Abstract
Of the Capsicum peppers (Capsicum spp.), cultivated C. annuum is the most commercially important, but has lacked an intraspecific linkage map based on sequence-specific PCR markers in accord with haploid chromosome numbers. We constructed a linkage map of pepper using a doubled haploid (DH) population derived from a cross between two C. annuum genotypes, a bell-type cultivar ‘California Wonder’ and a Malaysian small-fruited cultivar ‘LS2341 (JP187992)’, which is used as a source of resistance to bacterial wilt (Ralstonia solanacearum). A set of 253 markers (151 SSRs, 90 AFLPs, 10 CAPSs and 2 sequence-tagged sites) was on the map which we constructed, spanning 1,336 cM. This is the first SSR-based map to consist of 12 linkage groups, corresponding to the haploid chromosome number in an intraspecific cross of C. annuum. As this map has a lot of PCR-based anchor markers, it is easy to compare it to other pepper genetic maps. Therefore, this map and the newly developed markers will be useful for cultivated C. annuum breeding.
Keywords: pepper (Capsicum annuum L.), SSR markers, genetic map, 12 linkage groups
Introduction
Cultivated Capsicum fruits are used as a source of vegetables, spice, colorant and for some medical applications. The genus is native to Central and South America (Pickersgill 1991) and includes the species C. chinense, C. baccatum, C. frutescens, C. pubescens and C. annuum. Of these five species, C. annuum is the most important one because it is cultivated in both tropical and temperate area in the world and it is the most versatile of the five species. In contrast, the other four species are cultivated in limited regions in the world or only in tropical areas and they are mainly used as spices.
Linkage maps of Capsicum have been constructed using both intraspecific annuum populations and interspecific crosses such as C. annuum × C. chinense (Kang et al. 2001, Lee et al. 2004, Livingstone et al. 1999, Yi et al. 2006) and C. annuum × C. frutescens (Ben-Chaim et al. 2006, Rao et al. 2003, Wu et al. 2009). Interspecific crosses benefit from a high level of marker polymorphism but suffer from low fertility, segregation distortion and major structural rearrangements (Lanteri 1991, Lanteri and Pickersgill 1993, Wu et al. 2009), which limit the power of the linkage analysis and restrict their relevance to marker-assisted selection (MAS) applications (Lefebvre et al. 2002).
Several intraspecific maps of C. annuum have been reported (Barchi et al. 2007, Caranta et al. 1997a, 1997b, Lefebvre et al. 1995, Minamiyama et al. 2006, 2007, Ogundiwin et al. 2005, Sugita et al. 2006). RFLP and RAPD markers were used for constructing some of the maps. However, RFLP markers have been largely replaced by a new generation of molecular markers (e.g. SSR, AFLP and CAPS) which offer tremendous advances in cost, efficiency, throughput and sensitivity for plant genomics. RAPD markers also have problem with reproducibility. The map position of highly reproducible, locus-specific, co-dominant PCR-based markers is of particular value for the integration of genetic information from different populations and will underpin much applied research in pepper, including gene mapping, quantitative trait loci (QTL) analysis, and marker-assisted selection. Previously, Minamiyama et al. (2006, 2007) have constructed a pepper map mainly by using SSR markers with high polymorphism information content. Nevertheless, these studies have not resulted in complete genetic maps of the pepper genome in which 12 linkage groups correspond to the haploid chromosome numbers. The maps are also not comparable in marker position to any other maps in pepper, since they have few common markers with other pepper maps. We constructed an SSR-based map which involved several QTLs such as bacterial wilt (Ralstonia solanacearum) resistance and growth traits in a previous study (Mimura et al. 2009b, 2010). However, our earlier map also described several chromosomes as segmented short linkage groups.
In this study, we describe an SSR-based genetic map of cultivated C. annuum with the 12 pepper chromosomes by adding lots of reproducible markers in common with the maps of Minamiyama et al. (2006), Wu et al. (2009) and Yi et al. (2006). Moreover, we detected several QTLs related to economically important fruit traits. Therefore, the map developed through this study is useful for MAS and QTL in commercially important cultivated C. annuum.
Materials and Methods
Plant materials
Malaysian accession ‘LS2341 (JP187992)’ bearing small elongated, oval fruit and resistant to bacterial wilt (Mimura et al. 2009a) was used as the pollen donor. This accession was obtained from the National Institute of Agrobiological Sciences (NIAS) Genebank in Tsukuba, Japan. A sweet pepper cultivar, ‘California Wonder (CW)’ was employed as a seed parent. A segregating doubled haploid (DH) population (n = 94) was bred by anther culture of an F1 individual (Mimura et al. 2009b).
Marker analysis and map construction
AFLP and SSR polymorphisms were scored according to a method described by Minamiyama et al. (2006). The SSR primer pairs used in this study were developed from genomic libraries and/or registered sequences at the databases (Huang et al. 2001, Lee et al. 2004, Mimura et al. 2010, Minamiyama et al. 2006, 2007, Nagy et al. 2007, Yi et al. 2006).
In order to converge the expected 12 linkage groups and to assign a few, yet unknown linkage groups, we also tried to use Conserved Ortholog Set II (COSII) markers (Wu et al. 2009). COSII markers are PCR-based markers developed from a set of single-copy conserved orthologous genes. In pepper, map positions of COSII markers have already been shown (Wu et al. 2009). Since most of the markers had no polymorphism between the parental lines, the PCR products were sequenced and we detected SNPs for designing as original CAPS or dCAPS markers. Mapping was performed using JoinMap 3.0 software with a population type code, DH1 (Van Ooijen and Voorrips 2001). Markers were grouped at an LOD score of 4.0, where map distances were calculated using the Kosambi function (Kosambi 1944).
Fruit trait QTLs
The parents and the 94 F1DH lines were grown in a heated green house in Kyoto Prefectural Agriculture, Forestry and Fisheries Technology Research Centre, Seika, Kyoto, Japan, and the fruit traits were studied during two growth seasons (May–Sep. 2007 and Jan.–May 2009).
The following traits were evaluated for each fruit:
(1) fruit length (FL)—the distance (in millimetres) from the pedicel attachment to its apex; (2) fruit diameter (FD)—measured at the maximum width (in millimetres); (3) fruit shape (FS)—the ratio of fruit length to fruit diameter.
Average scores of 5 to 10 fruits for each line were treated as trait data.
QTL mapping was performed using Map QTL 6.0 software (Van Ooijen 2009) under the multiple QTL model, which is equivalent to composite interval mapping.
Results and Discussion
Genetic map construction
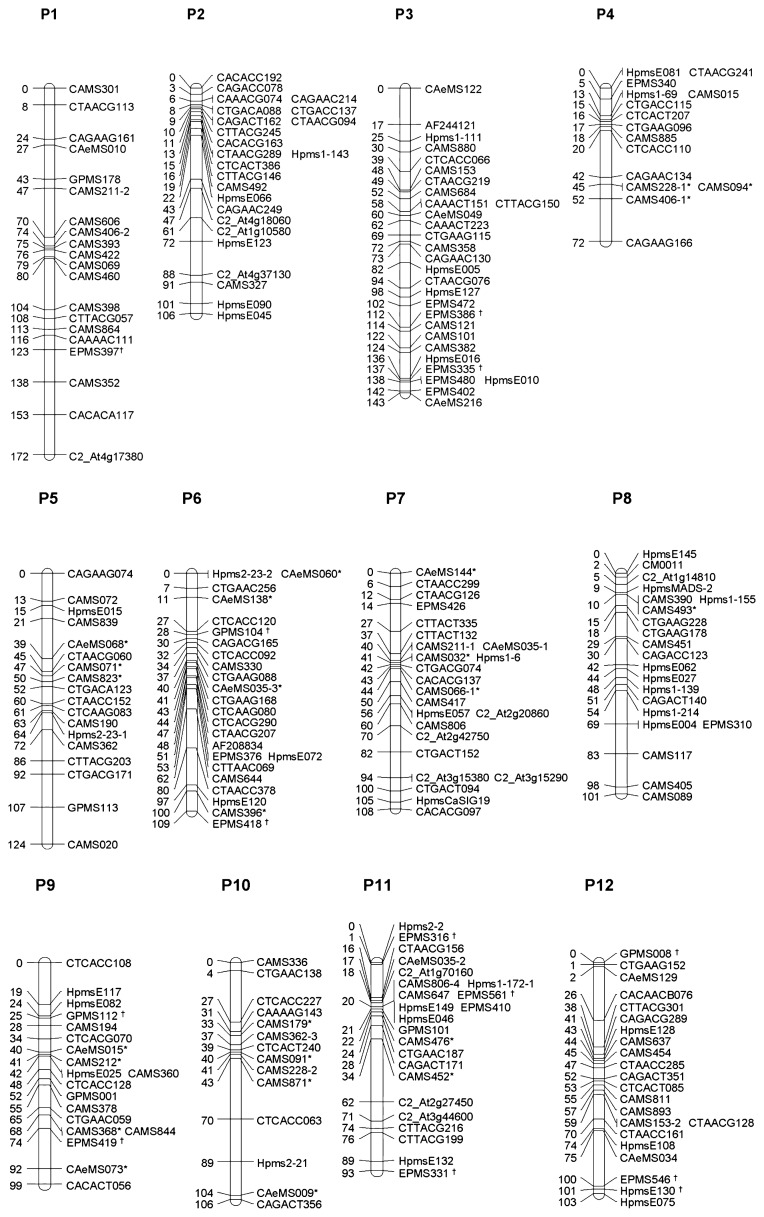
The map in this study contains 151 SSR, 90 AFLP, 10 CAPS/dCAPS and 2 STS markers in 12 linkage groups, and covers 1,336 cM (Fig. 1). As for COSII markers, we tried 84 markers, and obtained PCR products from two parents of this study in 61 markers. Then, 12 of 61 markers were able to be modified as CAPS/dCAPS or indel STS markers with polymorphism (Table 1). Moreover, new 24 SSR markers have been mapped in this study. Their unique primer sequences and other information are shown in Table 2. Furthermore, previously reported 13 SSR markers were firstly mapped in this study (Fig. 1).
Fig. 1.
A genetic linkage map of cultivated C. annuum genome. Nomenclature of linkage groups is referred to the consensus chromosome numbers (Wu et al. 2009). Marker names and the map distances (cM) are indicated on the right and left of linkage groups, respectively. Markers named AF__, CAeMS__, CAMS__, CM__, EPMS__, GPMS__, Hpms__ are SSR markers. COSII markers are represented by the name C2_At__ (Table 1). Others are AFLP markers. Newly used 24 SSR markers (Table 2) are indicated with asterisks (*). Previously reported but firstly mapped 13 SSR markers are indicated with daggers (†).
Table 1.
CAPS/dCAPS and STS markers modified and used in this study
| Marker namea | Forward primer (5′-3′) | Reverse primer (5′-3′) | Restriction enzyme | Chromosome | Expected product size (bp)b | |
|---|---|---|---|---|---|---|
|
| ||||||
| CW | LS | |||||
| C2_At4g17380 | caaggatgggaacaatggacag | gcaagttgaagaggtcaaactgcat | Tsp509 I | 1 | 140 | 150 |
| C2_At4g18060 | tcaagcagtttagtgcaactggttatg | tgccttaacaatctctttctgaaaatc | Mse I | 2 | 550, 300, 250, 100 | 550, 420, 250, 100 |
| C2_At1g10580 | agtaatgatggaagcaagtttttgac | agaagacaaacctccatcaggtgagaa | BsaB I | 2 | 250 | 300, 200 |
| C2_At4g37130 | ttacagcaaactgtagcaagatttgag | tgctgttttcattgattcaatgtactg | Alu I | 2 | 1000 | 600, 210, 190 |
| C2_At2g20860 | aaatgaggagctggtggtcacat | taggtatcgcttaactgatggtg | Rsa I | 7 | 180 | 100, 80 |
| C2_At2g42750 | gggaaaatggtgagatggcaaagttag | caagtataatcctccacgtgtcattg | Afa I | 7 | 110, 50 | 160 |
| C2_At3g15380 | ttgtttggcggctattgggc | agcattacgattcacagatttgatgg | Msp I | 7 | 380, 200 | 500 |
| C2_At3g15290 | tctgctattttggcttctaatacaag | acaatatgtgtcttctgatgtatctgc | Bsp1286 I | 7 | 1500 | 680 |
| C2_At1g14810 | gcattagtggtgttggaccaca | gacaggcaaggctatgtgacag | Indel | 8 | 150 | 140 |
| C2_At1g70160 | acatgtggaacgaagctctgaataa | tggaggtaaagaaggacaattctcattc | Alu I | 11 | 900, 200 | 600, 200 |
| C2_At2g27450 | gaatttctgtatctcatttggattc | acccctaataaaaaagagtcac | Taq I | 11 | 160 | 180 |
| C2_At3g44600 | tcctttataccgacttgaagctattg | agattctatgtttcttgaaagcacagc | Indel | 11 | 500 | 530 |
Restriction Sites were detected in PCR-amplified fragments from the population of this study and several primer pairs were newly designed. However, the marker names are the same as the original COSII markers to facilitate comparison with other maps.
CW = allele from California Wonder, LS = allele from LS2341.
Table 2.
Twenty four SSR markers newly used in this study
| Marker name | Forward primer (5′-3′) | Reverse primer (5′-3′) | Motif | Chromosome | Expected product size (bp)a | |
|---|---|---|---|---|---|---|
|
| ||||||
| CW | LS | |||||
| CAMS094 | tgtagctcacatcgtctccact | gcattgcatttcacttgcat | (ta)5(tg)13 | 4 | 190 | 188 |
| CAMS228 | gagggctaagcaaagcagaa | tgcatgtttcccttagtttcc | (ta)5(tg)13 | 4 | 241 | 239 |
| CAMS406 | taaaaatcgcggaaagttgc | gtcgttctatgcggcatttt | (ga)8 | 4 | 184 | 182 |
| CAeMS068 | atcaaatctcaacacatggtggct | gtttactgtatctccggccctgtca | (cct)5ctt(cct)3 | 5 | 169 | 166 |
| CAMS071 | aatgggatctgcatgagaca | ttccctaaaagatggtgattcc | (ac)11 | 5 | 172 | 166 |
| CAMS823 | tcctcctccttctcgtgttc | aaagaagcagcaggtgaaga | (ctt)5 | 5 | 226 | 228 |
| CAeMS035 | aggtctatcggaaacagcctttct | gtttgatcacatcccagtcgaatccta | (tgt)5 | 6 | 183 | 181 |
| CAeMS060 | atcaagacaacaacatcatgggga | gtttcgcctatcaacaatggcaaataca | (ta)10 | 6 | 286 | 292 |
| CAeMS138 | acacacacaatttccctcactcac | gtttctctcaaatccctccgttgttc | (ag)5...(ag)5...(ga)3...(ag)3 | 6 | 250 | 244 |
| CAMS396 | gtcggccgtcattcactatt | agcttgatgcacctggtctt | (ag)12 | 6 | 240 | 244 |
| CAeMS144 | ataactttgattcctagttcggcg | gtttgaacccccaatcatcatatcctca | (gaa)5 | 7 | 222 | 219 |
| CAMS032 | tgccacataggttggctttc | caaagccaatgcacataatca | (gt)13 | 7 | 233 | 245 |
| CAMS066 | aaaaacatgcaccagtcctt | caaccgcctgaattttctct | (ac)11 | 7 | 157 | 153 |
| CAMS493 | tcgatgacgaaaaagtgtgaa | agggcaaaagacccattctt | (ag)6 | 8 | 225 | 223 |
| CAeMS015 | atgccttggtggtggttaaatctg | gtttagcggtatggactgcgtacatctt | (caa)7 | 9 | 273 | 270 |
| CAeMS073 | atgcttctaagaaaccccacaaca | gtttctcataaaggggttgggattga | (tat)7 | 9 | 234 | 230 |
| CAMS212 | ttccctttcccaacatggta | acacccgaagatgggttaga | (tg)10 | 9 | 154 | 150 |
| CAMS368 | gagtggataagcaaggacgttt | tttgcttccctttttgcttc | (ag)23 | 9 | 206 | 180 |
| CAeMS009 | acgcaccaacgaatatctatctca | gtttccgtccagatctacttttcctgc | (ag)4...(ag)8 | 10 | 246 | 232 |
| CAMS091 | tgctaaacttggttccctatcc | cgaagatggattagcgggta | (ta)6(tg)10 | 10 | 180 | 172 |
| CAMS179 | catgtcatgaagttgataagacaatg | tgttccagtgaaaggcttctt | (ac)13(at)9 | 10 | 228 | 224 |
| CAMS871 | acaaagcatcggctgaaaat | gcgaccaagtaccaacaggt | (gaa)14 | 10 | – | 150 |
| CAMS452 | gaagtctgggacctcttttgg | ttcattttgatcttcacgaacg | (ga)11 | 11 | 161 | 163 |
| CAMS476 | ttttccctttccagttgttca | atgggtgaagtgtgaaaagaa | (tc)5 | 11 | 156 | 164 |
CW = Allele from California Wonder, LS = Allele from LS2341.
Comparison with other maps
The total map length of the present map is somewhat shorter than those of previous studies (Ben-Chaim et al. 2001, Livingstone et al. 1999, Wu et al. 2009, Yi et al. 2006). However, the map distance calculated by JoinMap is always shorter than that by Mapmaker (Bradeen et al. 2001). In addition, all the SSR markers, which had polymorphism in the DH population derived from F1 between CW and LS2341, were mapped in this study. Then, there was no unlinked the SSR markers. This result suggests that the present map covers the majority of the pepper genome. The map of this study had 26, 12 and 36 common SSR and/or STS markers with the maps of Minamiyama et al. (2006), Wu et al. (2009) and Yi et al. (2006), respectively. The order of the SSR and STS markers was in good agreement with the maps of previous studies (Barchi et al. 2007, Lee et al. 2004, Minamiyama et al. 2006, Wu et al. 2009, Yi et al. 2006). Only a discrepancy of the position in the linkage group between our map (P1) and the Minamiyama et al. (2006) map (LG7) was identified; the order of the SSR markers CAMS460 and CAMS606 was the converse in the two maps, although the distance of the two markers was estimated to be less than 10 cM.
Conflict of linkage groups P1 and P8 in cultivated Capsicum annuum
In the map of Yi et al. (2006), linkage group 8 was missing and fused with linkage group 1. As a result, the linkage group 1 represented two pepper chromosomes, P1 and P8. Such a pseudolinkage may occur resulting from reciprocal translocation of the two chromosomes between the parents of the mapping population (C. annuum and C. chinense), as proposed by Wu et al. (2009). These two chromosomes have been split into the expected linkage groups in the present map (Fig. 1, P1 and P8), though the linkage assignment was exchanged between P1 and P8 previously (Mimura et al. 2009b). This was because our previous assignment was done based on the integrated map by Paran et al. (2004) through the map by Yi et al. (2006), where Paran et al. (2004) made the assignments of P1 and P8 in a direction opposite to those of the more recent maps (Barchi et al. 2009, Wu et al. 2009). Here we concluded that the linkage group which was formerly expressed as P1 by Mimura et al. (2009b) was shifted to P8 in this map and vice versa.
Phenotypic variations and QTLs of fruit traits
The ranges of FL, FD and FS values were 32–137 mm, 19–61 mm and 0.89–4.91, respectively. The narrow sense heritabilities were higher than 94% in all traits.
A QTL for FL located on P3 had the largest effect in both years, explaining 51% and 52% of the total phenotypic variation in 2007 and 2009, respectively (Table 3). The ‘CW’ allele on P3 decreased the FL. Two additional QTLs were identified on P2. The QTL on P3 also brought about the largest effect for FD and FS, explaining 37–38% and 61–68% of total phenotypic variation, respectively (Table 3). Three additional QTLs for FD and one for FS were also detected. The major QTLs for the three traits were located on the same position of marker ‘CAAACT151’ on P3. The position may overlap with that of a QTL cluster of ‘fl3.1’, ‘fd3.1’ and ‘fs3.1’ (Ben-Chaim et al. 2001), because the cluster locus located at the 65 cM interval involves the ‘CAAACT151’ locus between the markers ‘AF244121’ and ‘HpmsE005’ on our map, when we compare the two maps using the map by Yi et al. (2006). Moreover, Ben-Chaim et al. (2001) and this study used similar C. annuum parent pairs, Bell type pepper and small elongated pepper from South-East Asia. Then, the FS QTLs of P3 in both studies also explained similar proportions of phenotypic variation, 63–67% and 61–68%, respectively. While the other study reported the high ratio of contribution in other chromosome (Ben-Chaim et al. 2003). However, the correspondence is unclear because of no PCR-based anchor marker in the vicinity of the QTL cluster.
Table 3.
QTLs detected for the fruit traits in this study
| Trait | Test | Markera | Chromosome | Positionb | LOD | R2c | Additived | Thresholde |
|---|---|---|---|---|---|---|---|---|
| Fruit length | 2007 | C2_At1g10580 | P2 | 63.7 | 3.0 | 6.7 | 6.6 | 3.0 |
| 2007 | C2_At4g37130 | P2 | 84.1 | 3.6 | 7.9 | 7.5 | 3.0 | |
| 2007 | CAAACT151 | P3 | 58.0 | 14.5 | 51.2 | −16.8 | 3.0 | |
|
| ||||||||
| 2009 | C2_At4g37130 | P2 | 82.1 | 3.6 | 8.1 | 6.6 | 2.9 | |
| 2009 | HpmsE045 | P2 | 105.7 | 3.7 | 8.2 | 5.8 | 2.9 | |
| 2009 | CAAACT151 | P3 | 58.0 | 14.4 | 52.2 | −14.3 | 2.9 | |
|
| ||||||||
| Fruit diameter | 2007 | GPMS178 | P1 | 38.4 | 4.2 | 11.7 | 3.2 | 3.0 |
| 2007 | CAAACT151 | P3 | 58.0 | 9.6 | 37.9 | 5.0 | 3.0 | |
|
| ||||||||
| 2009 | CAeMS010 | P1 | 33.4 | 5.0 | 14.2 | 2.8 | 3.1 | |
| 2009 | CAAACT151 | P3 | 58.0 | 9.1 | 37.1 | 4.0 | 3.1 | |
| 2009 | CAMS451 | P8 | 28.9 | 4.4 | 12.7 | 2.4 | 3.1 | |
| 2009 | CTCACC227 | P10 | 29.3 | 3.6 | 10.4 | 2.3 | 3.1 | |
|
| ||||||||
| Fruit shape | 2007 | CAAACT151 | P3 | 58.0 | 23.1 | 68.2 | −7.9 | 2.9 |
|
| ||||||||
| 2009 | CAAACT151 | P3 | 58.0 | 18.5 | 61.3 | −0.76 | 3.0 | |
| 2009 | CAMS493 | P8 | 11.6 | 3.9 | 6.9 | −0.27 | 3.0 | |
The marker on or in the vicinity of the LOD score peak.
Position of the LOD score peak in the linkage group in cM.
Percentage of phenotypic variation explained.
Additive effect of QTLs of the ‘California Wonder’ allele.
The significance threshold for detecting QTL by 1,000 permutations at P < 0.05.
Utility of the map in this study
Linkage groups P1 and P8 in cultivated C. annuum have important QTLs such as fruit related traits (Ben-Chaim et al. 2001), growth traits (Barchi et al. 2009, Ben-Chaim et al. 2001, Mimura et al. 2010) and several disease resistances (Mimura et al. 2009b, Ogundiwin et al. 2005, Sugita et al. 2006). The map in this study firstly revealed 12 linkage groups representing the 12 chromosomes in cultivated C. annuum with a lot of PCR-based anchor markers. Especially in P1 and P8, map length was comparable to those of previous studies (Wu et al. 2009, Yi et al. 2006). In addition, this map enables us to estimate a lot of CAMS (SSR) markers (Minamiyama et al. 2006) in other major maps. Moreover, the map has newly developed SSR and CAPS markers, and contains culturally important QTLs which affect fruits, growth and bacterial wilt resistance traits (Mimura et al. 2009b, 2010). In practice, breeding programmes involve lots of crossing between two cultivated C. annuum. Therefore, the map developed through this study is useful for MAS in breeding.
Acknowledgements
Pepper accession ‘LS2341 (JP187992)’ was provided by Genebank (Tsukuba, Japan) at NIAS. We appreciate some of the SSR primers design specifically for this study by Dr. H. Fukuoka at the National Institute of Vegetable and Tea Sciences.
Literature Cited
- Barchi L., Bonnet J., Boudet C., Signoret P., Nagy I., Lanteri S., Palloix A., Lefebvre V. (2007) A high-resolution, intraspecific linkage map of pepper (Capsicum annuum L.) and selection of reduced recombinant inbred line subsets for fast mapping. Genome 50: 51–60 [DOI] [PubMed] [Google Scholar]
- Barchi L., Lefebvre V., Sage-Palloix A.M., Lanteri S., Palloix A. (2009) QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor. Appl. Genet. 118: 1157–1171 [DOI] [PubMed] [Google Scholar]
- Ben-Chaim A., Paran I., Grube R.C., Jahn M. (2001) QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 102: 1016–1028 [Google Scholar]
- Ben-Chaim A., Borovsky Y., DeJong W., Paran I. (2003) Linkage of the A locus for the presence of anthoxyanin and fs10.1, a major fruit-shape QTL in pepper. Theor. Appl. Genet. 106: 889–894 [DOI] [PubMed] [Google Scholar]
- Ben-Chaim A., Borovsky Y., Falise M., Mazourek M., Kang B.C., Paran I., Jahn M. (2006) QTL analysis for capsaicinoid content in Capsicum. Theor. Appl. Genet. 113: 1481–1490 [DOI] [PubMed] [Google Scholar]
- Bradeen J.M., Staub J.E., Wye C., Antonise R., Peleman J. (2001) Towards an expanded and integrated linkage map of cucumber (Cucumis sativus L.) Genome 44: 111–119 [DOI] [PubMed] [Google Scholar]
- Caranta C., Lefebvre V., Palloix A. (1997a) Polygenic resistance of pepper to potyviruses consists of a combination of isolate-specific and broad-spectrum quantitative trait loci. Mol Plant-Microbe Interact. 10: 872–878 [Google Scholar]
- Caranta C., Palloix A., Lefebvre V., Daubeze A.M. (1997b) QTLs for a component of partial resistance to cucumber mosaic virus in pepper: restriction of virus installation in host-cells. Theor. Appl. Genet. 94: 431–438 [Google Scholar]
- Huang S., Zhang B., Milbourne D.D., Cardle L., Yang G., Guo J. (2001) Development of pepper SSR markers from sequence databases. Euphytica 117: 163–167 [Google Scholar]
- Kang B.C., Nahm S.H., Huh J.H., Yoo H.S., Yu J.W., Lee M.H., Kim B.D. (2001) An interspecific (Capsicum annuum × C. chinese) F2 linkage map in pepper using RFLP and AFLP markers. Theor. Appl. Genet. 102: 531–539 [Google Scholar]
- Kosambi D.D. (1944) The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175 [Google Scholar]
- Lanteri S. (1991) Lack of a karyotype class and skewed chromosome segregation in two back-cross progenies of Capsicum. J. Genet. Breed. 45: 51–58 [Google Scholar]
- Lanteri S., Pickersgill B. (1993) Chromosomal structural-changes in Capsicum annuum L. and C. chinense Jacq. Euphytica 67: 155–160 [Google Scholar]
- Lee J.M., Nahm S.H., Kim Y.M., Kim B.D. (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor. Appl. Genet. 108: 619–627 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Palloix A., Caranta C., Pochard E. (1995) Construction of an intraspecific integrated linkage map of pepper using molecular markers and doubled-haploid progenies. Genome 38: 112–121 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., Pflieger S., Thabuis A., Caranta C., Blattes A., Chauvet J.C., Daubèze A.M., Palloix A. (2002) Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome 45: 839–854 [DOI] [PubMed] [Google Scholar]
- Livingstone K.D., Lackney V.K., Blauth J.R., van Wijk R., Jahn M.K. (1999) Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152: 1183–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura Y., Yoshikawa M., Hirai M. (2009a) Pepper accession LS2341 is highly resistant to Ralstonia solanacearum strains from Japan. HortScience 44: 2038–2040 [Google Scholar]
- Mimura Y., Kageyama T., Minamiyama Y., Hirai M. (2009b) QTL analysis for resistance to Ralstonia solanacearum in Capsicum accession ‘LS2341’. J. Japan Soc. Hort. Sci. 78: 307–313 [Google Scholar]
- Mimura Y., Minamiyama Y., Sano H., Hirai M. (2010) Mapping for axillary shooting, flowering date, primary axis length, and number of leaves in pepper (Capsicum annuum). J. Japan Soc. Hort. Sci. 79: 56–63 [Google Scholar]
- Minamiyama Y., Tsuro M., Hirai M. (2006) An SSR-based linkage map of Capsicum annuum. Mol. Breed. 18: 157–169 [Google Scholar]
- Minamiyama Y., Tsuro M., Kubo T., Hirai M. (2007) QTL analysis for resistance to Phytophthora capsici in pepper using a high density SSR-based map. Breed. Sci. 57: 129–134 [Google Scholar]
- Nagy I., Stágel A., Sasvári Z., Röder M., Ganal M. (2007) Development, characterization, and transferability to other Solanaceae of microsatellite markers in pepper (Capsicum annuum L.). Genome 50: 668–688 [DOI] [PubMed] [Google Scholar]
- Ogundiwin E.A., Berke T.F., Massoudi M., Black L.L., Huestis G., Choi D., Lee S., Prince J.P. (2005) Construction of 2 intraspecific linkage maps and identification of resistance QTLs for Phytophthora capsici root-rot and foliar-blight diseases of pepper (Capsicum annuum L.). Genome 48: 698–711 [DOI] [PubMed] [Google Scholar]
- Paran I., vanderVoort J.R., Lefebvre V., Jahn M., Landry L., van Schriek M., Tanyolac B., Caranta C., Ben-Chaim A., Livingstone K., et al. (2004) An integrated genetic linkage map of pepper (Capsicum spp.). Mol. Breed. 13: 251–261 [Google Scholar]
- Pickersgill B. (1991) Cytogenetics and evolution of Capsicum L. In: Tsuchiya T., Gupta P.K. (eds.) Chromosome Engineering in Plants: Genetics, Breeding, Evolution. Part B, Elsevier, Amsterdam, pp. 139–160 [Google Scholar]
- Rao G.U., Ben-Chaim A., Borovsky Y., Paran I. (2003) Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens. Theor. Appl. Genet. 106: 1457–1466 [DOI] [PubMed] [Google Scholar]
- Sugita T., Yamaguchi K., Kinoshita T., Yuji K., Sugimura Y., Nagata R., Kawasaki S., Todoroki A. (2006) QTL analysis for resistance to Phytophthora bright (Phytophthora capsici Leon.) using an intraspecific doubled-haploid population of Capsicum annuum. Breed. Sci. 56: 137–145 [Google Scholar]
- VanOoijen J.W., Voorrips R.E. (2001) JOINMAP 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen, the Netherlands [Google Scholar]
- Van Ooijen J.W. (2009) MapQTL 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen, the Netherlands [Google Scholar]
- Wu F., Eannetta N.T., Xu Y., Durrett R., Mazourek M., Jahn M.M., Tanksley S.D. (2009) A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor. Appl. Genet. 118: 1279–1293 [DOI] [PubMed] [Google Scholar]
- Yi G., Lee J.M., Lee S., Choi D., Kim B.D. (2006) Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor. Appl. Genet. 114: 113–130 [DOI] [PubMed] [Google Scholar]