Portsmouth S, Wildfire A, Teo I, Nelson M, Bower M, Gotch F, Shaunak S, et al. The heat-shock protein receptor CD91 is up-regulated in monocytes of HIV-1-infected"true" long-term nonprogressors. Blood. 2003;101:4000–4004. doi: 10.1182/blood-2002-11-3353. [DOI] [PubMed] [Google Scholar]
- Streblow DN, Dumortier J, Moses AV, Orloff SL, Nelson JA. Mechanisms of cytomegalovirus-accelerated vascular disease: induction of paracrine factors that promote angiogenesis and wound healing. Curr Top Microbiol Immunol. 2008;325:397–415. doi: 10.1007/978-3-540-77349-8_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Whittaker GR. Role for influenza virus envelope cholesterol in virus entry and infection. Journal of virology. 2003;77:12543–12551. doi: 10.1128/JVI.77.23.12543-12551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrand J, Bruban V, Zhou L, Gong W, El AZ, May P, Zurhove K, Haffner P, Philippe C, Woldt E, et al. LRP1 controls intracellular cholesterol storage and fatty acid synthesis through modulation of Wnt signaling. JBiolChem. 2009;284:381–388. doi: 10.1074/jbc.M806538200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional Proteomics Establishes the Interaction of SIRT7 with Chromatin Remodeling Complexes and Expands Its Role in Regulation of RNA Polymerase I Transcription. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015156. M111 015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wal FJ, Kikkert M, Wiertz E. The HCMV gene products US2 and US11 target MHC class I molecules for degradation in the cytosol. Curr Top Microbiol Immunol. 2002;269:37–55. doi: 10.1007/978-3-642-59421-2_3. [DOI] [PubMed] [Google Scholar]
- Wang D, Bresnahan W, Shenk T. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc Natl Acad Sci U S A. 2004;101:16642–16647. doi: 10.1073/pnas.0407233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AP, Owens CN, Borysiewicz LK, Patel K. Down-regulation integrin alpha 1/beta 1 expression and association with cell rounding in human cytomegalovirus-infected fibroblasts. J Gen Virol. 1994;75(Pt 12):3319–3325. doi: 10.1099/0022-1317-75-12-3319. [DOI] [PubMed] [Google Scholar]
- Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod'homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, et al. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol. 2008;41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Womack A, Shenk T. Human cytomegalovirus tegument protein pUL71 required for efficient virion egress. MBio. 2010;1 doi: 10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. Journal of virology. 2011;85:1573–1580. doi: 10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Choi HY, Li WP, Xu F, Herz J. LRP1 controls cPLA2 phosphorylation, ABCA1 expression and cellular cholesterol export. PLoS One. 2009;4:e6853. doi: 10.1371/journal.pone.0006853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. Journal of virology. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.