Abstract
Deepwater rice possesses internode elongation ability to avoid drowning under deepwater conditions. Previous studies identified three QTLs regulating internode elongation ability on chromosomes 1, 3 and 12 using different populations. However, these QTLs only induce internode elongation in response to deepwater conditions from the 7-leaf stage and not during the early leaf stage. In this study, we detected two novel QTLs, qTIL2 and qTIL4 regulating deepwater response at the early leaf stage using an F2 population derived from the cross between NIL1-3-12 carrying the three QTLs regulating deepwater response in T65 (O. sativa ssp. japonica) genetic background and C9285 (O. sativa ssp. indica, deepwater rice). Plants of the BC2F2 population derived from NIL1-3-12/C9285 and the RILs of T65/Bhadua (O. sativa ssp. indica, deepwater rice) possessing these QTLs as well as the three QTLs previously identified also showed internode elongation during the early leaf stage. These results indicate that qTIL2 and qTIL4 regulate early internode elongation and function in coordination with the three major QTLs under deepwater conditions. The results presented here would not only help define the mechanism of deepwater response in rice but also contribute in the breeding of deepwater tolerant rice that is adapted to various water depths.
Keywords: deepwater rice, QTL, internode elongation
Introduction
Deepwater rice (floating rice) is an important crop in the flood-prone regions of Asia and West Africa. During the rainy season, the long and heavy rains bring floods that can reach several meters high. Non-deepwater rice (commonly cultivated rice) varieties cannot “breathe” in deepwater conditions and hence they “drown” and die. On the other hand, deepwater rice has adapted to periodic flooding by developing a capability for extreme internode elongation. Under dry or normal irrigated conditions, the leaves and internodes of deepwater rice do not elongate and behave similarly to non-deepwater rice. However, once subjected to flooding, the internodes of deepwater rice elongate and continue to do so with the rising water level (Vergara 1976). Internode elongation keeps the top leaves above the water surface, thereby facilitating respiration. Oxygen is also transported to the plant body through hollow structures in the internode, otherwise known as the aerenchyma. Daily internode elongation can reach up to 20–25 cm (Catling 1992).
Various genetic studies reported the monogenic, recessive (Eiguchi et al. 1993), monogenic, dominant (Tripathi and Balakrishana Rao 1985), and polygenic control of the deepwater trait in rice (Hamamura and Kuplanchankul 1979, Morishima et al. 1962). However, these reports have not been consistent. Tang et al. (2005) firstly reported that internode elongation ability is regulated by three QTLs on chromosomes 1, 3 and 12. Hattori et al. (2007) and Kawano et al. (2008) also reported the presence of three QTLs in the same chromosome regions using different populations. Nearly isogenic lines (NIL1, NIL3 and NIL12) and pyramiding lines (NIL1-12, NIL3-12, NIL1-3 and NIL1-3-12) carrying the C9285 (deepwater rice) genomic fragments covering the QTLs in the T65 (non-deepwater rice) genetic background demonstrated that the QTL in chromosome 12 is the major QTL controlling deepwater response in rice (Hattori et al. 2009). This QTL encodes an ethylene responsive factor, Snorkel1 (SK1) and Snorkel2 (SK2) (Hattori et al. 2009). NIL1-3-12 possessing all three QTLs showed the most responsive phenotype under deepwater conditions. However, the total internode length (TIL) of NIL1-3-12 did not reach that of C9285 (Hattori et al. 2009). This result indicates that other QTL(s), possibly minor QTLs, are also involved in the regulation of internode elongation in deepwater rice and are acting in concert with the three previously detected major QTLs. Possibly, the large effects of the three major QTLs mask the effects of the minor QTL(s). It is also possible that the other QTL(s) condition the deepwater response in rice during the early stages of growth. In this paper, we report the identification of novel QTLs that enhances internode elongation under deepwater conditions during the early vegetative stage.
Materials and Methods
Plant materials
The deepwater rice cultivars C9285 (O. sativa ssp. indica) and Bhadua (O. sativa ssp. indica) from Bangladesh and Tacihung 65 (T65; O. sativa ssp. japonica) were used in this study. C9285 was kindly provided by the National Institute of Genetics in Japan (http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp) whereas T65 was a cultivar maintained at Nagoya University. NIL1-3-12 possessing the three previously detected major QTLs regulating deepwater response in T65 genetic background was developed by repeated backcrossing with T65, accompanied by marker-assisted selection (MAS) (Hattori et al. 2009). An F2 population composed of 192 plants derived from a cross between NIL1-3-12 and C9285 was used for the QTL analysis. A BC2F2 population was developed by repeated backcrossing of the cross NIL1-3-12/C9285 with NIL1-3-12, accompanied by MAS. Recombinant inbred lines (RILs) were developed by 8-times selfing of the F1 derived from the cross between T65 and Bhadua.
Phenotypic evaluation of total internode length (TIL)
To determine the TIL, the main culm of each plant used in this study was cut at midline after deepwater treatment and the length of elongated internodes were measured (Supplemental Fig. 1). In a previous study, elongated internodes were defined as those more than 5 mm in length (Inouye 1987). Therefore, for this study, we considered elongated internodes as those measuring more than 5 mm. TIL was calculated by getting the sum of the measured internode lengths that are more than 5 mm. Plant height was measured from the leaf apex to the base of plant excluding the roots (Supplemental Fig. 1). Student’s t test was performed for each dataset to evaluate the statistical significance.
Evaluation of early internode elongation ability
To investigate elongation ability during the early vegetative stage, T65, NIL1-3-12 and C9285 were pre-germinated in water at 30°C for 72 h, individually transplanted in perforated pots (10 cm in diameter, 12 cm in height) and grown in dry conditions (i.e. normally watered condition achieved by irrigating the perforated pots in approximately 3 cm of water) (Supplemental Fig. 1). At the 10-leaf stage, these plants were put into the tank of 160 cm height and submerged in water at 70 cm depth. The water level was raised by 10 cm every two days for 20 days. The TIL and plant height of 5 plants each of T65, NIL1-3-12 and C9285 were measured every two days. To determine the onset of internode elongation, NIL1-3-12 and C9285 were submerged in water at the 3 to the 8-leaf stages for one week. TIL of 5 plants each of NIL1-3-12 and C9285 was measured at each leaf stage. To determine the frequency distribution for total internode length, F2 seeds generated from the cross NIL1-3-12/C9285 were pre-germinated at 30°C for 72 h, sown separately in pots (10 cm in diameter, 12 cm in height) and grown in dry conditions. At the 6-leaf stage, 192 F2 plants were submerged and TIL was measured. To determine the early internode elongation ability in the BC2F2 and RILs, seeds were pre-germinated at 30°C for 72 h and transplanted in pots. At the 6-leaf stage, 5 plants each of the BC2F2 and RILs were submerged in water for one week before gathering data on TIL.
Construction of linkage maps and QTL analysis
To construct linkage maps, genomic DNA was extracted from 192 F2 plants using the TPS method (Thomson and Henry 1995). Approximately 4 cm of leaves were sampled and ground in TPS buffer (100 mM Tris-HCl [pH 8.0], 1 M KCl and 10 mM EDTA) using the BMS ShakeMaster (Bio Medical Science). After centrifugation, the supernatant was recovered and an equal volume of isopropyl alcohol was added. The isopropanol-insoluble material was recovered by centrifugation and the pellet was washed with 75% ethanol. Thereafter, the pellet was dried and dissolved in TE (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). The extracted DNA samples were used for genotyping using SSRs (McCouch et al. 2002, Ware et al. 2002, http://www.gramene.org/) that was designed based on the comparative genomic sequence of each parent. PCR was performed as described by Chen et al. (1997). The amplified products were separated on 3% agarose gels in 0.5X TBE buffer and visualized using ethidium bromide. MAPMAKER/EXP, version 3.0 (Lander et al. 1987) was used to construct the linkage map. To identify QTLs for TIL, R/qtl (Broman and Sen 2009) was employed. Both standard interval mapping (SIM) and composite interval mapping (CIM) were used to detect QTLs. QTLs for TIL were considered significant at P < 0.01, with the LOD (logarithm of odds) score given by 1000 permutations (Churchill and Doerge 1994, Doerge and Churchill 1996). Multiple QTL mapping was conducted to detect epistatic interaction (Broman and Sen 2009). The region of the QTL for TIL enhanced by the deepwater allele was calculated using the lodint method in R/qtl (Broman and Sen 2009).
Results
Quantification of the effects of the three major QTLs in deepwater response
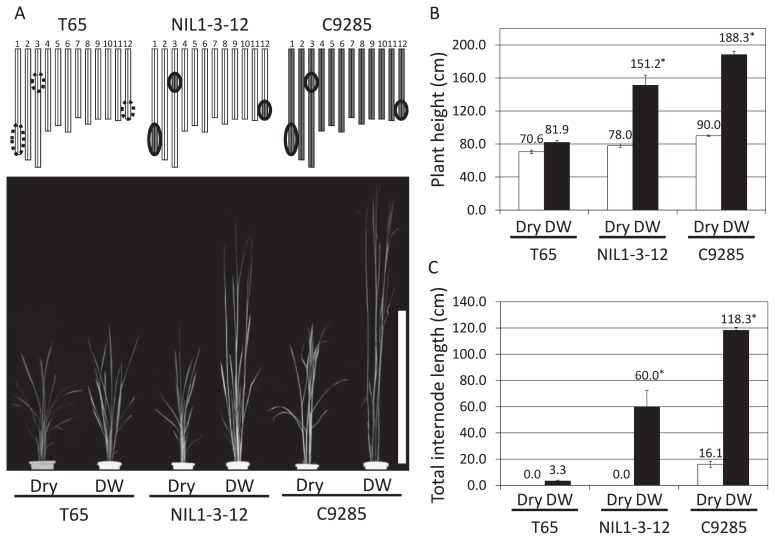
NIL1-3-12 was used to evaluate the effects of the three previously detected QTLs in response to deepwater conditions. (Hattori et al. 2009) (Fig. 1A). Upon deepwater treatment, NIL1-3-12 responded by an increase in plant height and elongation of internodes (Fig. 1B, 1C). Plant height (151.2 cm) and TIL (60.0 cm) of NIL1-3-12 reached 80% and 51%, respectively, of those of C9285 after two weeks of deepwater treatment.
Fig. 1.
Phenotypic and quantitative evaluation of the deepwater response. (A) Deepwater response in T65, NIL1-3-12 and C9285. Circles represent positions of major QTLs. Scale bar = 1 m. White and black bars indicate T65 and C9285 chromosomes, respectively. Dry and DW indicate dry conditions (normally watered conditions achieved by irrigating perforated pots in approximately 3 cm of water) and deepwater conditions (approximately 160 cm of water), respectively. (B) Plant height as affected by deepwater conditions. (C) Total internode elongation. White and black bars indicate the total internode length under dry and deepwater conditions, respectively. Plants were submerged for two weeks. Data represents mean value ± SD (n = 5). Asterisks indicate significant differences determined using Student’s t test (P < 0.05).
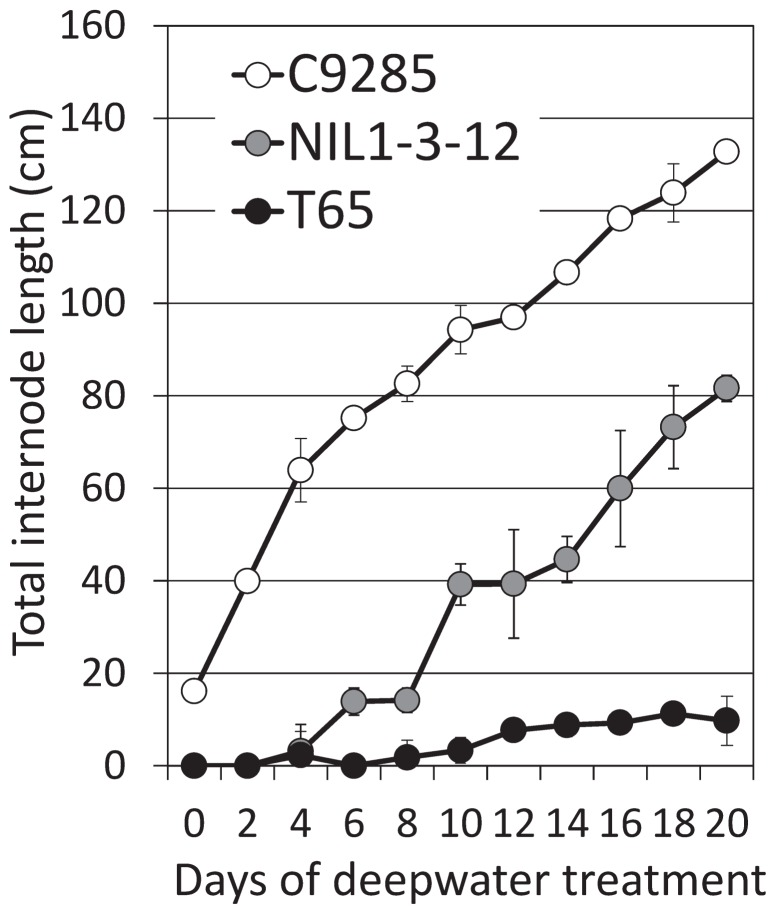
The onset of deepwater response measured in terms of TIL in T65, NIL1-3-12 and C9285 are presented in Fig. 2. Only slight internode elongation was observed in T65 under deepwater conditions. On the other hand, the TILs of NIL1-3-12 and C9285 increased with rising water levels, although the rate of internode elongation was not different between the two lines. Internode elongation in NIL1-3-12 averaged 11.2 cm every two days from the 8th to the 20th day of deep-water treatment. In a similar case, that of C9285 averaged 11.7 cm every two days from 0 to the 20th day of deepwater treatment. Whereas significant internode elongation was observed only from the 8th day of deepwater treatment in NIL1-3-12, C9285 responded to deepwater conditions even from the first day of treatment. The difference, therefore, between C9285 and NIL1-3-12 in terms of deepwater response is the timing or onset of internode elongation. These results indicate there are other factors controlling the elongation ability of deepwater rice at the early vegetative stage of the crop.
Fig. 2.
Time course of deepwater response. Plants were submerged in 70 cm water depth, and water level was raised 10 cm every two days for twenty days. White, gray and black dots indicate total internode length of C9285, NIL1-3-12 and T65, respectively. Data represents mean value ± SD (n = 5).
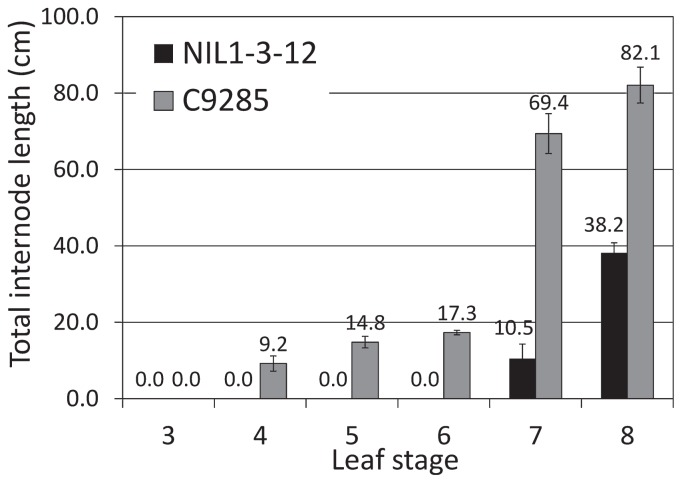
To assess the early elongation ability of C9285, we determined the leaf stage wherein NIL1-3-12 and C9285 would be able to elongate under deepwater conditions. We submerged NIL1-3-12 and C9285 plants at different leaf stages and measured the TIL for each line (Fig. 3). The results showed that internodes of C9285 have the ability to elongate from the 4-leaf stage, whereas those of NIL1-3-12 can elongate from the 7-leaf stage when subjected to deepwater conditions. These findings indicate that the previously identified QTLs on chromosomes 1, 3 and 12 do not regulate deep-water response during the early vegetative stage and that other genic factors exist between NIL1-3-12 and C9285 which regulate internode elongation in response to deep-water treatment during the early vegetative stage of rice.
Fig. 3.
Evaluation of internode elongation in NIL1-3-12 and C9285. NIL1-3-12 and C9285 were submerged in water at the 3 to the 8-leaf stages for one week. Black and gray bars indicate the total internode length of NIL1-3-12 and C9285, respectively. Data represents mean value ± SD (n = 5).
Distribution of TIL in NIL1-3-12/C9285 F2 population
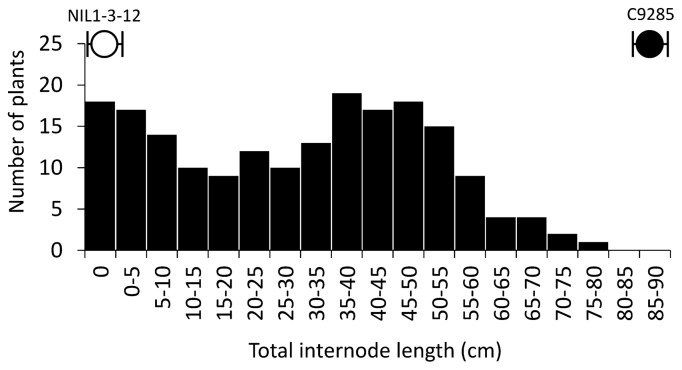
To identify other QTLs regulating internode elongation at the early vegetative stage, we selected NIL1-3-12 and C9285 as parental lines and used the F2 population developed from crosses between both lines for QTL analysis. By using these materials, we would be able exclude the effects of the three major QTLs because these regions will not be segregating in the F2 population. Fig. 3 shows that NIL1-3-12 cannot elongate before the 7-leaf stage, while C9285 can elongate at the 4-leaf stage. Thus, NIL1-3-12/C9285 F2 population were subjected to deepwater treatment at the 6-leaf stage for 1 week before TIL was measured. The 192 F2 plants showed a continuous frequency distribution for TIL (Fig. 4) ranging from 0 cm to 75–80 cm, which was not higher than the average values observed for C9285. Considering the environmental errors, the observed phenotypic segregation cannot be regarded as transgressive. A 3: 1 or 1: 2: 1 distribution was also not observed. Together, these observations indicate that internode elongation at the early vegetative stage of rice is controlled by QTLs.
Fig. 4.
Frequency distribution of the total internode length for 192 F2 individuals at the 6-leaf stage. After one week of submergence, the total internode length was measured. White and black dots indicate the total internode length of NIL1-3-12 and C9285, respectively.
Construction of linkage map for QTL analysis
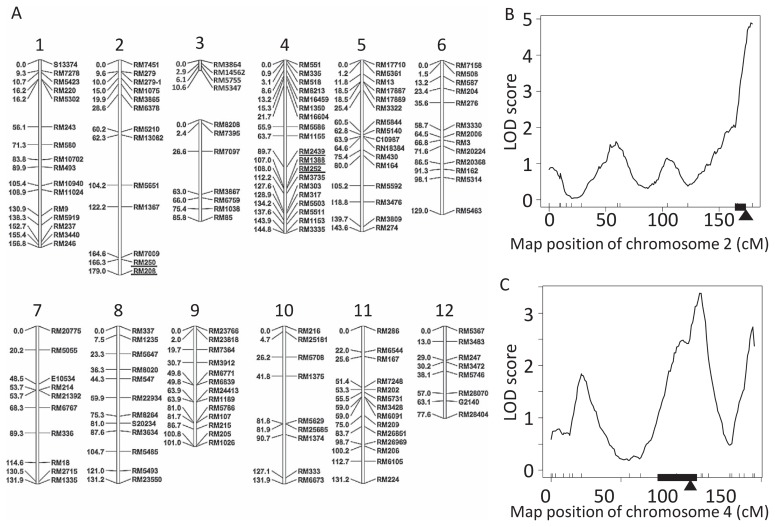
Linkage map was constructed using F2 plants of NIL1-3-12/C9285. We doubled the number of F2 plants used in this study from 92 individuals in our previous study to 192 individuals and also increased the number of markers from 92 to 154 (Hattori et al. 2007) (Fig. 5A). We produced a linkage map composed of 13 linkage groups. The linkage map spanned 1522.7 cM, with an average interval of 9.9 cM. Since the molecular markers used in this study covered the entire genome, it is expected that QTLs with minor effects and that may be located anywhere within the rice genome are likely to be detected by our analysis.
Fig. 5.
QTLs for internode elongation. (A) Linkage map of the F2 population. Crossed lines show the position of RM markers used for the whole genome survey. Numbers indicate the genetic distance (cM) of each RM marker. The RM markers on chromosomes 2 and 4 used in the selection of BC2F2 plants are underlined. Location of qTIL2 mapped by SIM technique on chromosome 2 (B) and qTIL4 mapped by CIM technique on chromosome 4 (C) constructed using an F2 population. Arrowhead indicates the position of peak LOD score. The region of the QTL for TIL enhanced by the deepwater allele was calculated using the lodint method in R/qtl (Broman and Sen 2009) (indicated black bars).
Two novel QTLs for TIL
Using the phenotypic data for the F2 population and linkage map, QTL analysis using R/qtl was performed for TIL. One QTL for TIL was detected in chromosome 2 (qTIL2) using SIM (Fig. 5B). The threshold value of LOD score given by 1000 permutation was 4.24 for TIL (P < 0.01). On the qTIL2 region, LOD score peaked from RM208 up to the end of chromosome 2. qTIL2 has the larger effect, explaining 14.4% of the total phenotypic variance (Table 1). CIM identified the peak for qTIL4 on chromosome 4 in the interval between RM2439 and RM1388 (Fig. 5C). In this region, LOD score is 3.4 (P < 0.01, threshold is 3.10). qTIL4 has a smaller effect, explaining 7.9% of the total phenotypic variance (Table 1). Any additional QTLs by CIM were not detected for TIL. Epistatic interaction between qTIL2 and qTIL4 was observed, with 23.3% total phenotypic variance (LOD score = 8.3; P-value = 1.71E-07) calculated using Multiple QTL mapping (Broman and Sen 2009).
Table 1.
Characteristics of the QTLs that affect TIL detected in the NIL1-3-12/C9285 F2 population
| QTL | Chr. | LOD | Pos. (cM) | Adda | Domb | %varc |
|---|---|---|---|---|---|---|
| qTIL2 | 2 | 4.9 | 179.0 | 10.8 | 5.1 | 14.4 |
| qTIL4 | 4 | 3.4 | 107.1 | 7.4 | 0.1 | 7.9 |
Additive effect of the C9285 allele.
Dominance effect.
Rate of contribution for phenotype.
QTL evaluation using the BC2F2 plants of NIL1-3-12/C9285 and RILs of T65/Bhadufa
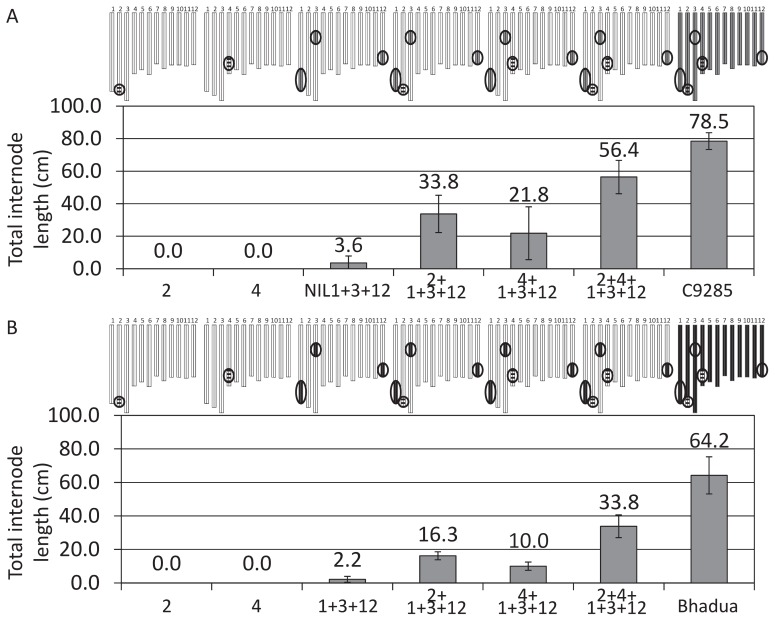
To evaluate the individual effects of qTIL2 and qTIL4, NIL1-3-12/C9285 BC2F2 population was produced and 6-leaf stage plants possessing qTIL2 or/and qTIL4, along with the three major QTLs on chromosomes 1, 3 and 12 (Fig. 6A) were screened for deepwater response for 1 week. Internodes of plants possessing the regions for only either qTIL2 (2), only qTIL4 (4) or NIL1-3-12 were not able to elongate at 6-leaf stage, whereas internodes of 2 + 1 + 3 + 12 (qTIL2 region + the three major QTLs) and 4 + 1 + 3 + 12 (qTIL4 region + the three major QTLs) elongated by 33.8 cm and 21.8 cm, respectively (Fig. 6A). Furthermore, 2 + 4 + 1 + 3 + 12, which has the regions of qTIL2, qTIL4 and the three major QTLs induced significant internode elongation (56.4 cm), which reached 71% of the internode elongation of C9285 (Fig. 6A). To investigate the effects of these QTLs, we screened plants possessing the regions of the three major QTLs (on chromosomes 1, 3 and 12) and qTIL2 or/and qTIL4 in RILs of T65/Bhadua (deepwater rice cultivar) (Fig. 6B). Internodes of plants possessing the regions for only either qTIL2 (2), qTIL4 (4) or the three major QTLs (1 + 3 + 12) did not elongate at the 6-leaf stage, while plants possessing the qTIL2 or qTIL4, alongside the three major QTLs (2 + 1 + 3 + 12 and 4 + 1 + 3 + 12) induced internode elongation at the 6-leaf stage (16.3 cm and 10.0 cm, respectively). Furthermore, plants having the regions for all QTLs (2 + 4 + 1 + 3 + 12), induced significant internode elongation (33.8 cm). Since these plants were developed using two different deepwater rice lines as parents, qTIL2 and qTIL4 clearly represents two novel QTLs that regulate deepwater response during the early vegetative stage of rice.
Fig. 6.
Evaluation of qTIL2 and qTIL4. Deepwater response of plants that were selected by marker assisted selection from the BC2F2 population of NIL1-3-12/C9285 (A) and RILs of Bhadua/T65 (B). Plants were submerged at 6-leaf stage for one week. In A, 2 + 1 + 3 + 12, 4 + 1 + 3 + 12 and 2 + 4 + 1 + 3 + 12 were significantly different (Student’s t test) compare with NIL1-3-12 (P < 0.05), whereas 2 and 4 were not. In B, 2 + 1 + 3 + 12, 4 + 1 + 3 + 12 and 2 + 4 + 1 + 3 + 12 were significantly different (Stutent’s t test) compare with 1 + 3 + 12 (P < 0.05), whereas 2 and 4 were not. Circles indicate major QTLs and QTL regions detected in this study. White, gray, black and striped box indicate T65 chromosomes, C9285 chromosomes and Bhadua chromosomes, and QTLs regions detected in this study, respectively. Data represents mean value ± SD (n = 5).
Discussion
Internode elongation in NIL1-3-12 and C9285
Plant height and TIL of NIL1-3-12 increased in response to deepwater conditions. Although the internodes of NIL1-3-12 elongated under deepwater conditions, the TIL of NIL1-3-12 did not reach that of C9285 (Figs. 1, 2). QTLs on chromosomes 3 and 12 were identified to control the lowest elongated internode (LEI), which is indicative of the onset of stem elongation. However NIL1-3-12 was not able to elongate before the 7-leaf stage (Fig. 3). Additionally, whereas the internodes of C9285 already elongated before submergence, those of NIL1-3-12 did not (Figs. 1, 2). These results indicate the presence of other QTL(s) that regulate elongation at early leaf stage under deepwater conditions and the possible interactive roles of these QTLs with the three major QTLs.
QTL analysis for early elongation ability
NIL1-3-12/C9285 F2 population was used for QTL analysis to separate the effects of the three major QTLs from other minor QTLs that may be involved in deepwater response during the early vegetative stage of rice. QTL analysis for TIL detected two QTLs on chromosomes 2 and 4 (Fig. 5). Multiple QTL analysis showed that qTIL2 and qTIL4 function in an additive epistatic manner. Sripongpangkul et al. (2000) reported on a QTL analysis for flood tolerance using an RIL population derived from IR74, a semidwarf non-elongating cultivar and Jalmagna, a deep-water rice cultivar from India. In their analysis, they measured internode increment (INI) during the rising of water level using 18-day-old seedlings. Their study showed that the major QTLs for INI were on chromosomes 1, 2 and 4. Because chromosome positions of these QTLs were different from those detected in this study, we regarded the QTLs reported here as new QTLs that regulate internode elongation as a deepwater response during the early vegetative stages of rice. In previous studies by three independent groups, the same three QTLs on chromosomes 1, 3 and 12 were detected, though they used different populations (Hattori et al. 2007, 2008, Kawano et al. 2008, Nemoto et al. 2004, Tang et al. 2005). However, qTIL2 and qTIL4 were not detected. The proper timing for deepwater treatment is necessary to determine the onset of internode elongation. Based on this factor, we confirmed whether the QTLs on chromosomes 2 and 4 can be detected using 192 F2 population derived from the cross between T65 and C9285 and same markers as this study. This population was not fixed in the region of the three major QTLs. In our study, we did not detect the QTLs on chromosomes 2 and 4 using this population at 6-leaf stage (data not shown). It is possible that the large effects of the three major QTLs mask the effects of the QTL on chromosomes 2 and 4. Therefore, exclusion of the effects of the major QTLs using a population that does not segregate for the previously detected loci is effective in detecting new QTLs. Selection of the appropriate leaf stage as phenotypic parameter would also help in the detection of novel QTLs.
Evaluation of QTLs on chromosomes 2 and 4
To characterize the roles of qTIL2 and qTIL4 in deep-water response, we screened plants carrying each QTL region from NIL1-3-12/C9285 BC2F2 population and RILs of T65/Bhadua (Fig. 6). BC2F2 and RILs carrying only either qTIL2 or qTIL4 did not show internode elongation in response to deepwater treatment at the 6-leaf stage. Similarly, plants carrying any of the three previously identified QTLs from chromosomes 1, 3 or 12 did not exhibit internode elongation at the 6-leaf stage. However, plants carrying qTIL2 or qTIL4 in addition to the three major deepwater QTLs from chromosomes 1, 3 and 12 showed internode elongation when subjected to deepwater conditions at the 6-leaf stage. Moreover, plants carrying both qTIL2 and qTIL4 along with the three major QTLs on chromosomes 1, 3 and 12 showed significant internode elongation. These results indicate that qTIL2 and qTIL4 act in concert with the three major QTLs for deep-water response at early vegetative stage and that these two QTLs function in coordination with the three major QTLs.
To understand the interaction among the several QTLs regulating the deepwater response, NILs and pyramiding lines possessing individual QTLs or different combinations of all the QTLs identified should be developed. Continuous studies using NILs and pyramiding lines will further elucidate the mechanism of deepwater response in rice. In addition, QTLs or combinations of QTLs conferring different extents of elongation abilities could be useful for breeding rice varieties adapted to various types of flooding regimes encountered in flood-prone areas.
Supplementary Material
Acknowledgement
We thank Professor Nori Kurata for providing C9285. We also thank Midori Ito for helping in the construction of linkage map. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated Research Project for Plants, Insects, and Animals using Genome Technology, QTL-4002), a research fellowship grant from the Japan Society for the Promotion of Science and Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 22119007). RILs of T65/Bhadua were provided by the Morning glory stock center of Kyushu University with support in part by the National Bio-Resource Project of the MEXT, Japan.
Literature Cited
- Broman K.W., Sen S. (2009) A guide to QTL mapping with R/qtl. Springer, New York [Google Scholar]
- Catling D. (1992) Rice in deep water. International Rice Research Institute, Macmillan [Google Scholar]
- Chen X., Temnykh S, Xu Y, Cho Y.G., McCouch S.R. (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 95: 553–567 [Google Scholar]
- Churchill G.A., Doerge R.W. (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge R.W., Churchill G.A. (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiguchi M., Hirano H.Y., Sano Y, Morishima H. (1993) Effects of water depth on internodal elongation and floral induction in a deepwater-tolerant rice line carrying the dw3 gene. Jpn. J. Breed. 43: 135–139 [Google Scholar]
- Hamamura K, Kupkanchankul T. (1979) Inheritance of floating ability of rice. Jpn. J. Breed. 29: 211–216 [Google Scholar]
- Hattori Y., Miura K, Asano K, Yamamoto E, Mori H, Kitano H, Matsuoka M, Ashikari M. (2007) A Major QTL confers rapid internode elongation in response to water rise in deepwater rice. Breed. Sci. 57: 305–314 [Google Scholar]
- Hattori Y., Nagai K, Mori H, Kitano H, Matsuoka M, Ashikari M. (2008) Mapping of three QTLs that regulate internode elongation in deepwater rice. Breed. Sci. 58: 39–46 [Google Scholar]
- Hattori Y., Nagai K., Furukawa S., Song X.J., Kawano R., Sakakibara H., Wu J., Matsumoto T., Yoshimura A., Kitano H., et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Inouye J. (1987) On floating rice and its ecological traits in Southeast Asia. Southeast Asia Studies 25: 51–61 [Google Scholar]
- Kawano R., Doi K, Yasui H, Mochizuki T, Yoshimura A. (2008) Mapping of QTLs for floating ability in rice. Breed. Sci. 58: 47–53 [Google Scholar]
- Lander E.S., Green P., Abrafamson J., Barlow A., Daly M.J., Lincoln S.E., Newbug L.A. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- McCouch S.R., Teytelman L, Xu Y., Lobos K.B., Clare K., Walton M., Fu B., Maghirang R., Li Z., Xing Y., et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 199–207 [DOI] [PubMed] [Google Scholar]
- Morishima H., Hinata K, Oka H.I. (1962) Floating ability and drought resistance in wild and cultivated species of rice. Indian J. Genet. 22: 1–11 [Google Scholar]
- Nemoto K., Ukai Y, Tang D.Q., Kasai Y, Morita M. (2004) Inheritance of early elongation ability in floating rice revealed by diallel and QTL analyses. Theor. Appl. Genet. 109: 42–47 [DOI] [PubMed] [Google Scholar]
- Sripongpangkul K., Posa G.B.T., Senadhira D.W., Brar D, Huang N, Khush G.S., Li Z.K. (2000) Genes/QTL affecting flood tolerance in rice. Theor. Appl. Genet. 101: 1074–1081 [Google Scholar]
- Tang D.Q., Kasai Y, Miyamoto N, Ukai Y, Nemoto K. (2005) Comparison of QTLs for elongation ability between two floating rice cultivars with a different phylogenetic origin. Breed. Sci. 55: 1–5 [Google Scholar]
- Thomson D., Henry R. (1995) Single-step protocol for preparation of plant tissue for analysis by PCR. BioTechniques 19: 394–400 [PubMed] [Google Scholar]
- Tripathi R.S., Balakrishna Rao M.J. (1985) Inheritance studies of characters associated with floating habit and their linkage relationship in rice. Euphytica 34: 875–881 [Google Scholar]
- Vergara B.S., Mazaredo A. (1979) Using the new standard evaluation system to measure elongation ability. Proc 1978 Intl Deep-water Rice Workshop, International Rice Research Institute, Manila, pp. 139–142 [Google Scholar]
- Ware D.H., Jaiswal P, Ni J, Yap I.V., Pan X, Clark K.Y., Teytelman L, Schmidt S.C., Zhao W, Chang K, et al. (2002) Gramene, a tool for grass genomics. Plant Physiol. 130: 1606–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.