Abstract
Total spikelet number per panicle (TSN) is one of the most important traits associated with rice yield potential. This trait was assessed in a set of 334 chromosomal segment introgression lines (ILs: BC3-derived lines), developed from new plant type (NPT) varieties as donor parents and having the genetic background of an indica-type rice variety IR64. Among the 334 ILs, five lines which had different donor parents and showed significantly higher TSN than IR64 were used for genetic analysis. Quantitative trait locus (QTL) analysis was conducted using F2 populations derived from crosses between IR64 and these ILs. As a result, a QTL for high TSN (one from each NPT donor variety) was detected on common region of the long arm of chromosome 4. The effect of the QTL was confirmed by an increase in TSN of five near-isogenic lines (NILs) developed in the present study. The variation in TSN was found among these NILs, attributing to the panicle architecture in the numbers of primary, secondary and tertiary branches. The NILs for TSN and the SSR markers linked to the TSN QTLs are expected to be useful materials for research and breeding to enhance the yield potential of rice varieties.
Keywords: total spikelet number per panicle, introgression lines, marker-assisted selection, near-isogenic lines, new plant type (NPT) rice
Introduction
Grain yield of rice (Oryza sativa L.) has four components: panicle number, total spikelet number per panicle (TSN), grain weight and spikelet fertility. There is wide variation in TSN among cultivated rice varieties and it is one of the targets of breeding programs to improve rice yield. However, genetic analysis of TSN is difficult because it is a complex trait controlled by multiple genes and influenced by environmental conditions. Quantitative trait locus (QTL) analysis using DNA markers has recently made it possible to understand the genetic basis of TSN and other complex traits. QTLs for TSN have been identified using various segregating populations, including F2 populations, recombinant inbred lines (RILs), and doubled haploid (DH) lines (Hittalmani et al. 2003, Kobayashi et al. 2004, Mei et al. 2005, Xing et al. 2002, Yagi et al. 2001, Zhuang et al. 1997, Zou et al. 2005). To identify the loci controlling panicle architecture, QTLs for panicle traits such as number of primary or secondary branches and spikelet number per primary or secondary branch have been mapped and studied (Ando et al. 2008, Yamagishi et al. 2002). Several QTLs for TSN in rice have also been identified from wild relatives O. rufipogon, O. nivara and O. glumaepatula (Brondani et al. 2002, Li et al. 2006, Moncada et al. 2001, Onishi et al. 2007, Septiningsih et al. 2003, Thomson et al. 2003, Xiao et al. 1998, Xiong et al. 1999). The QTLs detected in these studies, which were located throughout all 12 rice chromosomes, have provided useful information with which to survey the genes that govern TSN within different populations. Additionally, five QTLs for TSN—QSpp8 on chromosome 8, qSPP1 on chromosome 1, qSSP2 on chromosome 2, qSPP3 on chromosome 3 and qSPP7 on chromosome 7—have been mapped as single Mendelian factors (Zhang et al. 2006, 2009). Fine maps of three QTLs for grain number per panicle and TSN—SPP1 on chromosome 1 (Liu et al. 2009) and two QTLs (gpa7 and qSPP7) on chromosome 7 (Tian et al. 2006, Xing et al. 2008)—have been constructed. Furthermore, three QTLs for increasing grain number (Gn1a on chromosome 1, Ghd7 on chromosome 7 and WFP on chromosome 8) have been cloned (Ashikari et al. 2005, Miura et al. 2010, Xue et al. 2008).
Based on QTL information and linkage analyses, near-isogenic lines (NILs) for TSN have been developed (Zhang et al. 2009). These are suitable materials for precise genetic studies, including the evaluation of gene effects, selection of enhanced molecular markers that are tightly linked with the target gene, gene expression and gene isolation. The effects on grain number per panicle and TSN in several NILs were characterized in previous studies (Ando et al. 2008, Ashikari et al. 2005, Zhang et al. 2009). Ando et al. (2008) demonstrated the additive effects of SBN1 (a QTL for increasing secondary branch number on chromosome 1) and PBN6 (a QTL for increasing primary branch number on chromosome 6) on TSN by pyramiding using NILs containing these two QTLs.
Since the 1960s, IRRI-bred rice varieties have been distributed worldwide and used by both plant breeders and farmers. IR64, which was released in 1985, had been widely accepted as a high-quality rice variety in many countries (Khush 1987). Because of the wide adaptability of IR64, breeding materials with an IR64 genetic background, such as DH lines, RILs and thousands of mutant lines, have been developed for research and improvement of rice varieties (Guiderdoni et al. 1992, Wu et al. 2005). In the late 1980s, a breeding program to develop a new plant type (NPT) of rice was launched at IRRI with the goal of increasing yield potential under tropical environments. Unlike IR64, the NPT varieties have several agronomic traits inherited from tropical japonica-type varieties: low tiller number, low number of unproductive tillers, large panicle, thick culm, lodging resistance and large, dark green flag leaves (Khush 1995). Thus, the NPT varieties were chosen for experiments designed to improve the yield potential of IR64.
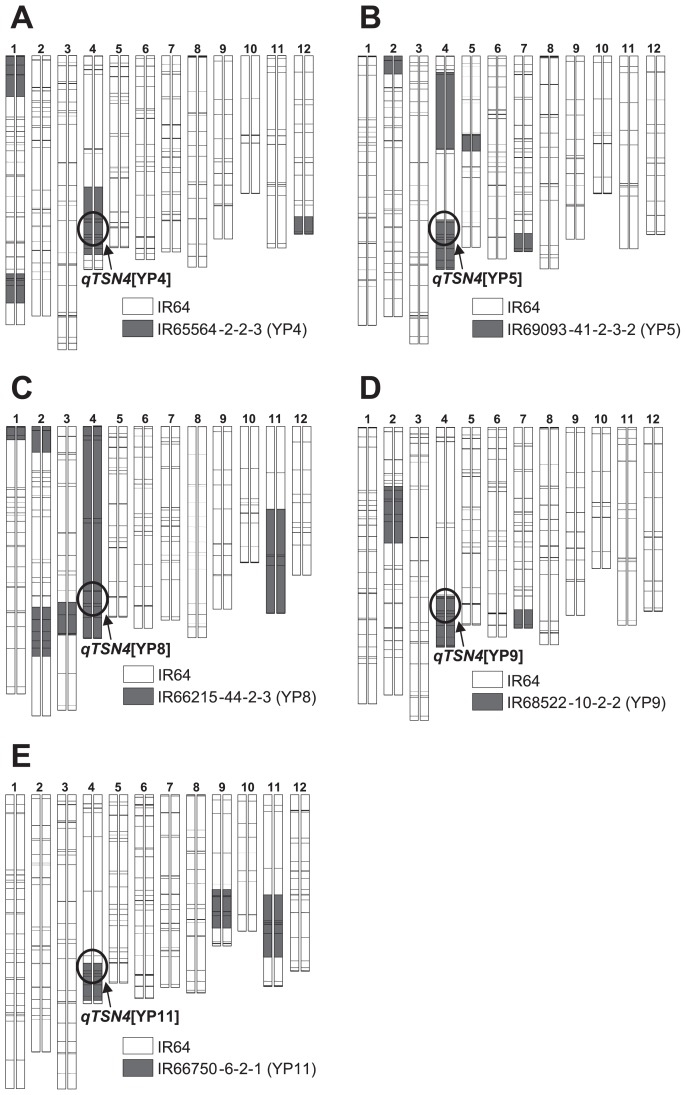
A total of 334 introgression lines (ILs) derived from crosses between IR64 and 10 donor varieties (mostly NPT varieties) have been developed as breeding materials for rice yield enhancement under the IRRI–Japan Collaborative Research Project (Fujita et al. 2009). Based on an association analysis between TSN and genotype of the 334 ILs, 11 chromosome regions were found to be associated with TSN. Five regions from five donor NPT varieties—IR65564-2-2-3 (here designated YP4), IR69093-41-2-3-2 (YP5), IR66215-44-2-3 (YP8), IR68522-10-2-2 (YP9) and IR66750-6-2-1 (YP11)—were associated with high TSN, while six regions from four donors were associated with low TSN. However, the detailed locations of genes for TSN have not been identified in previous study because of small population size.
To reveal the detailed genetic basis of high TSN in these ILs, QTL analysis was conducted using F2 populations derived from crosses between these ILs and IR64. By using simple sequence repeat (SSR) markers to follow these QTLs, five NILs, each carrying one of the QTLs for high TSN in the genetic background of IR64, were developed. The panicle architecture of these NILs was then characterized in the IRRI field.
Materials and Methods
Plant materials
A total of 334 ILs (BC3-derived lines) with the genetic background of indica-type rice variety IR64 was developed by recurrent backcross breeding, and the ILs were characterized for a number of agronomic traits (Fujita et al. 2009, 2010a, 2010b). Of the 334 ILs, approximately half of the lines in each of five sib IL groups showed high TSN. One representative line with high TSN was selected from each sib IL group based on other agronomic traits similar to IR64. Five ILs (YTH83, YTH155, YTH191, YTH288 and YTH326), each derived from a different NPT variety, were selected (Table 1 and Fig. 1). These five ILs were crossed with IR64 to generate F2 populations for QTL analysis.
Table 1.
Introgression lines for high spikelet number derived from new plant type (NPT) varieties with IR64 genetic background
| Introgression linea | Donorb | Parents of donor varietyc | No. of introgressed segments (chromosomal locationsa) | ||
|---|---|---|---|---|---|
|
|
|
||||
| Entry no. | IRRI accession no. | Entry no. | IRRI accession no. | ||
| YTH83 | IR84642-8-4-3-4-4-2-4-2-2-6-B | YP4 | IR65564-2-2-3 | NO 11, Bali Ontjer | 4 (1S, 1L, 4L, 12L) |
| YTH191 | IR84636-13-59-6-4-2-2-4-2-2-2-B | YP5 | IR69093-41-2-3-2 | Shen Nung 89-366, Ketan Lumbu, Gundil Kuning | 5 (2S, 4S, 4L, 5L, 7L) |
| YTH288 | IR84639-7-28-5-5-2-2-3-2-2-14-B | YP8 | IR66215-44-2-3 | Gaok, Chir 87-3-1, Moroberekan, Palawan | 8 (1S, 2S, 2L, 3L, 4S, 4L,7L, 11L) |
| YTH326 | IR84640-11-110-6-4-2-2-4-2-2-3-B | YP9 | IR68522-10-2-2 | Moroberekan, Shen Nung 89-366, Daringan | 3 (2S, 4L, 7L) |
| YTH155 | IR84643-11-105-8-7-2-2-4-3-2-3-B | YP11 | IR66750-6-2-1 | Shen Nung 89-366, Sri Kuning | 3 (4L, 9L, 11L) |
Information from Fujita et al. (2010a, 2010b).
Donors are NPT varieties bred at IRRI.
Pedigrees obtained from the International Rice Information System (IRIS; http://www.iris.irri.org/). Underlines indicate varieties from Indonesia (tropical japonica type).
Fig. 1.
Graphical genotypes of five ILs with high spikelet number per panicle derived from NPT varieties: A; YTH83, B; YTH191, C; YTH288, D; YTH326 and E; YTH155. The 12 pairs of vertical bars represent the chromosomes, numbered at the top. Horizontal lines across the bars show positions of SSR marker loci. White indicates chromosomal segments from IR64; gray indicates chromosomal segments from the donor parents.
QTL analysis for high TSN
The F2 populations derived from the crosses YTH288/ IR64 (166 plants), YTH83/IR64 (162 plants) and YTH326/ IR64 (187 plants) were grown at the IRRI experimental field, Los Baños, Laguna, Philippines, in the dry seasons (January to May) of 2005 or 2007. The other two F2 populations, from YTH155/IR64 (138 plants) and YTH191/IR64 (172 plants), were grown in the wet season (July to November) of 2007. Single plants were transplanted 21 days after sowing using a spacing of 20 cm between hills and 30 cm between rows. TSN was computed as the sum of filled and unfilled spikelets per panicle in the tallest culm.
Genomic DNA from individual plants in each of the five F2 populations was extracted from freeze-dried leaves using the CTAB method (Rogers and Bendich 1988). The DNA samples were analyzed using SSR markers corresponding to the known introgressed regions (i.e., from the donor parents) in each IL (Fujita et al. 2010a, McCouch et al. 2002). The genotypes of F2 population, YTH83/IR64 were detected using the SSR markers located on introgressed segments from the donor variety (YP4) on chromosomes 1, 4 and 12 (Fig. 1). To determine genotypes of other populations, we used SSR markers on chromosomes 2, 4, 5 and 7 for YTH191/IR64, chromosomes 1, 2, 3, 4, 7 and 11 for YTH288/IR64, chromosomes 2, 4 and 7 for YTH326/IR64 and chromosomes 4, 9 and 11 for YTH155/IR64. The SSR marker genotypes were determined by PCR amplification in a DNA Engine Dyad thermal cycler (Bio-Rad Laboratories, Hercules, CA). Each 10-μl PCR reaction mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 200 μM dNTP, 0.2 μM primer, 1 unit of Taq polymerase (SBS Genetech, Beijing, China) and 5–10 μg/ml of genomic DNA as template. The thermal cycler was programmed for a first denaturation step of 5 min at 95°C; followed by 35 cycles, each of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s. The PCR products were resolved in 4.0% agarose gel by electrophoresis at 250 V for 1 h in 0.5× TBE buffer. The gels were stained with ethidium bromide and photographed under ultraviolet light.
The genetic distances between SSR markers located on all introgression segments were determined based on genotypes of F2 individuals in each population using Kosambi centiMorgans (cM). Composite interval mapping was performed using Windows QTL Cartographer V2.5 (Wang et al. 2010). The proportion of observed phenotypic variation attributable to a particular chromosomal region was estimated by the coefficient of determination (R2). The critical threshold values of the LOD score for QTL identification were calculated by conducting 1000 permutation tests with significant level at P < 0.01. The critical threshold values of each population was equivalent to LOD = 3.0 for the F2 populations (YTH83/IR64, YTH288/IR64, YTH326/IR64 and YTH155/IR64) and 2.4 for YTH191/IR64.
Substitution mapping of QTL for high TSN using F4 lines derived from YTH326/IR64
We conducted detailed mapping using a population derived from YTH326, because this line had the fewest and smallest introgressed segments among the five ILs. YTH326 had one each introgression segment from a donor variety, YP9 on chromosomes 2, 4 and 7 (Fig. 1). Total length of introgressed segments was 15.4–28.8 Mbp based on the Nipponbare genome sequence.
To map the QTL more precisely, F4 lines derived from the cross combination YTH326/IR64 were surveyed for TSN, and detailed genotypes were obtained using SSR markers across the target region. To perform substitution mapping, 4 lines (HFP53, HFP56, HFP55 and HFP59) that had recombination between flanking markers of TSN QTL were chosen for delimitation of the location of QTL for high TSN.
Selection and phenotypic evaluation of NILs for TSN
From each of the five F2 mapping populations, we selected one F2 individual containing the detected QTLs for TSN and as few and small other introgressed segments as possible. F3 lines generated by self-pollination of the selected individuals were considered to be NILs for TSN. The five NILs were designated as IR64-NIL2 (derived from YTH83/ IR64), IR64-NIL3 (from YTH191/IR64), IR64-NIL4 (from YTH288/IR64), IR64-NIL5 (from YTH326/IR64) and IR64-NIL6 (from YTH155/IR64) and were characterized in detail.
The numbers of primary branches, secondary branches, and tertiary branches; number of spikelets on each branch; and TSN in the NILs were evaluated at the IRRI field in the 2009 dry season. Each line was represented by more than two rows of 12 F3 individuals. Twenty-one days after sowing, single plants were transplanted at 20 cm spacing between hills and at 30 cm between rows. Ten individual plants in the middle of each row were measured and a single value per plant was measured for each trait. Panicles in the tallest culms were collected for counting numbers of primary branches, secondary branches, and tertiary branches; number of spikelets on each branch; and TSN.
Results
Identification of QTLs for TSN in five F2 populations
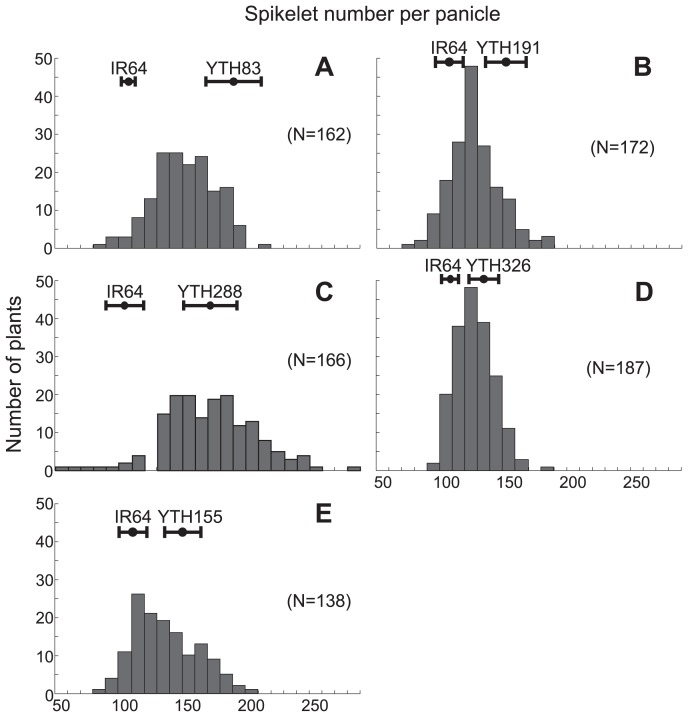
Five F2 populations derived from crosses between five high-TSN ILs and IR64 were investigated for the segregation of TSN. The TSN of each of the five populations showed a continuous frequency distribution with transgressive segregation (Fig. 2), although different patterns of frequency distribution were observed among the populations. The F2 segregating populations derived from the cross combinations YTH191/IR64 and YTH326/IR64 showed a relatively narrow variation in TSN, ranging from 70 to 180 (Fig. 2B, 2D), whereas the population derived from the cross combination YTH288/IR64 showed a wide range, from 50 to 270 (Fig. 2C). The distributions of TSN in the other two populations from YTH83/IR64 and YTH155/IR64 were intermediate, ranging from 70 to 200 (Fig. 2A, 2E).
Fig. 2.
Frequency distributions of spikelet number per panicle in the F2 populations derived from crosses between IR64 and five introgression lines: A; YTH83, B; YTH191, C; YTH288, D; YTH326 and E; YTH155. The dots and error bars at the top of each graph indicate the average and standard deviation of TSN for each parent.
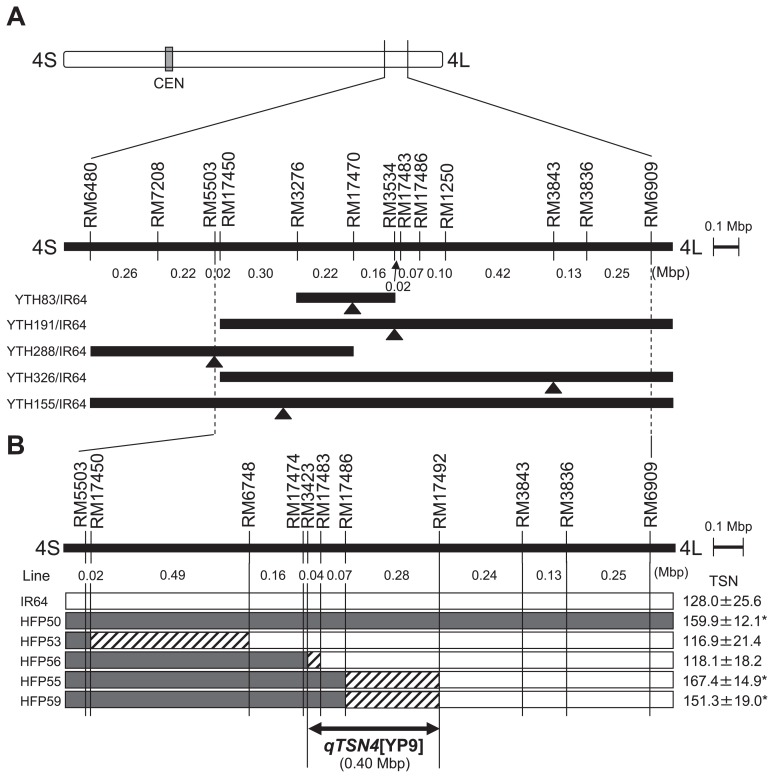
To find the detailed location of the QTLs for TSN, composite interval mapping was conducted in the five populations. A single QTL for TSN was detected in each segregating population. The QTL in YTH83 was identified within a 1.9-cM interval flanked by RM17470 and RM3534 (location of peak LOD score) on the long arm of chromosome 4, with a contribution of 8.4% to phenotypic variation and was designated as qTSN4[YP4] (Table 2 and Fig. 3A). (QTLs are designated by the letter “q” followed by the trait, then the chromosome number, then the donor parent; for example, qTSN4[YP4] indicates a QTL for trait TSN on chromosome number 4 from donor parent YP4.) QTL qTSN4[YP5], in YTH191, was identified within a 0.3-cM interval flanked by RM3534 and RM17486 on the long arm of chromosome 4 with a contribution of 15.8% to phenotypic variation. QTL qTSN4[YP8], in YTH288, was identified within a 3.0-cM interval flanked by RM6480 and RM5503 on the long arm of chromosome 4, with a contribution of 8.0% to phenotypic variation. QTL qTSN4[YP9], in YTH326, was identified within a 15.2-cM interval flanked by RM3843 and RM1113 on the long arm of chromosome 4, with a contribution of 26.6% to phenotypic variation. QTL qTSN4[YP11], in YTH155, was identified within a 4.4-cM interval flanked by RM17450 and RM17470 on the long arm of chromosome 4, with a contribution of 38.2% to phenotypic variation. For each of the QTL, the donor parent allele showed the effect of increasing TSN.
Table 2.
Effects of quantitative trait loci (QTLs) for spikelet number per panicle in five F2 populations derived from crosses between five intro-gression lines and IR64
| Cross combination | QTL | Marker intervala | Chr. | LOD scoreb | R2c | Additive effectd | Dominant effecte |
|---|---|---|---|---|---|---|---|
| YTH83/IR64 | qTSN4[YP4] | RM17470−RM3534 | 4 | 3.8 | 0.084 | −10.1 | 6.2 |
| YTH191/IR64 | qTSN4[YP5] | RM3534 −RM17486 | 4 | 8.2 | 0.158 | −11.5 | 8.0 |
| YTH288/IR64 | qTSN4[YP8] | RM6480 −RM5503 | 4 | 3.3 | 0.080 | −14.9 | −1.7 |
| YTH326/IR64 | qTSN4[YP9] | RM3843 −RM1113 | 4 | 12.8 | 0.266 | −10.8 | 2.7 |
| YTH155/IR64 | qTSN4[YP11] | RM17450 −RM17470 | 4 | 14.0 | 0.382 | −22.3 | 0.6 |
QTL analysis carried out by composite interval mapping with Windows QTL Cartographer ver. 2.5 (Wang et al. 2010).
Underlines show the nearest marker to each QTL.
The critical threshold value of the LOD score was equivalent to LOD at a significance level of 0.05.
Percentage of explained phenotypic variation.
A positive value indicates an increase in TSN associated with the IR64 allele.
A positive value indicates an increase in TSN caused by the heterozygote allele.
Fig. 3.
The location of QTLs for spikelet number per panicle on chromosome 4. A; QTLs detected in five F2 populations derived from cross combinations of YTH83/IR64, YTH191/IR64, YTH288/IR64, YTH326/IR64 and YTH155/IR64. The bars indicate the regions in which the LOD scores exceeded the critical threshold value. The arrowheads show the locations of peak LOD scores. B; The putative location of QTL qTSN4[YP9] estimated using F4 lines. Rectangles show the graphical genotypes of IR64 and five F4 lines (HFP50, HFP53, HFP56, HFP55 and HFP59) derived from YTH326/IR64. White indicates chromosomal segments from IR64; black indicates chromosomal segments from YP9; hatched box indicates the recombination region. Average ± standard deviation of spikelet number for each line is given to the right of the figure; value denoted by asterisk is significantly different from that of IR64 at P = 0.05 according to Dunnett’s test.
Detailed mapping of qTSN4[YP9]
All QTLs for TSN were detected in the same region of chromosome 4 (Fig. 3A). The peak LODs among the five populations in composite interval mapping were located between RM5503 and RM3836, with a distance of 1.45 Mbp. The QTL in YTH326, qTSN4[YP9], was mapped in more detail because the introgression segments in YTH326 are fewest and smallest among those in five ILs. When F4 lines with different size of introgressed segments were compared for TSN, HFP53 and HFP56 was similar to IR64 while HFP50, HFP55 and HFP59 were significantly higher than IR64 (Fig. 3B). These indicate that qTSN4[YP9] was located between RM3423 and RM17492, within a region of 398 kbp.
Characterization of NILs for TSN
Based on SSR genotype of F2 individuals, one plant containing the TSN QTL and the fewest and smallest introgressed chromosomal segments was selected from each of the five populations. F3 lines generated by self-pollination of the selected F2 plants were considered as NILs for TSN. The segments from YP4 in IR64-NIL2 were located on chromosomes 1 and 4. IR64-NIL3 had introgressed segments from YP5 on chromosomes 2, 4, 5 and 7. IR64-NIL4 had introgressed segments from YP8 on chromosomes 4 and 11. The introgressed segments of chromosomes 1, 2, 5, 7 and 11 in these three NILs were not associated with TSN, supporting the finding that a single QTL for TSN was present in each F2 population on chromosome 4. IR64-NIL5 and IR64-NIL6 had introgressed donor segments only on chromosome 4.
The five NILs for TSN showed differences in panicle architecture such as number of branches and spikelet number in each branch. All the NILs showed significantly higher TSN than IR64 (Table 3). The number of primary branches of IR64-NIL6 was the lowest among the five NILs and was not significantly different from that of IR64, whereas the number of secondary and tertiary branches was significantly higher than that of IR64. Two NILs, IR64-NIL-2 and IR64-NIL4, were similar in terms of numbers of primary and secondary branches; both were higher than the other lines. These two NILs differed in number of tertiary branches: IR64-NIL2 had significantly more tertiary branches than IR64-NIL4. Significant differences were not observed in either number of branches or number of spikelets between IR64-NIL3 and IR64-NIL5. These two NILs had a higher number of primary and secondary branches and a higher number of spikelets on each branch than IR64.
Table 3.
Panicle architecture of near-isogenic lines (NILs) with high spikelet number
| NIL | QTL | Number of brancha | Secondary branch no. per primary branch | Number of spikeleta | Filled spikelet number per panicle | Spikelet number per panicle | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Primary branch | Secondary branch | Tertiary branch | Primary branch | Secondary branch | Tertiary branch | |||||
| IR64 | – | 9.2 c | 28.6 c | 1.3 c | 3.1 c | 5.0 bc | 3.2 c | 2.0 b | 114.3 ± 11.5 b | 141.2 ± 17.8 c |
| IR64-NIL2 | qTSN4[YP4] | 10.6 ab | 46.8 a | 11.2 a | 4.4 a | 3.8 d | 3.5 ab | 2.6 a | 159.0 ± 15.9 a | 233.9 ± 22.6 a |
| IR64-NIL3 | qTSN4[YP5] | 11.3 a | 37.2 b | 1.9 c | 3.3 bc | 5.5 a | 3.5 b | 2.5 ab | 174.9 ± 31.3 a | 196.4 ± 19.1 b |
| IR64-NIL4 | qTSN4[YP8] | 10.9 a | 46.2 a | 6.8 b | 4.2 a | 4.9 bc | 3.6 a | 2.6 a | 149.0 ± 27.8 a | 239.4 ± 36.4 a |
| IR64-NIL5 | qTSN4[YP9] | 10.8 ab | 39.4 b | 2.3 c | 3.6 b | 5.2 ab | 3.4 b | 2.4 ab | 179.5 ± 16.4 a | 197.6 ± 19.6 b |
| IR64-NIL6 | qTSN4[YP11] | 9.8 bc | 43.1 ab | 6.9 b | 4.4 a | 4.7 c | 3.5 ab | 2.4 ab | 177.6 ± 33.3 a | 213.5 ± 25.3 ab |
The investigation was carried out in the dry season from January to June 2009.
Average values denoted by the same letter are not significantly different from one another at P = 0.05 according to the Tukey-Kramer test.
Discussion
In our previous study of 334 IR64-ILs, ILs derived from five donor varieties were found to contain regions on the long arm of chromosome 4 associated with high total spikelet number per panicle (Fujita et al. 2009). In the present study, we mapped the location of QTL in five ILs derived from different parent—qTSN4[YP4], qTSN4[YP5], qTSN4[YP8], qTSN4[YP9] and qTSN4[YP11]—by using F2 segregating populations (Table 2). The positions of peak LOD of this QTL were located between RM5503 and RM3836, with a distance of 1.44 Mbp (Fig. 3A). The most precisely mapped QTL, qTSN4[YP9], was delimited to the region between RM3423 and RM17492, with a distance of 398 kbp. In the region of chromosome 4 containing the QTL, several QTLs for TSN had been reported using various populations (Ashikari et al. 2005, Brondani et al. 2002, He et al. 2001, Hittalmani et al. 2003, Lafitte et al. 2002, Lanceras et al. 2004, Li et al. 2006, Mei et al. 2005, Zhuang et al. 1997, Zou et al. 2005). The relationships between the QTLs detected in previous studies and those found in the present study remain unknown. The QTL for panicle architecture that we detected on the long arm of chromosome 4 in five ILs might represent common loci, although there are differences in the locations of peak LOD among these populations (Fig. 3A).
The NPT varieties inherited several unique agronomic traits from tropical japonica-type varieties, such as low tiller number, large panicles and large, dark green flag leaves (Khush 1995). The five NPT varieties that were donor parents of the IR64-ILs were derived from the tropical japonica-type varieties Bali Ontjer, Ketan Lumbu, Gundil Kuning, Gaok, Daringan and Sri Kuning (Table 1). When the genotypes of the NPT varieties and these tropical varieties were analyzed using DNA markers located between RM5503 and RM3836, similar band patterns were observed (data not shown). The data show that the detected QTL for high TSN on the long arm of chromosome 4 might be derived from tropical japonica-type varieties.
The spikelet number is influenced by environmental factors. TSN of IR64 is different between Fig. 2 (90–110 spikelets) and Table 3 (140 spikelets) because the data were obtained in different seasons and years. Therefore, IR64 and five NILs for TSN QTLs were evaluated in same season and year, The effects of the TSN QTLs detected in the present study were confirmed by the increase in TSN of the five NILs (Table 3), although some differences in panicle architecture were observed among them. The high TSN in the five NILs was mainly attributed to the increased number of secondary branches. The high secondary branch number in IR64-NIL3 (containing the donor allele of qTSN4[YP5]) and in IR64-NIL5 (qTSN4[YP9]) was caused by the increase in primary branch number, whereas the secondary branch number per primary branch of these NILs was similar to that of IR64. The increased number of secondary branches in IR64-NIL6 (qTSN4[YP11]) was attributed to high secondary branch number per primary branch, whereas the number of primary branches was not significantly higher than that of IR64. The high secondary branch number in IR64-NIL2 (qTSN4[YP4]) and IR64-NIL4 (qTSN4[YP8]) was due to the increased number of both primary branches and secondary branches per primary branch. These two NILs showed higher TSN than the other NILs. The differences in panicle architecture of the NILs might be explained by differences in the genes or alleles contained within the QTLs detected in the five crosses.
IR64, which was released in 1985, has been widely grown in many countries because of its elite characteristics, such as high yield potential, short growth duration, good eating quality, and enhanced resistance to several diseases and insect pests (Khush 1987, Khush and Virk 2005). In this study, the yield potential of IR64 was enhanced by introducing QTLs for high TSN through backcross breeding aided by marker-assisted selection (MAS). The IR64-NILs carrying the qTSN4 QTLs are expected to be useful breeding materials to improve the yield potential of indica-type rice varieties. These NILs can also be used to reveal the effects of a single gene under various environments. These NILs in the genetic background of IR64, together with information on markers linked to the TSN QTLs, can serve as research materials as well as breeding materials for rice varietal improvement by MAS.
Acknowledgments
This paper reports results obtained from phases III, IV, V and VI of the IRRI–Japan Collaborative Research Project, which was supported by the Ministry of Foreign Affairs and the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Literature Cited
- Ando T., Yamamoto T., Shimizu T., Ma X.F., Shomura A., Takeuchi Y., Lin S.Y., Yano M. (2008) Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor. Appl. Genet. 116: 881–890 [DOI] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Brondani C., Rangel P.H.N., Brondani R.P.V., Ferreira M.E. (2002) QTL mapping and introgression of yield-related traits from Oryza glumaepatula to cultivated rice (Oryza sativa) using microsatellite markers. Theor. Appl. Genet. 104: 1192–1203 [DOI] [PubMed] [Google Scholar]
- Fujita D., Santos R.E., Ebron L.A., Telebanco-Yanoria M.J., Kato H., Kobayashi S., Uga Y., Araki E., Takai T., Tsunematsu H., et al. (2009) Development of introgression lines of an Indica-type rice variety, IR64, for unique agronomic traits and detection of the responsible chromosomal regions. Field Crops Res. 114: 244–254 [Google Scholar]
- Fujita D., Santos R.E., Ebron L.A., Telebanco-Yanoria M.J., Kato H., Kobayashi S., Uga Y., Araki E., Takai T., Tsunematsu H., et al. (2010a) Development and characterization of introgression lines of an Indica-type rice variety, IR64, for unique agronomic traits. JIRCAS Working Report 66: 1–95 [Google Scholar]
- Fujita D., Santos R.E., Ebron L.A., Telebanco-Yanoria M.J., Kato H., Kobayashi S., Uga Y., Araki E., Takai T., Tsunematsu H., et al. (2010b) Characterization of introgression lines for yield-related traits with Indica-type rice variety IR64 genetic background. JARQ 44: 277–290 [Google Scholar]
- Guiderdoni E., Galinato E., Luistro J., Vergara G. (1992) Anther culture of tropical japonica × indica hybrids of rice (Oryza sativa L.). Euphytica 62: 219–224 [Google Scholar]
- He P., Li J.Z., Zheng X.W., Shen L.S., Lu C.F., Chen Y., Zhu L.H. (2001) Comparison of molecular linkage maps and agronomic trait loci between DH and RIL populations derived from the same rice cross. Crop Sci. 41: 1240–1246 [Google Scholar]
- Hittalmani S., Huang N., Courtois B., Venuprasad R., Shashidhar H.E., Zhuang J.Y., Zheng K.L., Liu G.F., Wang G.C., Sidhu J.S., et al. (2003) Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia. Theor. Appl. Genet. 107: 679–690 [DOI] [PubMed] [Google Scholar]
- Khush G.S. (1987) Rice breeding: past, present and future. J. Genet. 66: 195–216 [Google Scholar]
- Khush G.S. (1995) Breaking the yield frontier of rice. GeoJournal 35: 329–332 [Google Scholar]
- Khush G.S., Virk P.S. (2005) Selection criteria. In: IR Varieties and their Impact, International Rice Research Institute, Los Banos, Philippines, pp. 6–15 [Google Scholar]
- Kobayashi S., Fukuta Y., Yagi T., Sato T., Osaki M., Khush G.S. (2004) Identification and characterization of quantitative trait loci affecting spikelet number per panicle in rice (Oryza sativa L.). Field Crops Res. 89: 253–262 [Google Scholar]
- Lafitte H.R., Courtois B., Arraudeau M. (2002) Genetic improvement of rice in aerobic systems: progress from yield to genes. Field Crops Res. 75: 171–190 [Google Scholar]
- Lanceras J.C., Pantuwan G., Jongdee B., Toojinda T. (2004) Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol. 135: 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhou A., Sang T. (2006) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol. 170: 185–194 [DOI] [PubMed] [Google Scholar]
- Liu T., Mao D., Zhang S., Xu C., Xing Y. (2009) Fine mapping SPP1, a QTL controlling the number of spikelets per panicle, to a BAC clone in rice (Oryza sativa). Theor. Appl. Genet. 118: 1509–1517 [DOI] [PubMed] [Google Scholar]
- McCouch S.R., Teytelman L., Xu Y., Lobos K.B., Clare K., Walton M., Fu B., Maghirang R., Li Z., Xing Y., et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 199–207 [DOI] [PubMed] [Google Scholar]
- Mei H.W., Li Z.K., Shu Q.Y., Guo L.B., Wang Y.P., Yu X.Q., Ying C.S., Luo L.J. (2005) Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theor. Appl. Genet. 110: 649–659 [DOI] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genet. 42: 545–549 [DOI] [PubMed] [Google Scholar]
- Moncada P., Martínez C.P., Borrero J., Chatel M., Gauch J.H., Guimaraes E., Tohme J., McCouch S.R. (2001) Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 102: 41–52 [Google Scholar]
- Onishi K., Horiuchi Y., Ishigoh-Oka N., Takagi K., Ichikawa N., Maruoka M., Sano Y. (2007) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed. Sci. 57: 7–16 [Google Scholar]
- Rogers S.O., Bendich A.J. (1988) Extraction of DNA from plant tissues. Plant Mol. Biol. Manual A6: 1–10 [DOI] [PubMed] [Google Scholar]
- Septiningsih E.M., Prasetiyono J., Lubis E., Tai T.H., Tjubaryat T., Moeljopawiro S., McCouch S.R. (2003) Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107: 1419–1432 [DOI] [PubMed] [Google Scholar]
- Thomson M.J., Tai T.H., McClung A.M., Lai X.H., Hinga M.E., Lobos K.B., Xu Y., Martinez C.P., McCouch S.R. (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493 [DOI] [PubMed] [Google Scholar]
- Tian F., Zhu Z., Zhang B., Tan L., Fu Y., Wang X., Sun C.Q. (2006) Fine mapping of a quantitative trait locus for grain number per panicle from wild rice (Oryza rufipogon Griff.). Theor. Appl. Genet. 113: 619–629 [DOI] [PubMed] [Google Scholar]
- Wang S., Basten C.J., Zeng Z.B. (2010) Windows QTL Cartographer 2.5. (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm) Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Wu J.L., Wu C., Lei C., Baraoidan M., Bordeos A., Madamba M.R.S., Ramos-Pamplona M., Mauleon R., Portugal A., Ulat V.J., et al. (2005) Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 59: 85–97 [DOI] [PubMed] [Google Scholar]
- Xiao J., Li J., Grandillo S., Ahn S.N., Yuan L., Tanksley S.D., McCouch S.R. (1998) Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y.Z., Tan Y.F., Hua J.P., Sun X.L., Xu C.G., Zhang Q. (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor. Appl. Genet. 105: 248–257 [DOI] [PubMed] [Google Scholar]
- Xing Y.Z., Tang W.J., Xue W.Y., Xu C.G., Zhang Q. (2008) Fine mapping of a major quantitative trait loci, qSSP7, controlling the number of spikelets per panicle as a single Mendelian factor in rice. Theor. Appl. Genet. 116: 789–796 [DOI] [PubMed] [Google Scholar]
- Xiong L.Z., Liu K.D., Dai X.K., Xu C.G., Zhang Q. (1999) Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251 [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genet. 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Yagi T., Nagata K., Fukuta Y., Tamura K., Ashikawa I., Terao T. (2001) QTL mapping of spikelet number in rice (Oryza sativa L.). Breed. Sci. 51: 53–56 [Google Scholar]
- Yamagishi M., Takeuchi Y., Kono I., Yano M. (2002) QTL analysis for panicle characteristics in temperate japonica rice. Euphytica 128: 219–224 [Google Scholar]
- Zhang Y., Luo L., Xu C., Zhang Q., Xing Y. (2006) Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza sativa). Theor. Appl. Genet. 113: 361–368 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Luo L., Liu T., Xu C., Xing Y. (2009) Four rice QTL controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds. Theor. Appl. Genet. 118: 1035–1044 [DOI] [PubMed] [Google Scholar]
- Zhuang J.Y., Lin H.X., Lu J., Qian H.R., Hittalmani S., Huang N., Zheng K.L. (1997) Analysis of QTL × environment interaction for yield components and plant height in rice. Theor. Appl. Genet. 95: 799–808 [Google Scholar]
- Zou G.H., Mei H.W., Liu H.Y., Liu G.L., Hu S.P., Yu X.Q., Li M.S., Wu J.H., Luo L.J. (2005) Grain yield responses to moisture regimes in a rice population: association among traits and genetic markers. Theor. Appl. Genet. 112: 106–113 [DOI] [PubMed] [Google Scholar]