Abstract
A total of 18 rainfed upland New Rice for Africa (NERICA) varieties were categorized as the heavy panicle and low tillering types and early heading, in compared with 32 different varieties. These chromosome components were clarified using 243 SSR markers which showed polymorphism among NERICA varieties and their parents, CG 14 (O. glaberrima Steud.) and one of the recurrent parents, WAB-56-104 (O. sativa L.). NERICA varieties were classified into three groups, which corresponded with these parents’ continuation including two exceptions, NERICAs 14 and 17, by a cluster analysis using polymorphism data of SSR markers and 14 differential markers among them were selected to classify NERICA varieties. However, three groups: NERICAs, 3 and 4, NERICAs, 8, 9 and 11 and NERICAs, 15 and 16 were not distinguishable. Association analysis was carried out for characterization of NERICA varieties by using SSR markers genotype and phenotype of agronomic traits. A total of 131 quantitative trait loci between SSR markers and 11 agronomic traits were detected. The characteristics of early maturity and heavy panicle of upland NERICA varieties were succeeded from Asian rice varieties and the characteristics of high dry matter production and late heading were introduced from CG 14 and the other varieties.
Keywords: rainfed upland NERICA, rice (Oryza sativa L.), O. glaberrima Steud., yield components, chromosome introgression
Introduction
Jones et al. (1997) started developing New Rice for Africa (NERICA) varieties for rainfed upland using interspecific hybridization between Asian rice (Oryza sativa L.) and African rice (Oryza glaberrima Steud.) at the African Rice Center (AfricaRice, Former name: Western Africa Rice Development Association, WARDA). The aim was to combine the high yield potential of O. sativa based on high spikelet number caused by secondary branches on the panicle and useful traits of O. glaberrima, such as rapid leaf canopy establishment and high nitrogen responsiveness. The BC2F1 progenies between some Asian Japonica-type rice (O. sativa) varieties, namely WAB 56-50, WAB 56-104 and WAB 181-18, which were bred in AfricaRice and used as the recurrent parents, and an African rice variety, CG 14, were established and rainfed upland NERICA varieties were selected. The first seven varieties, NERICAs 1–7, were released by AfricaRice in 2000 (Kaneda 2007a, WARDA 2006) and an additional 11 varieties were released, NERICAs 8–18, in 2005 (Africa Rice Center 2008). The recurrent parents, WAB 56-104 and WAB 56-50, were obtained from a cross between a variety, IDSA6, bred at the Institute of Savanna of Ivory Coast and a Brazilian upland variety, IAC164.
Kaneda (2007b) indicated that seven NERICAs, 1–7, were heavy panicle type with big panicles but fewer numbers of tillers, based on the data of yield component investigated at three locations in Ghana in 2004. Several lines among them were identified as tolerant to drought both at vegetative and reproductive stages. Kaneda (2007b) also indicated that NERICAs failed to succeed the target traits from the donor varieties, O. glaberrima, such as effective nitrogen use, plant growth in the young vegetative stage or weed competitiveness, phosphoric acid (P) absorption ability and highly responsive to its application. Ekeleme et al. (2009) examined the ability of weed competition in NERICAs 1, 2 and 4 together with WAB 56-104, CG 14 and ITA 150 as check varieties, but significant differences were not elucidated among NERICAs and WAB 56-104. Oikeh et al. (2008) found the differences in responses to nitrogen and phosphorus. In their study, NERICAs, 3 and 6, were categorized as the suitable varieties at low-input condition, and NERICA 1 showed the highest response to nitrogen. Oikeh et al. (2009) evaluated three NERICAs, 1, 2 and 4 and WAB 56-104 under different nitrogen and planting space. Sanni et al. (2009a) evaluated stability among 18 NERICAs based on the genotype by environment interaction study and indicated that four NERICAs, 3, 10, 11 and 18, could be considered stable in comparison with the others. Sanni et al. (2009b) reported a difference of ratooning formation among NERICAs, WAB 56-104 and CG 14. Ishizaki and Kumashiro (2008) evaluated the abilities of shoot differentiation from calluses and transformation, and found a variation among 18 NERICAs. These studies were carried out with NERICAs and a small number of check varieties. These characterization data for various kinds of agronomic and physiological traits related to the yield performance were still limited in these reports from AfricaRice and systematic analyses of upland NERICAs had not yet been carried out for comparison with various types of rice varieties. In addition, these characterizations of agronomical traits in NERICAs were not clearly evaluated and the contributions of each parent of O. sativa and O. graberrima for genetic improvement of them were not dicussed.
Semagn et al. (2006) tried to evaluate the relationships and genetic differences among 18 NERICAs using DNA markers and ten agronomic traits: days to heading, days to maturity, plant height, panicle length, number of primary branches, number of secondary branches, grain shattering, filled grain number, empty grain number and yield (kg/ha). Cluster analysis was carried out using the data of polymorphism of 102 SSR markers which covered all rice chromosomes and these varieties were classified into two groups, seven NERICAs 1–7 and eleven NERICAs 8–18. In this study, the number of SSR markers was limited and the evaluation of agronomic traits was carried out using only seven varieties, NERICAs 1–7. Moreover, the relationships between the genotypes of SSR markers and the phenotypes of ten traits in NERICAs had not been clarified. These NERICAs were expected to introduce many favorite traits from O. glaberrima into O. sativa genetic backgrounds; however, the detailed characterization of them and genetic factor(s) introgressed from O. glaberrima have not yet been clarified. Ikeda et al. (2007) reported that offtypes and segregation were included in the several varieties among NERICAs 1–7 based on observations for many morphological traits among the breeder and foundation seeds in the Genetic Resources Unit of the AfricaRice. The selections of homozygote lines in all NERICAs have been tried under the collaboration project between the Japan International Cooperation Agency (JICA) and Africa Rice (Personal communication).
Japan International Research Center for Agricultural Sciences (JIRCAS) introduced these seeds through the JICA project, AfricaRice and selected and confirmed the homozygosis of 18 NERICAs. By using these seeds, agronomic traits related with yield components and heading date were evaluated in comparison with various rice types to characterize NERICAs morphologically and physiologically. Chromosome components were also surveyed to identify the introgressions from O. glaberrima into NERICAs using SSR markers distributed in all genome chromosomes. Then, the association analysis between these introgressions and agronomic traits of 18 NERICAs was carried out. In this paper, the contributions of the chromosome segments from O. glaberrima in the performance of NERICAs were discussed. Differential DNA markers against NERICAs were also selected based on the polymorphism patterns of SSR markers to classify and distinguish each NERICA varieties.
Materials Methods
Rice varieties
A total of 18 rainfed upland NERICA varieties were used (Table 1). NERICAs 1–11 were bred with WAB 56-104, three upland NERICAs, 12, 13 and 14, were bred with WAB 56-50 and four NERICAs, 15 to 18, were bred with WAB 181-18 as the recurrent parents and adapted upland conditions (Jones et al. 1997, Kaneda. 2007a, WARDA 2006). The seeds of 18 NERICAs, which were selected and purified by the Japan International Cooperation Agency (JICA) project in Benin, were introduced to JIRCAS in 2005 and used for characterization.
Table 1.
Polymorphic SSR markers in upland NERICA varieties
| NERICA variety | Cross combinationa | No. of SSR markers (%) | ||
|---|---|---|---|---|
|
| ||||
| WAB 56-104b | CG 14b | Otherc | ||
| 1 | WAB 56-104/CG 14//2*WAB 56-104 | 211 (86.8) | 14 (5.8) | 18 (7.4) |
| 2 | WAB 56-104/CG 14//2*WAB 56-104 | 210 (86.4) | 17 (7.0) | 16 (6.6) |
| 3 | WAB 56-104/CG 14//2*WAB 56-104 | 226 (93.0) | 3 (1.2) | 14 (5.8) |
| 4 | WAB 56-104/CG 14//2*WAB 56-104 | 226 (93.0) | 3 (1.2) | 14 (5.8) |
| 5 | WAB 56-104/CG 14//2*WAB 56-104 | 202 (83.1) | 17 (7.0) | 24 (9.9) |
| 6 | WAB 56-104/CG 14//2*WAB 56-104 | 160 (65.8) | 26 (10.7) | 57 (23.5) |
| 7 | WAB 56-104/CG 14//2*WAB 56-104 | 213 (87.7) | 3 (1.2) | 27 (11.1) |
| 8 | WAB 56-104/CG 14//2*WAB 56-104 | 189 (77.8) | 13 (5.3) | 41 (16.9) |
| 9 | WAB 56-104/CG 14//2*WAB 56-104 | 189 (77.8) | 13 (5.3) | 41 (16.9) |
| 10 | WAB 56-104/CG 14//2*WAB 56-104 | 210 (86.4) | 16 (7.0) | 17 (7.0) |
| 11 | WAB 56-104/CG 14//2*WAB 56-104 | 189 (77.8) | 13 (5.3) | 41 (16.9) |
| 12 | WAB 56-50/CG 14//2*WAB 56-50 | 207 (85.2) | 17 (7.0) | 19 (7.8) |
| 13 | WAB 56-50/CG 14//2*WAB 56-50 | 208 (85.6) | 17 (7.0) | 18 (7.4) |
| 14 | WAB 56-50/CG 14//2*WAB 56-50 | 183 (75.3) | 16 (7.0) | 44 (18.1) |
| 15 | CG 14/WAB 181-18//2*WAB 181-18 | 194 (79.8) | 11 (5.3) | 38 (15.6) |
| 16 | CG 14/WAB 181-18//2*WAB 181-18 | 194 (79.8) | 11 (5.3) | 38 (15.6) |
| 17 | CG 14/WAB 181-18//2*WAB 181-18 | 196 (80.7) | 21 (8.6) | 26 (10.7) |
| 18 | CG 14/WAB 181-18//2*WAB 181-18 | 191 (78.6) | 12 (5.3) | 40 (16.5) |
|
| ||||
| Mean | – | (82.3) | (5.6) | (12.2) |
WAB and CG14 indicate the polymoriphism patterns of WAB 56-104 (O. sativa) and CG14 (O. glaberrima), respectively.
Other indicates the different polymoriphism patterns form both of WAB 56-104 and CG14.
A total of 243 SSR markers were used for the polymoriphism analysis.
The agronomic traits of NERICAs were evaluated in comparison with 32 controls selected from several categories, Japonica-and Indica-types, landrace and improved types and lowland and upland varieties. These varieties were selected as the representatives including almost all types of O. sativa and these made heading under the environmental conditions in Tsukuba city. Among the fourteen Indica-type varieties, four landrace varieties, Kasalath, Surjamkuhi, Tadukan and Tetep, are cultivated in tropical countries and the other ten improved varieties, IR 8, IR 24, IR 36, IR 64, IR 74, Hokuriku 143, Milyang 23, Taichung Native 1, Takanari and Mahsuri were bred from temperate and tropical areas. Nine landraces and nine improved varieties were included in the 18 Japonica-types. These nine land-races have been cultivated in lowland or upland areas as the leading varieties in several countries with tropical and temperate areas. Four varieties, Azucena and Davao from the Philippines, Trembese from Indonesia, and Moroberekan from Guinea, Africa, are landraces and cultivated in upland areas. The other four landraces, Basmati217 and Dular from India and Kamenoo and Kibi from Japan, are cultivated in irrigated areas. All nine improved Japonica varieties, Kotobukimochi, Owarihatamochi, Koshihikari, Reiho, Dontokoi, Aichiasahi, Nipponbare, Akihikari and Toride 1, were developed by crossbreeding in Japan. Kotobukimochi and Owarihatamochi were the upland varieties and the others are for irrigated lowlands.
Characterization of agronomic traits
A total of 50 varieties including 18 upland NERICAs and 32 control varieties were cultivated at an irrigated paddy field at JIRCAS, Tsukuba, Ibaraki, Japan, in the season from May to October 2007. Eleven agricultural traits, days to heading, panicle weight (PW), culm and leaf weight, total weight including panicle per plant, ratio of panicle against culm and leaf weight, culm length, panicle length and total length, panicle number per plant, spikelet number per plant and ratio of fertility seeds, were investigated in each variety.
Ten among the 40 plants were investigated with three duplications and the average of 30 plants was used as the representative value in each variety. Days to heading were defined as the time when half the plants showed exerted panicles in each variety.
Chromosomal components of NERICA
All 18 upland NERICAs, a donor variety, CG 14 (O. glaberrima), one of the recurrent parents, WAB 56-104 (O. sativa), and two controls of Asian rice varieties, Nipponbare and Kasalath, were used for genotyping with SSR markers. The recurrent parent, WAB 56-104, was selected as a repetitive control variety among three WAB lines, because it was used as the crossing parent in 11 NERICAs and the sister line, WAB 56-50, was also used the other three NERICAs.
A total of 295 SSR markers, which were distributed in whole genome chromosomes (McCouch et al. 2002, Temnykh et al. 2000), were used for genotyping these varieties.
Whole genomic DNA was extracted from the fresh leaves of each variety using a simple method. Approximately 1cm length of rice leaf chips was collected and ground in 100 μl NaOH using Mixer Mill MM200 (Retsch) and then added and mixed well with 400 μl Tris-HCL, pH 8.0. After centrifugation, the supernatant was recovered and stored in the freezer at −20°C. The extracted DNA was diluted with sterile water into 1/20 concentration and used as template DNA for PCR reaction.
These polymorphic data of SSR markers among WAB 56-104, CG 14 and 18 NERICAs were used for the investigations of chromosome components through developing of the graphical genotypes of each NERICA variety. The chromosome regions for introgression from the donor variety, CG 14, were identified finally and then specific SSR markers were selected to classify each NERICA variety from the others.
Classification of NERICA and control varieties based on agricultural traits and polymorphic data of SSR markers
First, data analysis for classification among 50 varieties was conducted using phenotypic values of five traits, culm length, panicle length, number of panicles, clum and leaf weight and panicle weight, which were related with plant type, performing Ward’s hierarchical analysis (Ward 1963) using a computer program, JMP6.0 (SAS Institute Inc., Cary, NC, USA) for Windows. Then, the variety groups classified by cluster analysis were characterized in more detail based on the comparison of differences in 13 agronomic traits: days to heading, panicle weight, leaf and culm weight, total weight per plant, ratio of panicle against culm and leaf weight, culm length, panicle length, total length, panicle number per plant, number of fertile seeds and number of sterile seeds per plant and ratio of fertility seeds.
A total of 127 SSR markers, which showed polymorphism among 18 NERICAs and four varieties, WAB 56-104, CG-14, Nipponbare and Kasalath, were used by the same method of cluster analysis as agronomic traits, and the differences and relationships among NERICA varieties were investigated.
These genotype data of SSR markers and agronomic traits were compared by association analysis. The association analysis was carried out between the agronomic traits and genotypes of SSR markers, which showed polymorphism among 18 NERICAs, to detect the QTLs for agronomic traits introgressed from CG 14. The associations were estimated by ANOVA test between the phenotypic values with two genotypes of WAB 56-104 and the other introgressed segment at each SSR markers in eleven traits among 18 NERICA varieties with the significant level, 1%. The plus and minus F values in ANOVA test indicated the increasing and decreasing of values in each trait, respectively, with the CG 14 allele, as these effects in each detected QTL.
Results
Agronomical characterization of NERICA varieties
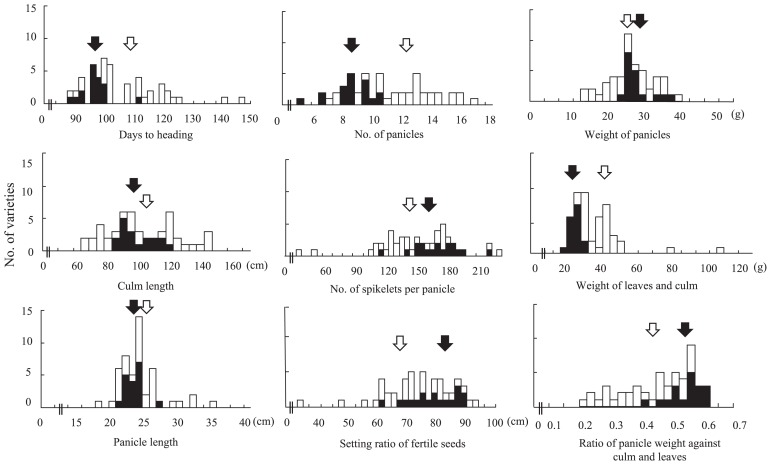
A total of 50 varieties including 18 upland NERICAs varied in the wide ranges in each trait. They showed several unique characters in comparison with the other 32 varieties, among nine agronomic traits: panicle weight (g), leaf and culm weight (g), ratio panicle weight against leaf and culm weight (%), panicle number, panicle length (cm), culm length (cm), days to heading, number of seeds per panicle and setting ratio of fertile seeds (%) (Fig. 1).
Fig. 1.
Variation of upland NERICA varieties for agronomic traits in compared with the 32 controls. Black and white arrows indicate the averages of 18 upland NERICA and the other 32 varieties, respectively.
The average of NERICAs for weight of panicle, panicle length and culm length was similar to those of the other 32 varieties. However the ranges of distribution were smaller. The values of three traits, days to heading, number of panicles and leaf and culm weight, of NERICAs were smaller than those of the other varieties. In contrast, number of seeds per panicle, ratio of panicle weight against leaf and culm weight and ratio of fertility seed per panicle, were higher. The results indicated that NERICAs had the characteristics for heavy panicle-type rice as compared with the other varieties used in this study. Thus, these dry matter productions of NERICAs were not high, but they had heavy panicles in compare with these stems.
Classification of NERICA based on agronomical traits
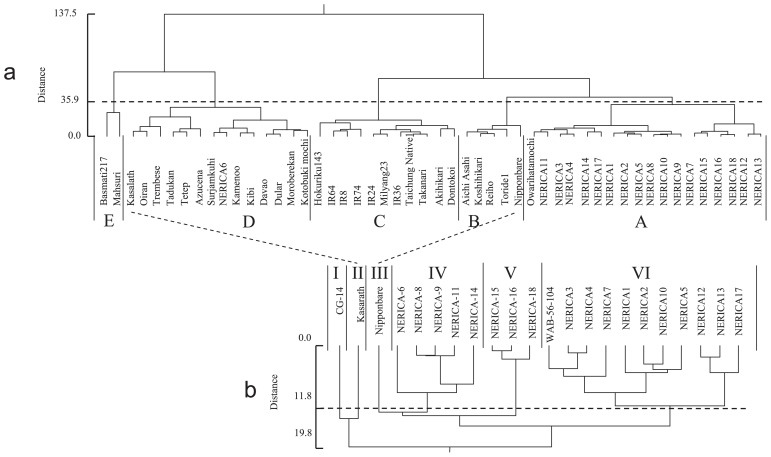
These 50 varieties were classified into five cluster groups, A–E, based on the characterizations of five agronomic traits, culm length, panicle length, number of panicles, clum and leaf weight and panicle weight, which were related with the plant types of rice (Fig. 2a and Table 2).
Fig. 2.
Classification of 18 upland NERICA varieties. Cluster analyses were carried out with genotype data of 243 SSR markers and nine agronomic traits following Ward’s hierarchal analysis (Ward 1963) using JMP6.0 software (SAS Institute Inc., Cary, NC, USA) for Windows in each. “a” and “b” indicate cluster analyses for agronomic traits and genotype of SSR markers, respectively. A total 50 varieties including 18 upland NERICAs were classified into 5 groups, A–E based on the cluster analysis for agronomic traits. A total 22 varieties unclosing CG 14, Kasalath, Nippaonbare, WAB 56-104 and 18 NERICAs were classified into six groups, I–VI. Dashed lines indicate position of Nipponbare and Kasalath.
Table 2.
Variations of agronomic traits among five cluster groups
| Trait | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| Cluster group | No. of varieties | Panicle weight (g) | Culm weight (g) | Total Weight (g) | P/CLa | Panicle length (cm) | Culm length (cm) | Total length (cm) | No. of panicles | No. of seeds | No. of fertile seeds | No. of sterile seeds | Ratio of fertile seeds (%) | Days to heading |
| A | 18 | 25.1 ab | 23.0 d | 48.1 c | 0.52 a | 22.2 a | 92.7 b | 114.9 b | 8.1 b | 165.1 a | 126.3 a | 38.8 a | 77.0 a | 96 c |
| B | 5 | 17.3 c | 42.1 b | 59.4 b | 0.29 b | 20.7 a | 89.2 b | 109.9 b | 13.1 a | 123.0 a | 77.1 a | 45.9 a | 63.0 b | 110 b |
| C | 11 | 30.4 a | 31.3 c | 61.6 b | 0.49 a | 23.7 a | 69.2 c | 92.9 c | 11.9 ab | 156.0 a | 112.6 a | 43.4 a | 72.5 ab | 108 b |
| D | 14 | 21.3 bc | 36.2 bc | 57.5 b | 0.37 b | 24.6 a | 121.5 a | 146.1 a | 10.5 ab | 136.3 a | 103.8 a | 32.5 a | 74.9 ab | 105 b |
| E | 2 | 23.6 abc | 79.9 a | 103.4 a | 0.23 b | 26.7 a | 129.4 a | 156.2 a | 12.1 ab | 152.2 a | 131.1 a | 21.1 a | 86.7 a | 144 a |
Ratio of panicle against culm and leaf weight.
The Tukey test was carried out for comparing differences among five cluster groups at 5% significant level.
Cluster A included a Japanese upland variety, Owarihatamochi and all NERICAs except for NERICA 6 and these were all upland Japonica varieties. This variety group was characterized as low dry culm weight and matter production (Total weight), high ratio of panicle against culm and leaf (P/CL) weight, small panicle number, and short days for heading, in comparison with the other groups. Thus, this group was categorized as a heavy panicle type variety which showed low tiller, many seeds (spikelets) per panicle, high harvest index, low dry matter production and early heading.
Cluster B had five improved Japanese Japonica-type lowland varieties: Aichiasahi, Koshihikari, Reiho, Toride 1 and Nipponbare. These varieties were characterized by low panicle weight, low ratio of P/CL weight, low number of total seeds (spikelets) per plant, low number of seeds per plant and low number and ratio of fertile seeds per plant, short panicle length and high panicle number. This variety group showed the most panicle number types among the five.
Cluster C included 11 improved lowland varieties. All varieties were semidwarf type except for Akihikari. Five varieties, IR 64, IR 8, IR 74, IR 24 and IR 36 were bred through introducing a semidwarf gene, sd-1, from a Chinese variety, Dee-geo-woo-gen, at the International Rice Research Institute (IRRI). Three varieties, Milyang 23, Tanaknari (Imbe et al. 2004) and Dontkoi (Tabuchi et al. 2000, Uehara 2001) also harbor the sd-1 from IR24 (Dee-geo-woo-gen). Hokuriku 143 is a non-shattering mutant line from Nan-jing 11 (Fukuta 1995, Fukuta et al. 1994) and Nan-jing 11 introduced a semidwarf gene from Ai-Jio-Nan-Te (Lin and Min 1991). Oba et al. (1990) confirmed that Ai-Jio-Nan-Te harbored the same allele of sd-1. These varieties harbor the common semidarf gene, sd-1, and were characterized as the other heavy panicle type, which had the higher values in these traits, weights of panicle, culm and total, panicle length and number of panicles and lower ratio of P/CL weight and shorter culm length, than those in group A.
Cluster D included a total of 14 landraces, four Indica-type varieties, Kasalath, Tadukan, Tetep and Surjamkuhi and eight Japonica-type upland varieties, Oiran, Trembese, Azusena, Kamenoo, Kibi, Davao, Kotobukimochi and Moroberekan and one Japonica-type lowland variety, Dular and NERICA 6. These varieties showed the intermediate values in all traits among the five groups.
Cluster E had two lowland varieties, Basmati 217 and Mahsuri, and showed the higher weights of culm and total of panicle, culm and leaf and longer lengths of panicle, culm and total of panicle and culm, longer days to heading, and lower sterile and high ratio of fertile seeds and lower ratio of P/CL weight than those in the other groups. This group was characterized by the long vegetative stage and low harvest index.
These results indicated that almost all NERICAs were heavy panicle type, but the type was different from that of the improved lowland varieties, however three recurrent parents of NERICAs and a similar type variety, tropical Japonica, were not included in the investigation of agricultural traits. It is necessary to characterize these NERICAs in detail by comparison with many different rice varieties.
Chromosome component of upland NERICAs
A total of 243 among 295 SSR markers showed the polymorphism among Asian rice, O. sativa variety, WAB 56-104, O. glaberrima line, CG 14 and 18 NERICAs. The chromosomal components of NERICAs were clarified using these SSR markers, which were distributed in rice 12 whole chromosomes (Table 1).
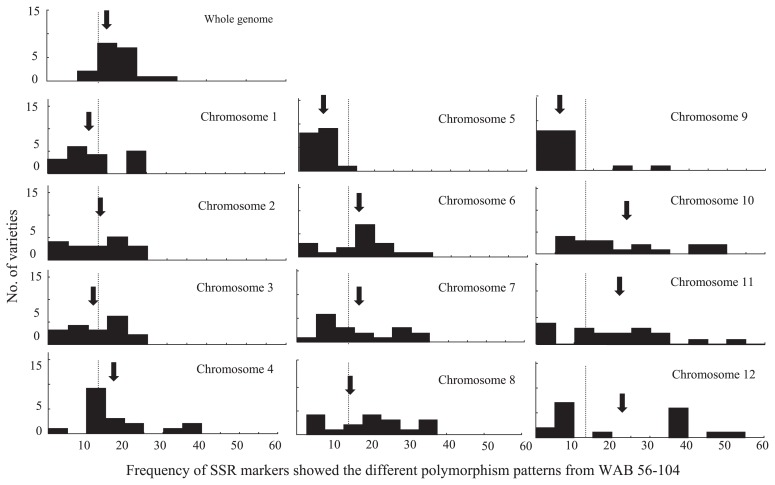
The theoretical frequency for introgression from CG 14 was 12.5% in each NERICAs genetic background, according to the breeding of two times’ backwoods with Asian varieties. However, these frequencies of SSR markers with the different types of polymorphic patterns from WAB 56-104 were varied from 7.0 to 34.2%, among 18 NERICAs. The introgression included some polymorphism types, CG 14 and different types from CG 14 and WAB 56-104. The frequencies of CG 14 and other type SSR marker introgression varied in the ranges from 1.2 to 10.7% and from 5.8 to 23.5%, respectively. NERICA 6 showed that a total of 34.2% SSR markers were not WAB 56-104 types and was the highest introgression value among NERICAs. Nine NERICAs, 5, 8, 9, 11, 14, 15, 16, 17 and 18, also showed higher frequencies of introgression in the range from 16.9 to 24.7% in comparison with the expected value, 12.5%. Two NERICAs, 3 and 4, were only 7.0% with quite low introgression frequencies.
The average introgression frequencies in each chromosome also varied among NERICAs (Fig. 3). Chromosomes 4, 6, 7, 10, 11 and 12 were higher frequencies than 12.5%, and chromosome 9 was lower. The distributions of introgression frequencies in 18 NERICAs were also different in 12 chromosomes. Almost all chromosomes showed variations in the ranges from 0 to approximately 40%, but three chromosomes, 10, 11 and 12, varied the introgressions with the widest ranges from 0 to approximately 60%.
Fig. 3.
Frequencies of SSR markers showed different polymorphism patterns form WAB 56-104 of upland NERICA varieties in each chromosome. Arrows indicate the means of 18 upland NERICAs. Dotted lines indicate the theoretical frequency, 12.5%, of different polymorphism markers in the progenies derived from the two times’ backcrossing by recurrent parents, WAB-56-104.
These results indicated that the introgressions varied among NERICAs and also in each chromosome.
Differential DNA maker among upland NERICAs
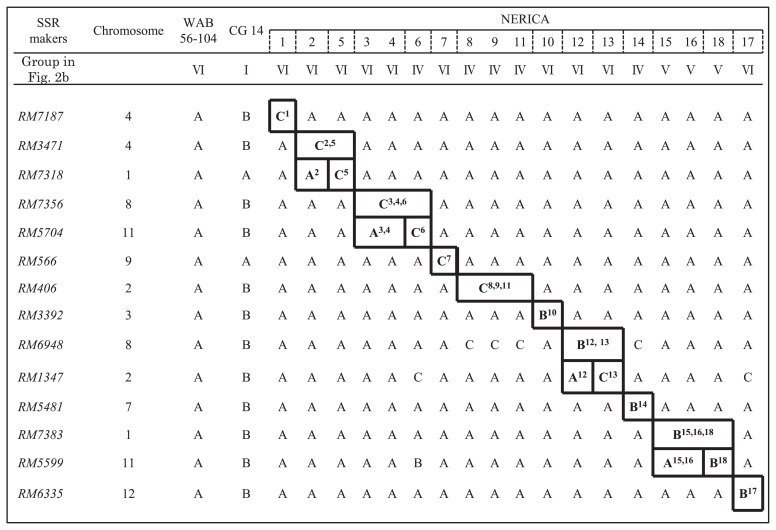
A total of 14 SSR markers were selected as a set of differential tools for 18 upland NERICAs (Fig. 4).
Fig. 4.
Differential markers for the 18 upland NERICA varieties and its capability of differentiation. A: WAB 56-104 type, B: CG 14 type, C: Other types. Polymorphism patterns enclosed by a heavy line are the unique pattern to differentiate NERICA varieties. Superior numbers indicates the number of upland NERICA varieties showing specific polymorphic pattern. Three variety groups of two NERICAs, 3 and 4, three NERICAs, 8, 9 and 11 and two NERICAs, 15 and 16, were not differentiated from each other in the same groups.
Seven SSR makers, RM7187for NERICA 1, RM7318 for NERICA 5, RM5704 for NERICA 6, RM566 for NERICA 7, RM3392 for NERICA 10, RM5481 for NERICA 14 and RM6335 for NERICA 17, showed the specific polymorphic patterns in each variety. These DNA markers could be identified in each among the 18 NERICAs.
NERICA 2 was differentiated by two SSR makers, RM3471 and RM7318, from the others. RM3471 showed the specific polymorphism patterns to two NERICAs, 2 and 5 and could differentiate from the other varieties. RM7318 was the specific marker for NERICA 5 and NERICA 2 was differentiated from NERICA 5.
Two NERICAs, 3 and 4, were differentiated from the other varieties by two SSR markers, RM7356 and RM5704. RM7356 showed the specific polymorphism pattern to three NERICAs, 3, 4 and 6, and NERICA 6 was differentiated from two NERICAs, 3 and 4, by RM5704. However no marker was found to distinguish between NERICAs 3 and 4.
Three NERICAs, 8, 9 and 11, were also differentiated from the others by RM406, but could not differentiate in each other using any markers.
Two NERICAs, 12 and 13, were differentiated from the other varieties by two markers, RM6948 and RM1347. These varieties showed the specific polymorphism pattern of RM6948 and differentiated from the other varieties. The polymorphism pattern of NERICA 12 to RM1347 was different from that of NERICA 13.
Two NERICAs, 15 and 16, were differentiated from the other varieties by two SSR markers, RM7383 and RM5599. RM7383 was a specific marker for three NERICAs, 15, 16 and 18, and two NERICAs, 15 and 16, showed different polymorphism pattern from that of NERICA 18 in RM5599. These two, NERICAs, 15 and 16, were identified using the two makers. However, they could not be differentiated from each other.
NERICA 18 was identified by two SSR markers, RM5599 and RM5704, on chromosome 11. RM5599 showed the different polymorphism pattern to two, NERICAs 18 and 6, from the other varieties and the specific marker, RM5704, for NERICA 6 could differentiate both varieties.
Classification of upland NERICAs based on the genotypes of DNA markers
A total 22 varieties, 18 upland NERICAs, and four controls, CG 14, Kasalath, Nipponbare and WAB 56-104, were classified into six groups, I–VI, by cluster analysis using polymorphism data of 127 SSR markers, which consisted of the complete matrix and no missing data between markers and varieties (Fig. 2b).
Three control varieties, CG 14, Kasalath and Nipponbare, were classified as a different group and into clusters I, II and III, respectively. Cluster VI included eleven NERICAs, 1, 2, 3, 4, 5, 7, 10, 12, 13 and 17, and one recurrent parent, WAB 56-104. Cluster V consisted of three NERICAs, 15, 16 and 18. NERICAs 6, 8, 9, 11 and 14 were included in cluster IV. Among them, variety groups IV, V and VI for NERICAs were close to group III (Nipponbare), in comparison with the other groups, I (CG 14) and II (Kasalath).
Association analysis between SSR markers and agronomic traits
Three NERICAs, 15, 16 and 18, in group V and two NERICAs, 3 and 4, four NERICAs, 1, 2, 5 and 10 and two NERICAs, 12 and 13, in group VI, were categorized as similar varieties in each by the classification of SSR markers. The same results were shown in the subgroups in group A classified based on agronomic traits. (Fig. 2). These results suggested that the genome chromosomal segments associated with the agronomic traits in NERICAs.
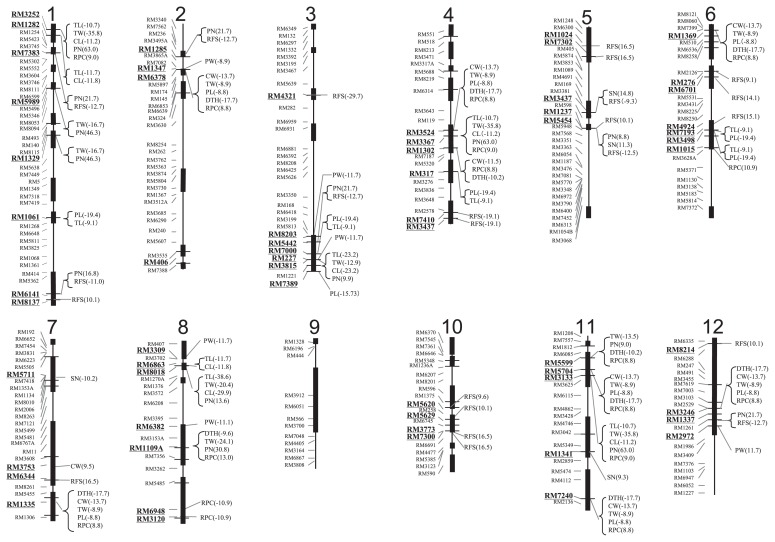
A total of 131 QTLs were detected in the combinations between 61 SSR markers and 11 agronomic traits on all chromosomes except for chromosome 9, and several traits were co-associated at several SSR markers (Fig. 5).
Fig. 5.
Association analysis between SSR markers and agronomic traits in 18 NERICA varieties. Thick bars on chromosomes were regions showing the different polymorphism patterns from WAB 56-104 among 18 NERICAs. SSR markers associated with agronomic traits are indicated with underlines. ANOVA test were carried out between traits’ values of WAB 56-104 and the other including CG-14 genotype at 1% significant level. The F values are indicated the combination between association markers and traits in each chromosome’s right side. Minus and plus of F indicate the increasing and decreasing of the values of traits with the other including CG-14 genotype, respectively. PN: Panicle No., DTH: Days to heading, CW: Culm and leaves weight, PW: Panicle weight, TW: Total weight of panicle, culm and leaves, CL: Culm length, PL: Panicle length, TL: Total length, RFS: Ratio of fertility seeds per panicle, SN: Spikelet No. per panicle, RPC: Ratio of panicle against culm and leaves’ weights.
Days to heading among NERICAs varied from 92.0 to 110.0 with the mean value of 96.6, and NERICA 6 was the latest. A total of nine SSR markers, RM6378 on chromosome 2, RM3524 and RM1302 on chromosome 4, RM1369 on chromosome 6, RM1335 on chromosome 7, RM1109A on chromosome 8, RM5599, RM5704 and RM1341 chromosome 11, and RM3246 on chromosome 12, were associated for the agronomic traits in NERICAs. The F values varied among nine markers from −9.2 to −17.7 and all QTLs showed the late heading with introgression genotypes, CG 14 or the other types of CG 14 and WAB 56-104.
Number of panicles per plant varied from 5.9 to 9.1 with the mean of 7.9 among NERICAs. A total of 15 QTLs were detected at these SSR markers, RM3252, RM7383, RM5989, RM1329 and RM6141 on chromosome 1, RM1285 on chromosome 2, RM5442 and RM3815 on chromosome 3, RM3524 on chromosome 4, RM5454 on chromosome 5, RM8018, RM1109A on chromosome 8, RM5599 and RM3133 on chromosome 11 and RM1337 on chromosome 12. These F values varied from 6.8 to 63.0, and the biggest values were shown at RM3252, RM3367 and RM3133. All QTLs showed the effects of decreasing for panicles with introgression genotypes.
Culm, panicle and total lengths varied from 82.2 to 108.8 cm, from 19.8 to 26.6 and from 103.6 to 135.0. NERICA 6 had the longest culm, panicle and total length among NERICAs. Five QTLs for culm length (CL) were detected on chromosomes 1(2), 4, 8(2) and 11. Thirteen QTLs for panicle length (PL) were detected on chromosomes 1, 2, 3(2), 4(2), 6(3), 7, 11(2) and 12. These QTLs for culm and panicle lengths were found at different SSR markers’ loci. Eleven QTLs for total length (TL) were detected on chromosomes 1(2), 3 (2), 4(2), 6(2), 8(2) and 11. Among them, these seven SSR markers, RM3252 and RM7383, on chromosome 1, RM3815 on chromosome 2, RM3367 on chromosome 4, RM6863 and RM8018 on chromosome 8 and RM3133 on chromosome 11 and five markers, RM1061 on chromosome 1, RM8203 on chromosome 2, RM317 on chromosome 4 and RM7193 and RM3498 on chromosome 6, were found as the common QTLs for culm and panicle length, respectively. These two markers, RM3815 on chromosome 2 and RM8018 on chromosome 8, showed the highest F values: −23.2 for culm and total length and −29.9 for culm and −38.6 for total length, respectively. All QTLs for three traits showed the increasing of length with the introgression genotypes.
A total of five QTLs for panicle weight were found on the SSR markers on chromosomes 2, 3(2), 5 and 8, and nine associations on chromosome 2, 4(2), 6, 7(2), 11(2) and 12 were for culm weight. The QTLs of total weight detected a total of 16 SSR markers on chromosomes 1(3), 2, 3, 4(2), 6, 7, 8(2), 11(4) and 12. All QTLs for them showed the increased effect of weight with the introgression genotypes, expect for panicle weight on RM2972 of chromosome 12. The QTLs for ratio of P/CL weights were detected on a total of 15 SSR markers on chromosomes 1, 2, 4(3), 6(2) 7, 8(3), 11(3) and 12(1), and all QTLs were effective to decrease of ratio of P/CL weights, expect for RM6948 and RM3120 on chromosome 8. Among them eleven SSR markers, RM3252, on chromosome 1, RM6378 on chromosome 2, RM3524 and RM1302 on chromosome 4, RM1369 on chromosome 6, RM1335 on chromosome 7, RM1109A on chromosome 8, RM5599, RM5704 and RM7240 on chromosome 11 and RM3246 on chromosome 12, were commonly detected for at least two traits among them.
Four QTLs for number of seeds (spikelets) were detected on chromosomes 5(2), 7 and 11 and only RM5711 on chromosome 7, showed the effect of increasing the number of seeds. The ratio of fertility seeds per panicle was detected at 25 SSR makers on chromosomes 1(2), 2, 3(2), 4 (5), 5(5), 6(2), 7, 8, 10(4) and 12(2). The increase and decrease effects were shown at 10 and 16 SSR markers, respectively. The expectation was that the ratio of fertility seeds per panicle was a complex trait and related with many genetic factors in the NERICAs, but the two SSR markers, RM3437 and RM5454 on chromosome 5 were common with these for the number of spikelets. The decrease of spikelets might influence an increase in fertile seeds.
Discussion
The 18 rainfed upland NERICAs were developed for targeting the accumulation of favorite traits, tolerant of drought, pest and disease resistances, early maturing, high nitrogen responses and high yielding potentials using the an African rice (O. glaberrima), CG 14 and three Asian rice (O. sativa) varieties, WAB 56-104, WAB 56-50 and WAB 181-18 (WARDA 2001). NERICAs were characterized as early heading, low tiller (panicle) number, low dry matter production, and heavy panicle, in comparison with the other 32 control varieties, with the variation in each trait among NERICAs in this study.
While the genetic characterization of NERICAs was carried out based on genotyping with DNA markers, investigation of agronomic traits related to the yield components, and association analysis between the introgressed chromosome segments and agronomic traits genetically. A total of 131 SSR markers found associations with agronomic traits. Days to heading of CG 14 was two weeks later than that of WAB 56-104 at Tsukuba in 2008 (Data was not shown). These NERICAs were characterized as early maturity varieties, but all nine QTLs detected by association analysis affected late heading in the genetic background. The introgressions from donor varieties including CG 14 affected late heading and the early maturities of NERICAs were contributed by the genetic background of donor varieties of Asian cultivate rice (O. sativa). A total of 15 QTLs for number of panicles showed effects of decreasing in NERICAs. The QTLs for panicle weight and panicle length were detected at five and thirteen SSR markers loci, respectively and these showed positive effects of increasing these values with the introgression genotypes, except for one for panicle weight on chromosome 12. All four QTLs for number of seeds (spikelets) also showed the effect of increasing. These QTLs might contribute to heavy panicles. Kobayashi et al. (2003) detected the QTLs for number of panicles on chromosomes 1 and 11. Xu et al. (2001) also detected QTLs on chromosomes 3 and 12. Zhu et al. (2000) reported a QTL for panicle weight located in the terminal region of chromosome 3. Li et al. (1997) found a QTL for number of spikelets on the middle region of chromosome 10. Thus, QTLs related to heavy panicle type in NERICAs as the architectural traits were detected in the same chromosome region as in other previous studies using different hybrid populations. These results demonstrated that the association analyses in this study were useful to detect the genetic factors from O. glaberrima, CG 14 and to characterize the 18 NERICAs.
Several limitations for missing and over counting might be included in this association analysis. When the segments of CG 14 or Asian rice varieties were introduced commonly at same chromosome region in all NERICA varieties, no polymorphism was detected among NERICAs and no QTLs were detected. When the same patterns of introgression among 18 NERICAs were observed in several different chromosome regions, and several QTLs might be detected all chromosome regions had the same effects. Only one QTL among them directly associated to the agronomic traits, and the other QTLs detected might be the shades of the original one. The need is to confirm these associations between SSR marker on introgression and agronomic traits by co-segregation analysis with the hybrid populations derived from the crosses between NERICAs and the recurrent parents or the other Asian variety. For this study, upland NERICAs were cultivated at irrigated lowlands with temperate conditions in Japan. These varieties are cultivating in the uplands in Africa with tropical conditions and the performance in upland conditions might be different from those in this study. It will also be necessary to investigate at the uplands in Africa and other conditions, to confirm the detail characters of upland NERICAs. Upland NERICAs were bred aiming to introduce drought resistance, pest and disease resistance and high yield potentials (WARDA 2001), but these were not well elucidated in this study. These evaluations for biotic and abiotic resistances also need physiological and genetic analyses to elucidate and understand NERICAs.
Based on the analysis using SSR markers, the graphical genotypes and 14 differential SSR markers were clarified and selected for the 18 upland NERICAs, respectively. These differential SSR markers and genome chromosome information will be used for the identification and purification check of these varieties. The higher frequencies in comparison with the theoretical ratio and additional chromosomal introgression from foreign rice varieties, CG 14 and the third parents(s) of NERICAs, had been reported using 18 NERICAs, by Africa Rice Center (Semagn et al. 2006), and our results agreed with those results. We can expect some reasons for the origins of the third parent(s): (1) out-crossing in the breeding process, (2) differences in the genetic backgrounds of CG 14 and the other recurrent parents of WAB lines between our materials used in this study and those of the Africa Rice Center and (3) differences in genotype information between the other two recurrent parents, WAB 56-104 and WAB 181-18. These NERICAs used by Semagn et al. (2006) might be different from our materials, which were provided from JICA and used in this study, because NERICAs segregated in several traits and the JICA project selected and purified them again.
Based on the field observation in Japan, several NERICAs were still segregating in some agricultural traits (Data were not shown). These relationships between genotypes and phenotype of segregants in these NERICAs need to be clarified with respect to these related chromosome regions. In three variety groups, NERICAs 3 and 4, NERICAs 8, 9 and 11 and NERICAs 15 and 16, polymorphic SSR markers were not found, and differences in these phenotypes of varieties in the same group were not found in field observations (Data not shown). Semagn et al. (2006) also reported that these varieties were close to each other in three groups based on the cluster analysis using 37 SSR markers. Further investigation is necessary to confirm differences among these varieties by using the genotypes of DNA markers and evaluation of agricultural traits, considering the different source between Semagn et al. (2006) and this study, and segregation of NERICAs.
This study compared agricultural traits of upland NERICAs with a number of rice varieties, clarified graphical genotypes using SSR markers and selection of differential SSR markers, and conducted association analyses between SSR markers and agronomic traits. The result was successful NERICA characterizations and these information may be used for genetic and agronomic analyses for understanding NERICAs and further genetic improvements in rice for Africa.
Acknowledgements
We would like to thank Dr. Ryouichi Ikeda, who is currently at Tokyou Agricultural University for providing these seeds of upland NERICA varieties under a project of Japan International Cooperation Agency (JICA)/African Rice Center (AfricaRice), Cotonou, Benin.
Literature Cited
- Africa Rice Center (WARDA)/FAO/SAA(2008) NERICA: the New Rice for Africa—a Comprndium. Cotonou, Benin: Africa Rice Center (WARDA); Rome, Italy: FAO, Tokyo, Japan: Sasakawa Africa Association; p. 15 [Google Scholar]
- Ekeleme F., Kamara A.Y., Oikeh S.O., Omoigui L.O., Amaza P., Abdoulaye T., Chikoye D. (2009) Response of upland rice cultivars to weed competition in the savannas of West Africa. Crop Protection 28: 90–96 [Google Scholar]
- Fukuta Y. (1995) RFLP mapping of a shattering-resistance gene in the mutant line, SR-1, induced from an indica rice variety, Nan-jing 11. Breed. Sci. 45: 15–19 [Google Scholar]
- Fukuta Y., Yoshida H., Fukui K., Kobayashi A. (1994) Analysis of shattering and abscission layer development in shattering-resistance mutant lines induced from an indica rice (Oryza sativa L.) variety, Nan-jing 11. Breed. Sci. 44: 195–200 [Google Scholar]
- Ikeda R., Sokei Y., Akintayo I. (2007) Reliable multiplication of seed for NERICA varieties of rice, Oryza sativa L. Genet. Resour. Crop Evol. 54: 1637–1644 [Google Scholar]
- Imbe T., Akama Y., Nakane A., Hata T., Ise K., Ando I., Uchiyamada H., Nakagawa N., Furutachi H., Horisue N., et al. (2004) Development of a multipurpose high-yielding rice variety “Takanari.” Bulletin of the National Institute of Crop Sciencs 5, National Institute of Crop Science, pp. 35–51 [Google Scholar]
- Ishizaki T., Kumashiro T. (2008) Genetic transformation of NERICA, interspecific hybrid rice between Oryza glaberrima and O. sativa, mediated by Agrobacterium tumefaciens. Plant cell Rep. 27: 319–327 [DOI] [PubMed] [Google Scholar]
- Jones M.P., Dingkuhn M., Aluko G.K., Semon M. (1997) Interspecific Oryza sativa L. × O. glaberrima Steud. Progenies in upland rice improvement. Euphytica 92: 237–246 [Google Scholar]
- Kaneda C. (2007a) Breeding and dissemination efforts of NERICA (1) Breeding of upland rice. Jpn. J. Trop. Agr. 51: 1–4 [Google Scholar]
- Kaneda C. (2007b) Breeding and dissemination efforts of NERICA (2) Evaluation of important characteristics. Jpn. J. Trop. Agr. 51: 41–45 [Google Scholar]
- Kobayashi S., Fukuta Y., Sato T., Osaki M. (2003) Molecular marker dissection of rice (Oryza sativa L.) plant architecture under temperate and tropical climates. Theor. Appl. Genet. 107: 1350–1356 [DOI] [PubMed] [Google Scholar]
- Li Z.K., Pinson S.R., Park W.D., Paterson A.H., Stansel J.W. (1997) Epistasis for three grain yield components in rice (Oryza sativa L.). Genetics 145: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Min S.K. (1991) Rice varieties and their genealogy in China. Shanghai Science and Technology Press, Shanghai, China [Google Scholar]
- McCouch S.R., Teytelman L., Xu Y., Lobos K.B., Clare K., Walton M., Fu B., Maghirang R., Li Z., Xing Y., et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 199–207 [DOI] [PubMed] [Google Scholar]
- Oba S., Kikuchi F., Maruyama K. (1990) Genetic analysis of semi-dwarfness and grain shattering of Chinese rice variety “Ai-Jio Nan-Te”. Jpn. J. Breed. 40: 13–20 [Google Scholar]
- Oikeh S.O., Nwilene F., Diatta S., Osiname O., Toure A., Okeleye K.A. (2008) Responses of upland NERICA rice to nitrogen and phosphorus in forest agroecosystems. Agron. J. 100: 735–741 [Google Scholar]
- Oikeh S.O., Toure A., Sidibe B., Niang A., Semon M., Sokei Y., Mariko M. (2009) Responses of upland NERICA rice varieties to nitrogen and plant density. Arch. Agro. Soil Sci. 55: 301–304 [Google Scholar]
- Sanni K.A., Ojo D.K., Adebisi M.A., Somado E.A., Ariyo O.J., Sie M., Akintayo L., Tia D.D., Ogunbayo S.A., Cisse B., et al. (2009a) Ratooning potential of interspecific NERICA rice varieties (Oryza glaberrima × Oryza sativa). Int. J. Botany 5: 112–115 [Google Scholar]
- Sanni K.A., Ariyo O.J., Ojo D.K., Gregorio G., Somado E.A., Sanchez I., Sie M., Futakuchi K., Ogunbayo S.A., Guei R.G., et al. (2009b) Additive main effects and multicative interaction analysis of grain yield performances in rice genotype across environments. Asian J. Plant Sci. 8: 48–53 [Google Scholar]
- Semagn K., Ndjiondjop M.N., Cissoko M. (2006) Microsatellites and agronomic traits for assessing genetic relationships among 18 New Rice for Africa (NERICA) varieties. Afr. J. Biotec. 5: 800–810 [Google Scholar]
- Tabuchi H., Hashimoto N., Takeuchi A., Terao T., Fukuta Y. (2000) Genetic analaysis of semidwarfism of the japonica rice cultivar Kinuhikari. Breed. Sci. 50: 1–7 [Google Scholar]
- Temnykh S., Park W.D., Ayres N., Cartinhour S., Hauck N., Lipovich L., Cho Y.G., Ishii T., McCouch S.R. (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100: 697–712 [Google Scholar]
- Uehara Y. (2001) Breeding, diffusion and adoption as parents of rice varieties, Kinuhikari and Dontokoi with a superior eating quality, excellent grain quality, and stable high yielding ability. Breed. Res. 3: 157–167 [Google Scholar]
- Ward J.H. (1963) Hieralchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58: 236–244 [Google Scholar]
- WARDA(2001) Rice Interspecific Hybridization Project: Research Highlights 2000. WARDA, Bouake, Cote d’Ivoire: 36 pp. [Google Scholar]
- WARDA(2006) WARDA: A concise history. http:/www.Warda.org/warda/history.asp
- Xu J.L., Xua Q.Z., Luo L.J., Li Z.K. (2001) QTL dissection of panicle number per plant and spikelet number per panicle in rice (Oryza sativa L.). Chin. J. Genet. 28: 752–759 [PubMed] [Google Scholar]
- Zhu J., Wu P., Benmoussa M., Yan J., He C. (2000) QTL mapping for developmental behavior of panicle dry weight in rice. Scientia Agricultura Sinica 33: 24–32 [Google Scholar]