Abstract
Tea is one of the most popular beverages in the world and the tea plant, Camellia sinensis (L.) O. Kuntze, is an important crop in many countries. To increase the amount of genomic information available for C. sinensis, we constructed seven cDNA libraries from various organs and used these to generate expressed sequence tags (ESTs). A total of 17,458 ESTs were generated and assembled into 5,262 unigenes. About 50% of the unigenes were assigned annotations by Gene Ontology. Some were homologous to genes involved in important biological processes, such as nitrogen assimilation, aluminum response, and biosynthesis of caffeine and catechins. Digital northern analysis showed that 67 unigenes were expressed differentially among the seven organs. Simple sequence repeat (SSR) motif searches among the unigenes identified 1,835 unigenes (34.9%) harboring SSR motifs of more than six repeat units. A subset of 100 EST-SSR primer sets was tested for amplification and polymorphism in 16 tea accessions. Seventy-one primer sets successfully amplified EST-SSRs and 70 EST-SSR loci were polymorphic. Furthermore, these 70 EST-SSR markers were transferable to 14 other Camellia species. The ESTs and EST-SSR markers will enhance the study of important traits and the molecular genetics of tea plants and other Camellia species.
Keywords: Camellia sinensis, tea plants, expressed sequence tags, EST-SSR
Introduction
Tea is one of the most popular non-alcoholic beverages and is drunk all around the world. The tea plant, Camellia sinensis (L.) O. Kuntze, is a woody evergreen plant of the genus Camellia in the family Theaceae and is native to southern China. In 2007, the harvested area totaled about 2.8 million hectares, with China, India, Sri Lanka, Kenya and Indonesia being major producers (http://faostat.fao.org/).
Tea has a genome of 4.0 Gb (Tanaka et al. 2006) with a basic chromosome number of n = 15. The genome size is larger than that of human (3.1 Gb; International Human Genome Sequencing Consortium 2004), 33 times that of Arabidopsis thaliana (120 Mb; Arabidopsis Genome Initiative 2000), 10 times that of rice (389 Mb; International Rice Genome Sequencing Project 2005) and one-quarter that of wheat (16 Gb; Arumuganathan and Earle 1991). An efficient first step for the analysis of the large-genome species such as tea is to survey the expressed genes. Expressed sequence tag (EST) analysis in which partial sequences of a large number of cDNA clones are isolated, is a useful approach to reveal expressed sequences in the genome and it enables the identification of many genes responsible for important traits. In addition, ESTs can be used as a resource for functional genomics experiments, such as gene expression analysis using microarrays.
Several EST analyses of tea plants have been reported. Chen et al. (2005) reported 1,684 ESTs generated from tender shoots. Park et al. (2004) reported 588 ESTs isolated by suppression subtractive hybridization. Sharma and Kumar (2005) reported three drought-responsive ESTs obtained by differential display. Shi et al. (2011) reported details of the transcriptome of C. sinensis that were generated by RNA-seq analysis using a high-throughput Illumina GA IIx sequencer. The ESTs reported in the first three studies were derived from green tissues, such as young shoots and mature leaves, but not roots. The RNA-seq data reported by Shi et al. (2011) were generated from seven different organs, including young roots, flower buds, and immature seeds, but the RNAs were mixed before analysis, and thus the origin of each transcript could not be identified.
DNA markers such as microsatellites (Becher 2007, Gupta and Prasad 2009, Hanai et al. 2007, Heesacker et al. 2008, Laurent et al. 2007) and single-nucleotide polymorphisms (SNPs) (Chagne et al. 2008, Choi et al. 2007, Deleu et al. 2009, Lijavetzky et al. 2007, Sato et al. 2009) can be developed by using sequence information from ESTs. Those ESTs that harbor simple sequence repeat (SSR) motifs, referred to as EST-SSRs, show a high level of transferability to closely related species because they originate from transcribed regions, which are often conserved. Therefore EST-SSRs of C. sinensis should be useful for genome analysis in many other Camellia species as well.
Sharma et al. (2009) developed 61 EST-SSRs of C. sinensis and demonstrated the polymorphism of these marker loci. However, to construct linkage maps of C. sinensis, several hundred markers are necessary.
In this paper, we report 17,458 ESTs derived from seven cDNA libraries of young shoots, mature leaves and roots of tea plants. To facilitate gene identification and functional studies, we performed Gene Ontology (Ashburner et al. 2000) annotation of tea unigenes. Furthermore, we developed EST-SSR markers developed using the EST data, and show them to be highly polymorphic and transferable to many Camellia species.
Materials and Methods
Plant material
Organs for RNA isolations were collected from tea plants growing at Makurazaki Tea Research Station, NARO Institute of Vegetable and Tea Science, Kagoshima, Japan. Young roots (RT) came from 15-d-old seedlings derived from natural crosses of C. sinensis cv. ‘Sayamakaori’. Tap roots (TR) and lateral roots (LR) were harvested from 30-d-old seedlings. Young leaves (YL), terminal buds (TB) and young stems (YS) of growing shoots with two leaves and a bud were harvested from field-grown ‘Sayamakaori’ in April of the first flush (first harvest) season. Mature leaves (ML) that developed the previous year were harvested from field-grown ‘Sayamakaori’ during the first flush season. The 16 accessions of C. sinensis and the 14 other Camellia species used for EST-SSR analysis are listed in Tables 1, 2, respectively.
Table 1.
Plant materials used in investigation of polymorphisms of EST-SSR loci
| Accession | Origin | Origin | IDa |
|---|---|---|---|
| Sayamakaori | selected from seedlings of Yabukita | Japan | 27029293 |
| KanaCk17 | introduced from Keemun, China | Japan | 27001948 |
| Minamisayaka | MiyaA6 × NN27 | Japan | – |
| Yabukita | selected from indigenous seedlings in Japan | Japan | 27027257 |
| ShizuInzatsu131 | selected from hybrids of var. sinensis and var. assamica | Japan | – |
| Asatsuyu | selected from indigenous seedlings in Japan | Japan | 27027248 |
| Miyamakaori | KyoKen283 × Saitama No. 1 | Japan | – |
| ME52 | selected from indigenous seedlings in Japan | Japan | 27025724 |
| ShizuZai16 | selected from indigenous seedlings in Japan | Japan | – |
| Shizu7132 | selected from seedlings of Yabukita | Japan | – |
| KaCp1 | introduced from Pingshui, China | China | – |
| Z1 | selected from seedlings of Tamamidori | Japan | – |
| Benifuki | Benihomare × MakuraCd86 | Japan | – |
| Ak1699 | introduced from Darjeeling, India | India | 27002929 |
| MakuraNo.1 | introduced from India | India | 27003028 |
| TaiwanYamacha95 | introduced from Taiwan | Taiwan | 27003335 |
Accession ID of the NIAS (National Institute of Agrobiological Sciences) Genebank, Japan.
Table 2.
Species used in investigation of transferability of EST-SSRs
| Names of accession | Species | Subgenus |
|---|---|---|
| Taliensis Midorime | C. taliensis | subgenus Thea |
| Irrawadiensis | C. irrawadiensis | subgenus Thea |
| Suzukayama | C. japonica | subgenus Camellia |
| Pitardii | C. pitardii | subgenus Camellia |
| Hongkongensis | C. hongkongensis | subgenus Camellia |
| Chekiangoleosa | C. chekiangoleosa | subgenus Camellia |
| Saluenensis | C. saluenensis | subgenus Camellia |
| Kissi | C. kissi | subgenus Camellia |
| Oleifera | C. oleifera | subgenus Camellia |
| Sasanqua Matsumoto 1 | C. sasanqua | subgenus Camellia |
| Furfuracea | C. furfuracea | subgenus Camellia |
| Cuspidata | C. cuspidata | subgenus Metacamellia |
| Salicifolia | C. salicifolia | subgenus Metacamellia |
| Granthamiana | C. granthamiana | subgenus Protocamellia |
Preparation of total RNA and cDNA library construction
Total RNAs from above-ground tissues (YL, ML, YS and TB) were extracted using TRIzol reagent (Life Technologies, USA). Total RNAs from young root tissues (RT, TR and LR) were extracted using an RNeasy Plant mini kit (Qiagen, Germany).
For cDNA library construction from the RT RNA sample, total RNA was dephosphorylated and decapped with a GeneRacer kit (Life Technologies). The decapped RNA was ligated with GeneRacer RNA Oligo and reverse-transcribed with SuperScript II reverse transcriptase (Life Technologies). After first-strand cDNA synthesis, the RNA was degraded with RNase H. cDNA was amplified by PCR with 5′ (5′-CGACTGGAGCACGAGGACACTGA-3′) and 3′ (5′-GCTGTCAACGATACGCTACGTAACG-3′) primers for 2 min at 94°C; followed by 20 cycles at 94°C for 20 s, 56°C for 30 s and 72°C for 10 min; followed by 10-min extension at 72°C. To enrich the content of long cDNAs, the PCR products were separated by agarose gel electrophoresis and products longer than 1,000 bp were isolated and cloned into the pGEM-T Easy vector (Life Technologies) and then transformed into Escherichia coli strain DH5α cells.
To construct cDNA libraries from the other organs, double-stranded cDNA was synthesized with a SMART cDNA Library Construction Kit (Clontech, USA), digested with restriction enzyme SfiI and size-fractionated in a CHROMA-SPIN 400 column (Clontech). The cDNA fragments were directionally ligated into an SfiI-digested pTriplEx2 vector. The ligation mixture was electroporated into E. coli DH5α competent cells.
DNA sequencing
Both ends of cDNAs from the RT library were sequenced using T7 (5′-TAATACGACTCACTATAGGG-3′) or SP6 (5′-ATTTAGGTGACACTATAGAA-3′) primers and the 5′ ends of cDNAs from the YL, TB, YS, ML, LR and TR libraries were sequenced using the 5′ λTriplEx2 sequencing primer (5′-TCCGAGATCTGGACGAGC-3′). Cycle sequencing reactions were performed using a BigDye Terminator Cycle Sequencing Kit (Life Technologies) and capillary electrophoresis was done using an ABI 3730xl or ABI 3130xl sequencer (Life Technologies).
Sequence analysis
Base-calling of sequence reads was performed using the KB basecaller program (Life Technologies). Ambiguous sequences were removed using the Sequencing Analysis program (Life Technologies) and vector sequences were trimmed using the cross_match program (http://www.phrap.org/). Sequences of less than 100 bp were then eliminated from the analysis. A total of 17,458 ESTs were generated and submitted to the DDBJ database (accession numbers AB361047 to AB361052, AB461364 to AB461372, AB485966 to AB505873 and FS943336 to FS960759). The 17,458 ESTs were assembled using the phrap program (http://www.phrap.org/). If the 5′ read and 3′ read derived from the same clone in the RT library belonged to different contigs, or both reads were singletons, or one read was a member of a contig and the other was a singleton, then the contigs or singletons were treated as a single scaffold.
The nucleotide sequences of the unigenes were searched using the BLASTX program (Gish and States 1993) against the non-redundant protein sequences in GenBank (http://www.ncbi.nlm.nih.gov/genbank/), the UniProt database (http://www.uniprot.org/), the Arabidopsis proteome database (TAIR8; http://www.arabidopsis.org/) and amino acid sequences deduced from the rice genome sequence (IRGSP/RAP build 5; http://rapdb.dna.affrc.go.jp/download/). Functional annotation of unigenes was performed using the Blast2GO program (Conesa et al. 2005). GO Slim annotations of unigenes were also generated with Blast2GO using the plant GO Slim mapping program provided by TAIR (http://www.arabidopsis.org/).
Digital analysis of expression
We selected 144 unigenes that were generated by assembly of 10 or more independent ESTs and used them for expression profiling based on the number of ESTs within each library. Differential expression levels were tested with the Audic and Claverie statistical test in IDEG6 software (Romualdi et al. 2003). To eliminate false positives, we used Bonferroni’s correction for the adjustment of multiple comparisons. Sixty-seven unigenes that were expressed differently among the seven libraries were clustered by using Hierarchical Clustering Explorer v. 3.0 software (http://www.cs.umd.edu/hcil/hce/).
Identification and analysis of EST-SSRs
Using the tea unigene set as a target, we identified micro-satellites that the total number of repeats were six or more, with each repeat unit being at least three times repeats of di-nucleotides or trinucleotides by using the Read2Marker program (Fukuoka et al. 2005). We also designed PCR primers for amplification of EST-SSRs using Read2Marker.
PCR was performed in a 10-μl reaction mix including 20 ng of genomic DNA, 10× PCR Gold buffer (Life Technologies), 0.8 μl of 8 mM dNTP, 0.1 U of AmpliTaq Gold polymerase, 0.8 μl of 25 mM MgCl2 and 1 μM forward and reverse primers. The PCR reactions were carried out in a GeneAmp 9600 thermal cycler (Life Technologies) according to the following “touchdown PCR” cycling program: 95°C for 5 min; 95°C for 1 min, 62°C for 30 s and 72°C for 1 min; 13 cycles at decreasing annealing temperatures in decrements of 0.5°C per cycle; 25 cycles of 95°C for 1 min, 62°C for 30 s and 72°C for 1 min and a final 72°C for 10 min. PCR products were directly labeled with fluorescence-labeled R110-ddUTP by the single-tube method (Inazuka et al. 1996). The labeled PCR products were analyzed with an ABI Prism 3130xl Genetic Analyzer, and the resulting allele data were analyzed with GeneMapper v. 3.7 software (Life Technologies). Polymorphism information content and heterozygosity information were calculated in PowerMarker software (Liu and Muse 2005).
Results
Sequencing and assembly
Seven cDNA libraries were constructed from tea plant organs (Table 3). From the young roots (RT) cDNA library, 3,072 clones were randomly selected and single-pass-sequenced from both ends. From each of the other six libraries, 2,880 clones were sequenced from their 5′ ends. After removal of low-quality sequences and vector trimming, the resulting data set contained 17,458 sequences with an average length of 481 bp (Table 4). The GC content of the 17,458 ESTs (8,391,523 bases) was 44.0%. These 17,458 high-quality ESTs were assembled into contigs in phrap, which resulted in 2,227 contigs and 3,477 singletons. Some 5′ and 3′ reads from the same clones from RT library were not assembled into the same contigs; in such cases, the contigs and singletons that contained such 5′ and 3′ pair reads were treated as scaffolds. As a result, 442 scaffolds, 1,851 contigs and 2,969 singletons were generated. Together, the 5,262 sequences were used for further analysis as a 5.3-k tea unigene set. Among these sequences, 3,372 unigenes (64.1%) were longer than 500 bp. The assembly of ESTs contained in each cDNA library generated 846 to 1,587 unique transcripts per library (Table 3).
Table 3.
cDNA library statistics
| Library | Source of RNA | No. of clones | ESTs | Unique transcripts |
|---|---|---|---|---|
| RT | young roots | 3,072 | 4,529 | 1,587 |
| TR | tap roots | 2,880 | 1,927 | 1,013 |
| LR | lateral roots | 2,880 | 2,316 | 1,230 |
| YL | young leaves | 2,880 | 2,233 | 1,090 |
| TB | terminal buds | 2,880 | 2,221 | 1,066 |
| YS | young stems | 2,880 | 2,147 | 1,187 |
| ML | mature leaves | 2,880 | 2,085 | 846 |
|
| ||||
| total | – | 20,352 | 17,458 | 5,262a |
number of unigenes generated from 17,458 ESTs.
Table 4.
Tea plant EST summary
| Feature | Value |
|---|---|
| Sequence information | |
| Total number of sequences | 17,458 |
| Total nucleotides (bp) | 8,391,523 |
| Average read length (bp) | 481 |
| GC content (%) | 44.0 |
| Unigene information | |
| Number of scaffolds | 442 |
| Number of contigs | 1,851 |
| Number of singletons | 2,969 |
| Number of sequences in unigenes | |
| 2 ESTs | 958 |
| 3–5 ESTs | 823 |
| 6–10 ESTs | 335 |
| 11–15 ESTs | 83 |
| >16 ESTs | 94 |
| Unigene length distribution | |
| Length | No. of unigenes |
| 100–500 bp | 1,890 |
| 501–1000 bp | 2,900 |
| >1000 bp | 472 |
On 5 August 2011, the NCBI GenBank database contained 14,246 ESTs and 34.5 million RNA-seqs from tea. Similarity searches of the 5.3-k unigene set were performed against the 14,246 ESTs and the 76,159 assembled sequences from the RNA-seq analysis by Shi et al. (2011), which had been deposited in the Transcriptome Shotgun Assembly Sequence Database (TSA) at NCBI with accession numbers HP701085–HP777243. The searches were performed by using BLASTN with a cutoff value of 1e-10. Within the 5.3-k unigene set, 3,340 unigenes (63.5%) had no matches among the 14,246 tea plant ESTs in GenBank and 1,118 unigenes (21.2%) had no matches among the 76,159 assembled sequences of RNA-seqs; 732 unigenes (13.9%) had no significant matches within either data set.
Similarity search and functional annotation
Before annotation of the tea unigenes, sequence homology searches were performed. By a BLASTX search against the GenBank non-redundant database (cutoff of ≤1e–6), 3,055 in the 5.3-k unigene set (58.1%) returned significant hits. Out of the 3,055 unigenes, 762 (24.9%) were annotated as hypothetical, predicted, putative, unknown, or unnamed proteins. A BLASTX search was also performed against the UniProt database and the complete protein sets of Arabidopsis thaliana and Oryza sativa. We found that 2,484 (47.2%) of the 5.3-k unigene set encoded peptides that were significantly similar to those in the UniProt database, 3,417 (64.9%) were similar to Arabidopsis proteins and 3,673 (64.4%) were similar to rice proteins, all with a cutoff value of 1e–6.
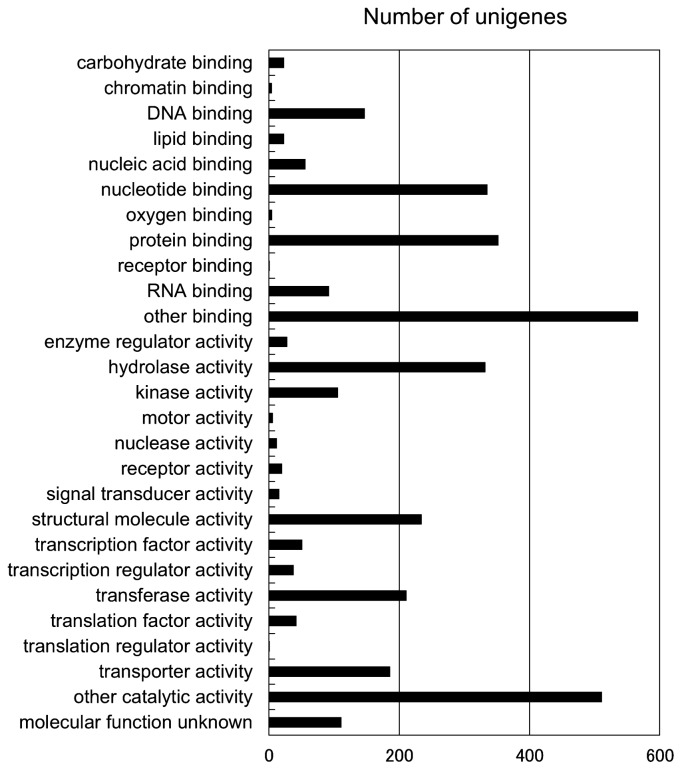
Subsequently, Gene Ontology annotation was performed with Blast2GO. A total of 2,639 unigenes were annotated with 11,260 annotations, distributed among the main Gene Ontology categories: Biological Process (4,582), Molecular Function (3,509) and Cellular Component (3,169) (Fig. 1 and Supplemental Table 1). There were 1,191 unigenes annotated for all three Gene Ontology categories.
Fig. 1.
GO Slim term annotation of tea unigenes in the 5.3-k set. Each unigene was assigned GO Slim terms by Blast2GO software. Unigenes could be assigned more than one GO Slim term.
To evaluate the usefulness of the 5.3-k unigene set as a gene resource for tea, we searched for unigenes involved in important agricultural and biological processes of tea, such as nitrogen assimilation and amino acid metabolism, catechin and caffeine biosynthesis, photoresponse and aluminum response (Table 5). For nitrogen assimilation, we found unigenes involved in primary assimilation of inorganic nitrogen, such as nitrate transporter, ammonium transporter and glutamate synthetase, and in amino acid metabolism. For catechin biosynthesis, we found unigenes encoding 10 enzymes were found, including phenylalanine ammonialyase and leucoanthocyanidin reductase. In addition, we identified unigenes encoding caffeine synthetase and several involved in aluminum response and photoresponse.
Table 5.
Unigenes related to important biological processes in tea
| Classification | Function | No. of unigenes | No. of ESTs |
|---|---|---|---|
| Aluminum response | aluminum-induced protein | 2 | 17 |
| citrate synthetase | 1 | 1 | |
|
| |||
| Caffeine biosynthesis | caffeine synthase | 1 | 9 |
|
| |||
| Catechin biosynthesis | 4-coumarate CoA: ligase | 2 | 5 |
| anthocyanidin reductase | 2 | 6 | |
| chalcone isomerase | 4 | 16 | |
| chalcone synthase | 3 | 20 | |
| cinnamate 4-hydroxylase | 2 | 3 | |
| dihydroflavonol 4-reductase | 2 | 41 | |
| flavonoid 3-hydroxylase | 4 | 5 | |
| flavonol synthase | 3 | 9 | |
| leucoanthocyanidin reductase | 1 | 2 | |
| phenylalanine ammonialyase | 2 | 4 | |
|
| |||
| Nitrogen assimilation and amino acid metabolism | 2-oxoglutarate malate translocator | 2 | 2 |
| alanine aminotransferase | 3 | 3 | |
| amino acid channel protein | 1 | 1 | |
| amino acid transporter | 4 | 4 | |
| ammonium transporter | 2 | 2 | |
| aspartate aminotransferase | 2 | 2 | |
| glutamate dehydrogenase | 1 | 2 | |
| glutamate synthetase | 1 | 1 | |
| glutamine dumper | 1 | 1 | |
| glutamine synthetase | 3 | 12 | |
| glycine decarboxylase | 3 | 7 | |
| NAD+-dependent isocitrate dehydrogenase | 1 | 1 | |
| NADP+-dependent isocitrate dehydrogenase | 1 | 2 | |
| nitrate transporter | 1 | 1 | |
| serine hydroxymethyltransferase | 1 | 1 | |
|
| |||
| Photoresponse | cryptochrome | 1 | 1 |
| CIP8 (COP1-interacting protein 8 ) | 1 | 1 | |
Digital analysis of gene expression
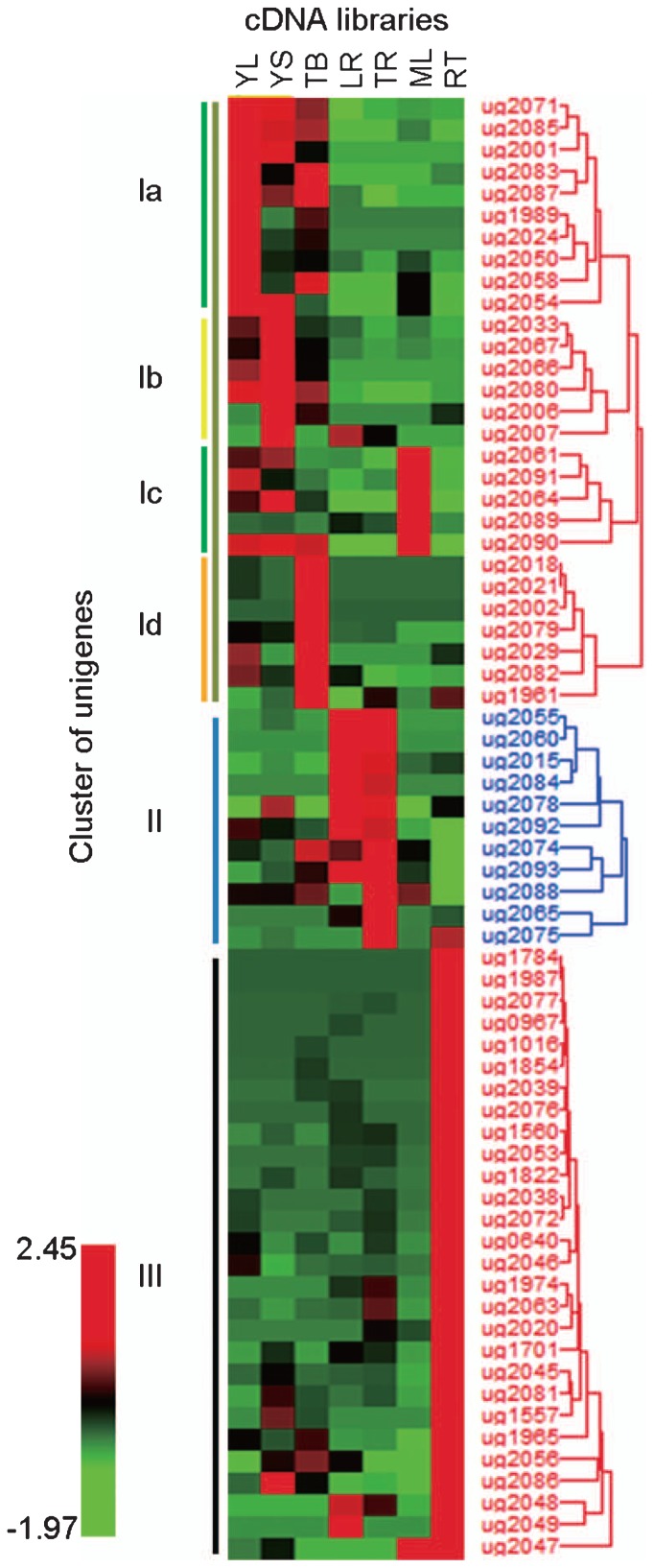
To reveal patterns of gene expression and correlations of expression patterns between organs, we analyzed the EST data using R statistics of the IDEG6 web tool to identify unigenes that were differentially expressed. From the 5.3-k unigene set, 144 unigenes that consisted of more than 10 EST sequences were selected for analysis; of these, 67 showed significant differences in expression profile among the libraries (Supplemental Table 2). Cluster analysis using Hierarchical Clustering Explorer 3.0 divided the 67 unigenes into three major clusters (Fig. 2 and Supplemental Table 2). Cluster I was divided into four subclusters, Ia–Id, which contained unigenes highly expressed in the YL, YS, ML and TB libraries, respectively. Clusters II and III showed high expression in root: specifically, cluster II in the LR and TR libraries and cluster III in the RT library.
Fig. 2.
Hierarchical clustering of 67 unigenes showed differential expression among the seven cDNA libraries. These 67 unigenes were grouped into three major clusters (indicated by vertical color bars) and cluster I was further subdivided into four subclusters. Red bars represent normalized expression values greater than the mean for that gene; green bars represent lower expression than the mean.
Clusters Ia and Ic contained a number of photosynthesis-related genes, including chlorophyll-a/b-binding protein and photosystem I reaction center subunit, respectively (Supplemental Table 2). Cluster II contained a unigene that encodes dihydroflavonol 4-reductase; this enzyme synthesizes leucoanthocyanidin, which is the direct precursor to (+)-catechin and (+)-gallocatechin. In cluster III, 10 out of 28 unigenes encoded stress-response proteins, including manganese superoxide dismutase and glutathione S-transferase.
Identification and analysis of EST-SSRs
An SSR motif search within the 5.3-k unigene set identified 1,835 unigenes (34.9%) that harbored SSR motifs of six or more repeat units. Among these 1,835 SSR-containing unigenes, the most frequent repeat motif was AT/GC repeat, which was found in 24.4% of all unigenes, followed by AC/GT repeat (6.5%) (Table 6).
Table 6.
Number and motif distribution of EST-SSRs
| Motif | Number of unigene sequences containing the number of repeats specified | |||
|---|---|---|---|---|
|
| ||||
| ≥6 times | ≥10 times | |||
|
|
|
|||
| n | (%) | n | (%) | |
| AG/TC | 1,284 | 24.4 | 608 | 11.0 |
| AT/TA | 271 | 5.2 | 127 | 2.3 |
| AC/GT | 344 | 6.5 | 156 | 2.8 |
| GC/CG | 17 | 0.3 | 13 | 0.2 |
| AAC/GTT | 45 | 0.9 | 24 | 0.4 |
| AAG/CTT | 88 | 1.7 | 42 | 0.8 |
| ACC/GGT | 120 | 2.3 | 60 | 1.1 |
| ACG/CGT | 26 | 0.5 | 15 | 0.3 |
| ACT/AGT | 57 | 1.1 | 24 | 0.4 |
| AGC/GCT | 37 | 0.7 | 24 | 0.4 |
| AGG/CCT | 81 | 1.5 | 35 | 0.6 |
| ATC/GAT | 52 | 1.0 | 28 | 0.5 |
| TAT/ATA | 24 | 0.5 | 13 | 0.2 |
| CGC/GCG | 23 | 0.4 | 10 | 0.2 |
|
| ||||
| Any SSR motifsa | 1,835 | 34.9 | 878 | 16.0 |
number of unigenes containing any SSR motifs listed in this table.
We selected the 100 EST-SSRs with the highest numbers of repeat units and designed primer sets to amplify them (Supplemental Table 3). Out of the 100, three (MSE0049, MSE0066 and MSE0089) had high homology to EST-SSRs reported by Sharma et al. (2009), but the other 97 EST-SSRs were novel.
The 100 EST-SSRs were tested for their ability to amplify fragments within 16 tea (C. sinensis) accessions (Table 1). Of these, 71 produced well-amplified fragments, and 70 revealed polymorphism among the 16 accessions (Table 7). For 61 markers, only one or two fragments were amplified in each accession and these were considered single-locus markers. For the other 10 markers, more than three amplified fragments were observed in some accessions and these were considered multi-locus markers. For the single-locus markers, the number of alleles per locus ranged from 1 to 15, with an average of 8.2 alleles. Observed hetero-zygosities (Ho) ranged from 0 to 1.0, with an average of 0.64. Expected heterozygosities (He) ranged from 0 to 0.91, with an average of 0.72. Polymorphism information content ranged from 0 to 0.90, with an average of 0.69.
Table 7.
Features of EST-SSRs and polymorphism information in 16 tea accessions
| Marker name | SSR motif | Position of repeat motifsa | Approximate size range (bp) | No. of accessions with amplification | Number of transferable species | No. of locib | No. of alleles | Heterozygosityc | PIC valued | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| He | Ho | |||||||||
| MSE0019 | (ac)23(tc)12 | 5′ | 105–150 | 16 | 13 | m | ||||
| MSE0021 | (tc)20(ta)10 | 5′ | 265–310 | 13 | 7 | s | 10 | 0.80 | 0.46 | 0.78 |
| MSE0022 | (tc)19(ta)9 | 5′ | 165–210 | 16 | 14 | s | 14 | 0.89 | 0.88 | 0.88 |
| MSE0023 | (tc)13, (tc)7 | unknown | 180–240 | 16 | 12 | s | 8 | 0.83 | 0.63 | 0.81 |
| MSE0024 | (ag)11 | 5′ | 255–285 | 16 | 14 | s | 9 | 0.82 | 0.56 | 0.80 |
| MSE0025 | (tc)14 | unknown | 260–305 | 16 | 14 | m | ||||
| MSE0026 | (ag)7, (ag)6 | 5′ | 275–300 | 16 | 14 | s | 7 | 0.79 | 0.56 | 0.75 |
| MSE0027 | (tc)19 | 5′ | 105–135 | 15 | 6 | s | 5 | 0.56 | 0.47 | 0.52 |
| MSE0029 | (ag)14, (ag)7 | 5′ | 365–340 | 16 | 11 | m | ||||
| MSE0030 | (tc)11 | 5′ | 245–270 | 16 | 14 | s | 10 | 0.78 | 0.81 | 0.75 |
| MSE0035 | (tc)13 | 5′ | 210–250 | 16 | 13 | s | 10 | 0.82 | 0.81 | 0.80 |
| MSE0037 | (tc)12 | TR, 3′ | 200–250 | 16 | 14 | m | ||||
| MSE0038 | (ag)8 | 5′ | 300–320 | 16 | 13 | m | ||||
| MSE0039 | (ag)21 | unknown | 135–180 | 16 | 14 | m | ||||
| MSE0040 | (tc)18 | 5′, TR | 125–155 | 16 | 14 | s | 9 | 0.73 | 0.75 | 0.71 |
| MSE0042 | (tc)16 | unknown | 100–115 | 16 | 12 | s | 8 | 0.77 | 0.50 | 0.74 |
| MSE0043 | (ta)7, (ag)10 | unknown | 170–210 | 16 | 12 | s | 10 | 0.79 | 0.69 | 0.77 |
| MSE0044 | (ta)11, (ag)6 | 5′ | 120–145 | 16 | 10 | s | 9 | 0.75 | 0.56 | 0.72 |
| MSE0045 | (ag)14 | unknown | 215–230 | 16 | 14 | s | 6 | 0.75 | 0.88 | 0.71 |
| MSE0047 | (tc)13 | 5′ | 245–275 | 16 | 14 | s | 11 | 0.79 | 0.75 | 0.77 |
| MSE0049 | (ag)14 | unknown | 220–250 | 16 | 14 | s | 10 | 0.84 | 0.81 | 0.83 |
| MSE0050 | (tc)14 | unknown | 265–285 | 16 | 14 | s | 11 | 0.85 | 0.88 | 0.83 |
| MSE0051 | (tc)15 | 5′ | 185–215 | 16 | 14 | s | 11 | 0.87 | 0.81 | 0.86 |
| MSE0052 | (tc)9 | tr | 260–285 | 16 | 13 | s | 12 | 0.86 | 0.81 | 0.85 |
| MSE0053 | (tc)16 | 5′ | 250–275 | 16 | 12 | s | 11 | 0.83 | 0.81 | 0.81 |
| MSE0054 | (tc)15 | 5′ | 165–195 | 16 | 14 | m | ||||
| MSE0055 | (ta)6 | unknown | 235–265 | 16 | 14 | s | 3 | 0.22 | 0.25 | 0.21 |
| MSE0056 | (tc)10 | 5′, tr | 220–240 | 13 | 13 | s | 6 | 0.75 | 0.77 | 0.71 |
| MSE0058 | (tc)12 | unknown | 185–275 | 16 | 14 | m | ||||
| MSE0059 | (gaa)11 | 5′ | 195–225 | 16 | 14 | s | 7 | 0.71 | 0.63 | 0.67 |
| MSE0061 | (tc)9 | 5′ | 135–165 | 16 | 13 | m | ||||
| MSE0062 | (ag)18 | tr | 110–140 | 16 | 14 | s | 9 | 0.84 | 0.81 | 0.82 |
| MSE0063 | (ag)9 | 5′, tr | 230–255 | 16 | 14 | s | 10 | 0.79 | 0.81 | 0.77 |
| MSE0066e | (tc)4, (tc)4, (tc)3, (tc)4, (tc)3 | 5′, tr | 240–260 | 16 | 5 | s | 1 | 0.00 | 0.00 | 0.00 |
| MSE0067 | (tc)18 | 5′ | 135–165 | 16 | 4 | s | 11 | 0.88 | 0.69 | 0.87 |
| MSE0068 | (tc)9 | 5′ | 255–275 | 16 | 14 | s | 10 | 0.80 | 0.88 | 0.77 |
| MSE0069 | (tc)17 | unknown | 195–225 | 15 | 11 | s | 11 | 0.85 | 0.40 | 0.83 |
| MSE0070 | (tc)8,(tc)6 | 5′ | 125–140 | 16 | 11 | s | 6 | 0.72 | 0.56 | 0.68 |
| MSE0071 | (tc)14 | 5′ | 110–135 | 16 | 13 | s | 8 | 0.80 | 0.63 | 0.78 |
| MSE0072 | (tc)14 | 5′, tr | 295–320 | 16 | 14 | s | 9 | 0.81 | 0.75 | 0.79 |
| MSE0074 | (tc)7 | unknown | 200–210 | 16 | 13 | s | 4 | 0.62 | 0.56 | 0.54 |
| MSE0075 | (tc)12 | unknown | 140–200 | 16 | 14 | m | ||||
| MSE0076e | (ca)3, (ac)4, (tat)3, (gta)3, (tgg)3 | tr | 240–245 | 16 | 13 | s | 3 | 0.36 | 0.19 | 0.33 |
| MSE0077 | (tc)10 | 5′ | 120–140 | 16 | 14 | s | 8 | 0.68 | 0.56 | 0.64 |
| MSE0078 | (ag)16 | 5′ | 140–160 | 16 | 14 | s | 7 | 0.80 | 0.69 | 0.77 |
| MSE0079 | (ag)16 | 5′ | 100–115 | 16 | 14 | s | 11 | 0.76 | 0.81 | 0.74 |
| MSE0080e | (ac)4, (ta)4, (tca)3, (tc)5 | unknown | 225–245 | 16 | 13 | s | 5 | 0.52 | 0.38 | 0.48 |
| MSE0081 | (tc)9 | 5′ | 100–140 | 16 | 8 | s | 9 | 0.82 | 0.31 | 0.79 |
| MSE0082 | (ta)13 | unknown | 155–185 | 16 | 13 | s | 8 | 0.81 | 0.81 | 0.78 |
| MSE0083 | (tct)6 | 5′ | 235–265 | 16 | 14 | s | 9 | 0.84 | 0.69 | 0.83 |
| MSE0084 | (tc)16 | 5′ | 395–420 | 16 | 14 | s | 11 | 0.83 | 0.81 | 0.81 |
| MSE0087 | (ag)13 | 5′ | 265–285 | 16 | 14 | s | 7 | 0.79 | 0.75 | 0.76 |
| MSE0088 | (tc)9 | unknown | 185–195 | 15 | 3 | s | 4 | 0.46 | 0.33 | 0.42 |
| MSE0089 | (ag)7 | 5′, tr | 290–310 | 16 | 14 | s | 7 | 0.61 | 0.63 | 0.59 |
| MSE0094 | (ta)8 | 3′ | 180–220 | 16 | 14 | s | 8 | 0.70 | 0.56 | 0.67 |
| MSE0096 | (tc)12 | 3′ | 235–260 | 16 | 14 | s | 11 | 0.74 | 0.75 | 0.72 |
| MSE0098 | (cca)6 | tr | 245–270 | 16 | 14 | s | 7 | 0.62 | 0.81 | 0.58 |
| MSE0099 | (ag)6 | 5′ | 285–300 | 16 | 14 | s | 7 | 0.73 | 1.00 | 0.70 |
| MSE0100 | (tc)15 | 5′ | 235–260 | 15 | 7 | s | 10 | 0.86 | 0.93 | 0.84 |
| MSE0101 | (cca)6 | 5′ | 240–270 | 16 | 14 | s | 7 | 0.77 | 0.75 | 0.73 |
| MSE0102e | (tc)5(ctc)3, (tc)4, (tc)3 | tr | 305–345 | 16 | 12 | s | 9 | 0.78 | 0.63 | 0.76 |
| MSE0103 | (tc)14 | 5′ | 170–195 | 16 | 13 | s | 8 | 0.74 | 0.81 | 0.72 |
| MSE0106 | (tc)6 | unknown | 145–175 | 16 | 14 | s | 8 | 0.80 | 0.44 | 0.77 |
| MSE0107 | (tc)8 | 5′, tr | 290–315 | 15 | 14 | s | 10 | 0.78 | 0.87 | 0.76 |
| MSE0108 | (tc)6(ta)8 | unknown | 245–270 | 16 | 14 | s | 9 | 0.79 | 0.81 | 0.77 |
| MSE0109 | (tc)10 | unknown | 105–125 | 13 | 5 | s | 6 | 0.67 | 0.31 | 0.63 |
| MSE0112e | (ag)5, (ag)3, (ag)3, (tg)3 | unknown | 280–285 | 16 | 14 | s | 2 | 0.48 | 0.56 | 0.37 |
| MSE0113 | (tc)14 | 5′ | 350–380 | 16 | 0 | s | 9 | 0.84 | 0.75 | 0.82 |
| MSE0114 | (tc)14 | unknown | 195–205 | 16 | 2 | s | 5 | 0.61 | 0.94 | 0.53 |
| MSE0116 | (tc)10 | 5′ | 175–195 | 16 | 14 | s | 6 | 0.65 | 0.38 | 0.61 |
| MSE0117e | (aca)3, (ag)4, (ag)3(tg)3 | tr, 3′ | 105–125 | 16 | 14 | s | 7 | 0.59 | 0.31 | 0.55 |
5′, 5′-UTR; 3′, 3′-UTR; tr, translated region.
s, single locus; m, multi-locus.
He, expected heterozygosity; Ho, observed heterozygosity.
PIC, polymorphism information content.
All SSR motifs in these markers are less than 6 times repeats, but these markers were included in the analysis, because the total number of repeats are more than 10.
Using 14 Camellia species (Table 2), we investigated transferability of the EST-SSRs developed in this study. Seventy of the 71 markers usable in C. sinensis were amplified in more than one species (Table 7). The average proportion of C. sinensis markers amplified in each of the 14 species was 87.1%. In C. irrawadiensis, a member of the same subgenus (Thea) as C. sinensis, 68 markers (95.8%) showed amplification (Table 7 and Supplemental Table 4).
Discussion
Before this study, the NCBI GenBank database held 14,246 ESTs and 34.5 million RNA-seqs from C. sinensis. In this study, we report the identification of 17,458 ESTs from seven cDNA libraries. Within the 5.3-k unigene set developed here, 732 unigenes had no significant matches by BLASTN homology searches against the tea ESTs and assembled sequences from RNA-seqs previously deposited in GenBank, indicating that these unigenes are novel mRNA sequences from tea. The lengths of 64.1% of the sequences in the 5.3-k unigene set were more than 500 bp, whereas in the unigenes generated by RNA-seq analysis, only 17.9% were longer than 500 bp. In general, EST analysis using Sanger sequencing generates longer sequence reads than RNA-seqs using a high-throughput Illumina GA IIx sequencer, so the difference in unigene length distribution is attributed to the difference in sequencing technique.
The data presented here are expected to become a useful gene resource for research aimed at understanding physiological processes important for tea cultivation and quality, such as nitrogen assimilation and amino acid metabolism. In Japan, large amounts of nitrogen fertilizers are used in tea plantations, causing pollution of groundwater, rivers and lakes. To improve this situation, it is important to develop tea cultivars with high nitrogen use efficiency (Tanaka and Taniguchi 2007). Therefore, we searched for unigenes related to nitrogen assimilation within the unigene set and found several that were homologous to genes related to nitrogen assimilation, such as glutamine synthetase, glutamate de-hydrogenase, ammonium transporter and nitrate transporter. In addition, the unigene set contains theanine synthase and several unigenes related to the metabolism of 2-oxoglutarate, a key component of the interaction of nitrogen and carbon metabolism.
In addition to nitrogen compounds such as amino acids, secondary metabolites such as catechins and caffeine are important for tea quality. Among our ESTs, we found several unigenes related to synthesis of these secondary metabolites. The metabolisms of nitrogen compounds and secondary metabolites are regulated by environmental status. For example, in young tea leaves, catechins increase under high light intensity (Saijo 1980). In contrast, shading of young tea leaves leads to an increase in total nitrogen content, as well as enhancement of theanine (Anan and Nakagawa 1974, Karasuyama and Matsumoto 1988). In the future, it will be important to decipher the mechanism of photoresponsive regulation of genes related to the metabolism of nitrogen compounds and secondary metabolites to enable improvement of these traits. Two unigenes related to photoresponse were found in our ESTs, providing us with tools to analyze the associated regulatory mechanisms.
Tea is well known as an aluminum-accumulating plant that grows well in very acidic soils containing high levels of Al3+; this is of interest because aluminum toxicity limits the growth of many other species in acidic soils (Morita et al. 2004, 2008) and the aluminum in the xylem sap of tea is complexed with citrate (Morita et al. 2004). Three unigenes potentially related to aluminum response were found in this study: one citrate synthetase and two aluminum-response proteins. Further analyses, such as expression analysis of the response of tea to aluminum, might reveal whether these genes have roles in aluminum resistance or response.
Using the EST data derived from seven different organs of the tea plant, digital northern analysis was performed to identify unigenes with different expression levels among different organs; 67 such unigenes were identified out of a sample of 144. Cluster analysis showed that the groups of unigenes highly expressed in each organ were related to different physiological functions. For example, several photosynthesis-related genes were highly expressed in the YL and ML libraries. Cluster III, which showed high expression in the RT library, was the largest cluster (25 unigenes), indicating that the physiological and developmental status of young root is considerably different from that of other organs. Interestingly, dihydroflavonol 4-reductase (DFR) was highly expressed in tap roots and lateral roots. Although catechins are not contained in tea roots (Forrest and Bendall 1969), leucoanthocyanidin, which is the product of DFR and the precursor of (+)-catechin, is contained in roots. Thus, we assume this DFR in roots to be involved not in catechin biosynthesis, but in other metabolic processes such as lignin or anthocyanin biosynthesis. One more unigene encoding DFR was found in the 5.3-k unigene set. This unigene was expressed in young stem, and the sequence similarity between the two DFRs was 52%. We think that the DFR from young stem is involved in catechin biosynthesis.
Ellis and Burke (2007) surveyed EST data from 33 species and showed that the proportion of unigenes containing SSRs was 2.5% to 21.1% (9.0% ± 0.1%, mean ± SEM). Based on this survey, the percentage of SSR-containing unigenes in this study (34.9%) is relatively high compared to that in other plant species.
The proportion of multi-locus markers in this study was higher than that reported by Sharma et al. (2009). We used a capillary sequencer for fragment analysis, whereas Sharma et al. (2009) used autoradiography of PAGE gels, which has lower resolution. Thus, the difference in the proportion of multi-locus markers might have been caused by the difference in the analysis method. Because of the paleopolyploidy of C. sinensis (Shi et al. 2010), it is not surprising that many multi-locus markers are contained in the set of EST-SSRs reported here.
The 16 accessions used in this study include major tea cultivars in Japan, parental cultivars and several foreign germplasms. These materials are representative of the genetic diversity of breeding materials in Japan. The EST-SSRs developed in this study were highly polymorphic among the 16 accessions. They should be very useful for many genetic studies in tea, such as construction of linkage maps, analysis of genetic diversity and cultivar identification. For example, using 377 EST-SSRs and other co-dominant markers, we recently constructed a reference linkage map of tea (Taniguchi et al. 2012).
Most of the EST-SSR markers developed here were applicable to other Camellia species. Species of Camellia other than C. sinensis contain useful traits that have been utilized in tea breeding; for instance, a parental line containing a high level of anthocyanin (Ogino et al. 2005) and a caffeineless tea plant (Ogino et al. 2009) were developed from interspecific crosses. EST-SSR markers will enable genetic analysis of important agronomic traits of various Camellia species, thus expanding the usefulness of these species in tea breeding.
In conclusion, the tea ESTs obtained in this study are valuable resources for analysis of gene function and for development of SSR markers. The 5.3-k tea unigene set contains novel transcripts from tea, and 67 out of 144 unigenes tested showed specific expression patterns among a set of seven organs. The SSR markers developed in this study are highly polymorphic in C. sinensis and many other Camellia species. Further studies using the tea EST dataset are expected to accelerate functional genomics and genetic breeding research in tea.
Supplementary Material
Acknowledgments
We would like to thank N. Rokutan and M. Iwata for their technical assistance. This work was supported by NARO Research Project No. 211, ‘Establishment of Integrated Basis for Development and Application of Advanced Tools for DNA Marker-Assisted Selection in Horticultural Crops.’
Literature Cited
- Anan T., Nakagawa M. (1974) Effect of light on chemical constituents in the tea leaves. Nippon Nogeikagaku Kaishi 48: 91–98 [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle E.D. (1991) Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218 [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D, Butler H, Cherry J.M., Davis A.P., Dolinski K, Dwight S.S., Eppig J.T., et al. (2000) Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher R. (2007) EST-derived microsatellites as a rich source of molecular markers for oats. Plant Breed. 126 : 274–278 [Google Scholar]
- Chagne D., Gasic K., Crowhurst R.N., Han Y., Bassett H.C., Bowatte D.R., Lawrence T.J., Rikkerink E.H., Gardiner S.E., Korban S.S. (2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92: 353–358 [DOI] [PubMed] [Google Scholar]
- Chen L., Zhao L.P., Gao Q.K. (2005) Generation and analysis of expressed sequence tags from the tender shoots cDNA library of tea plant (Camellia sinensis). Plant Sci. 168: 359–363 [Google Scholar]
- Choi I.Y., Hyten D.L., Matukumalli L.K., Song Q., Chaky J.M., Quigley C.V., Chase K., Lark K.G., Reiter R.S., Yoon M.S., et al. (2007) A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics 176: 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J., Terol J., Talón M., Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Deleu W., Esteras C, Roig C, Gonzalez-To M, Fernandez-Silva I, Gonzalez-Ibeas D, Blanca J, Aranda M.A., Arus P, Nuez F, et al. (2009) A set of EST-SNPs for map saturation and cultivar identification in melon. BMC Plant Biol. 9: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.R., Burke J.M. (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99: 125–132 [DOI] [PubMed] [Google Scholar]
- Forrest G.I., Bendall D.S. (1969) The distribution of polyphenols in the tea plant (Camellia sinensis L.). Biochem. J. 113: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka H., Nunome T., Minamiyama Y., Kono I., Namiki N., Kojima A. (2005) Read2Marker: a data processing tool for micro-satellite marker development from a large data set. Biotechniques 39: 472–476 [DOI] [PubMed] [Google Scholar]
- Gish W, States D.J. (1993) Identification of protein coding regions by database similarity search. Nature Genet. 3: 266–272 [DOI] [PubMed] [Google Scholar]
- Gupta S., Prasad M. (2009) Development and characterization of genic SSR markers in Medicago truncatula and their transferability in leguminous and non-leguminous species. Genome 52: 761–771 [DOI] [PubMed] [Google Scholar]
- Hanai L., deCampos T., Camargo L., Benchimol L., de Souza A., Melotto M., Carbonell S., Chioratto A., Consoli L., Formighieri E., et al. (2007) Development, characterization, and comparative analysis of polymorphism at common bean SSR loci isolated from genic and genomic sources. Genome 50: 266–277 [DOI] [PubMed] [Google Scholar]
- Heesacker A., Kishore V, Gao W, Tang S, Kolkman J, Gingle A, Matvienko M, Kozik A, Michelmore R, Lai Z, et al. (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor. Appl. Genet. 117: 1021–1029 [DOI] [PubMed] [Google Scholar]
- Inazuka M., Tahira T, Hayashi K. (1996) One-tube post-PCR fluorescent labeling of DNA fragments. Genome Res. 6: 551–557 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Karasuyama M, Matsumoto J. (1988) Effect of shade treatment on the utilization of absorbed nitrogen in tea plants. Jpn. J. Soil Sci. 59: 486–492 [Google Scholar]
- Laurent V., Devaux P, Thiel T, Viard F, Mielordt S, Touzet P, Quillet M.C. (2007) Comparative effectiveness of sugar beet micro-satellite markers isolated from genomic libraries and GenBank ESTs to map the sugar beet genome. Theor. Appl. Genet. 6: 793–805 [DOI] [PubMed] [Google Scholar]
- Lijavetzky D., Cabezas J.A., Ibanez A., Rodriguez V., Martinez-Zapater J.M. (2007) High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genomics 8: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Muse S.V. (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129 [DOI] [PubMed] [Google Scholar]
- Morita A., Horie H., Fujii Y., Takatsu N., Watanabe N., Yagi A., Yokota H. (2004) Chemical forms of aluminum in xylem sap of tea plants (Camellia sinensis L.). Phytochemistry 65: 2775–2780 [DOI] [PubMed] [Google Scholar]
- Morita A., Yanagisawa O., Takatsu S., Maeda S., Hiradate S. (2008) Mechanism for the detoxification of aluminum in roots of tea plant (Camellia sinensis (L.) Kuntze). Phytochemistry 69: 147–153 [DOI] [PubMed] [Google Scholar]
- Ogino A., Tanaka J, Yoshida K, Taniguchi F, Omae H, Nesumi A, Saba T, Takyu T, Takeda Y. (2005) New parental line ‘Cha Chuukanbohon Nou 6’ for anthocyanin-rich tea. Bull. Natl. Inst. Veg. Tea Sci. 4: 77–85 [Google Scholar]
- Ogino A., Tanaka J, Taniguchi F, Yamamoto M.P., Yamada K. (2009) Detection and characterization of caffeine-less tea plants originated from interspecific hybridization. Breed. Sci. 59: 277–283 [Google Scholar]
- Park J., Kim J, Hahn B, Kim K, Ha S, Kim Y. (2004) EST analysis of genes involved in secondary metabolism in Camellia sinensis (tea), using suppression subtractive hybridization. Plant Sci. 166: 953–961 [Google Scholar]
- Romualdi C., Bortoluzzi S., D’Alessi F., Danieli G.A. (2003) IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol. Genomics 12: 159–162 [DOI] [PubMed] [Google Scholar]
- Saijo R. (1980) Effect of shade treatment on biosynthesis of catechins in tea plants. Plant Cell Physiol. 21: 989–998 [Google Scholar]
- Sato K., Nankaku N., Takeda K. (2009) A high-density transcript linkage map of barley derived from a single population. Heredity 103: 110–117 [DOI] [PubMed] [Google Scholar]
- Sharma P, Kumar S. (2005) Differential display-mediated identification of three drought-responsive expressed sequence tags in tea [Camellia sinensis (L.) O. Kuntze]. J. Biosci. 30: 231–235 [DOI] [PubMed] [Google Scholar]
- Sharma R., Bhardwaj P, Negi R, Mohapatra T, Ahuja P. (2009) Identification, characterization and utilization of unigene derived microsatellite markers in tea (Camellia sinensis L.). BMC Plant Biol. 9: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., Huang H, Barker M.S. (2010) Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Ann. Bot. 106: 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.Y., Yang H., Wei C.L., Yu O., Zhang Z.Z., Jiang C.J., Sun J., Li Y.Y., Chen Q., Xia T., et al. (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J., Taniguchi F, Hirai N, Yamaguchi S. (2006) Estimation of the genome size of tea (Camellia sinensis), camellia (C. japonica), and their interspecific hybrids by flow cytometry. Tea Res. J. 101: 1–7 [Google Scholar]
- Tanaka J, Taniguchi F. (2007) Tea. In: Kole C. (ed.) Genome Mapping and Molecular Breeding in Plants, vol. 6 Technical Crops, Springer-Verlag, Berlin Heidelberg, pp. 119–125 [Google Scholar]
- Taniguchi F., Furukawa K, Ota Metoku S, Yamaguchi N, Ujihara T, Kono I, Fukuoka H, Tanaka J. (2012) Construction of high-density reference linkage map of tea (Camellia sinensis). Breed. Sci. 62 (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.