Abstract
Novel mutant alleles of an ethylene receptor Solanum lycopersicum ETHYLENE RESPONSE1 (SlETR1) gene, Sletr1-1 and Sletr1-2, were isolated from the Micro-Tom mutant library by TILLING in our previous study. They displayed different levels of impaired fruit ripening phenotype, suggesting that these alleles could be a valuable breeding material for improving shelf life of tomato fruit. To conduct practical use of the Sletr1 alleles in tomato breeding, genetic complementation analysis by transformation of genes carrying each allele is required. In this study, we generated and characterized transgenic lines over-expressing Sletr1-1 and Sletr1-2. All transgenic lines displayed ethylene insensitive phenotype and ripening inhibition, indicating that Sletr1-1 and Sletr1-2 associate with the ethylene insensitive phenotype. The level of ethylene sensitivity in the seedling was different between Sletr1-1 and Sletr1-2 transgenic lines, whereas no apparent difference was observed in fruit ripening phenotype. These results suggested that it is difficult to fine-tune the extent of ripening by transgenic approach even if the weaker allele (Sletr1-2) was used. Our present and previous studies indicate that the Micro-Tom mutant library combined with TILLING could be an efficient tool for exploring genetic variations of important agronomic traits in tomato breeding.
Keywords: ethylene receptor, tomato fruit shelf-life, Micro-Tom, molecular breeding, TILLING, transgenic tomato
Introduction
Tomato (Solanum lycopersicum) is an important vegetable crop in the world and is a significant source of phytonutrients and micronutrients such as vitamins, minerals, fiber and other beneficial compounds for human diet. It is also a central model for studying fruit biology including fruit development and ripening, softening, as well as fruit metabolism (Brummell and Harpster 2001, Carrari and Fernie 2006, Giovannoni 2004). With the progress of genome sequencing by the International Solanaceae Genomics Project (SOL), a great number of tomato gene sequences could be retrieved from the databases (Mueller et al. 2005a, 2005b, 2009).
Fruit ripening and softening are major factors contributing to perishability of fleshy or climacteric fruits (e.g. tomato, banana, mango and avocado) (Bapat et al. 2010). The attribute of ripening has both positive and negative aspects from the standpoint of agriculture. Among the positive aspects, ripening results in desirable flavor, color and texture. In contrast, crop loss results from negative ripening characteristics, such as ripening-associated increase in fruit pathogen susceptibility. In addition, over-ripening causes excessive softening, changes in taste, aroma and skin color. These are unavoidable processes bringing significant losses to both farmers and consumers (Bapat et al. 2010, Giovannoni 2001).
Targeting Induced Local Lesions In Genomes (TILLING) has been known as a high-throughput reverse genetic approach which allows for the identification of allelic series of mutants with a range of modified functions for a desired gene (Colbert et al. 2001, Comai and Henikoff 2006, McCallum et al. 2000a, 2000b). To efficiently obtain a broad range of variation in desired agronomic traits from a mutant population is important for crop improvement in mutation breeding. Hence, TILLING is considered as a useful method for accelerating conventional mutation breeding. Previous studies have demonstrated the availability of TILLING for crop improvement in several plant species (Kurowska et al. 2011). For example, novel waxy mutant alleles in wheat showed altered amylose content (Slade et al. 2005) and GmFAD2-1b mutant alleles in soybean resulted in the elevated content of oleic acid in the seed oil (Hoshino et al. 2010). Also in tomato, the SleIF4E1 G1485A mutant showed potyvirus resistance (Piron et al. 2010) and in melon, namely the CmACO1 G194D mutant produced fruit with an enhanced shelf life (Dahmani-Mardas et al. 2010). Our previous study also provided evidence that TILLING approach is an effective tool for isolating mutants of interest (Okabe et al. 2011). We isolated novel mutant alleles of an ethylene receptor Solanum lycopersicum ETHYLENE RESPONSE1 (SlETR1) gene, Sletr1-1 and Sletr1-2, from the Micro-Tom EMS mutant library by TILLING. They displayed ethylene-insensitive phenotypes. The Sletr1-1 allele exhibited strong ethylene insensitivity, which resulted in orange mature fruit, while the Sletr1-2 allele exhibited moderate ethylene insensitivity, which resulted in red mature fruit with increased shelf-life compared to the wild-type fruit (Okabe et al. 2011).
Currently, Micro-Tom has been focused as a model variety for accelerating functional genomic research in tomato for its characteristic features. Micro-Tom possesses several attractive features as a research material, such as its small plant size (15–20 cm), short life cycle (70–90 days) which enables to produce three or four generations during a year (Matsukura et al. 2008), and technical platforms and associated information i.e., highly efficient transformation protocols, a TILLING platform, a mutant database, full-length cDNA and EST databases, BAC libraries and DNA markers (Aoki et al. 2010, Okabe et al. 2011, Saito et al. 2011, Shirasawa et al. 2010, Sun et al. 2006). Such advantages of Micro-Tom could be applied to mutation breeding of tomato. Indeed, recent works have revealed that Micro-Tom can be used for studying fruit ripening, fruit set and sugar metabolism, that are major important traits in tomato (Okabe et al. 2011, Serrani et al. 2008, Yin et al. 2010). Thus, Micro-Tom is regarded as a beneficial material to conduct mutation breeding.
To date, several ripening mutants have been identified in tomato, such as ripening-inhibitor (rin), Colorless non-ripening (Cnr), non-ripening (nor), Green-ripe (Gr) and Never-ripe (Nr), which are spontaneous variants selected from production fields or breeding programs. The causative genes were isolated and shown to act as upstream regulators of the ethylene signaling network or in ethylene perception (Barry et al. 2005, Barry and Giovannoni 2007, Giovannoni 2004, Manning et al. 2006, Vrebalov et al. 2002, Wilkinson et al. 1995). Among these mutants, only rin has been used for improving the shelf life of tomato fruit as demonstrated in a previous study that F1 hybrid lines of rin exhibited prolonged fruit shelf life and these lines were practically used as a breeding material (Kitagawa et al. 2005). Fruit shelf life is one of the important agronomic traits in tomato, since improved shelf life provides commercial and industrial values in various aspects (e.g. fruit harvest, shipping and quality retention). These findings clearly indicate that an effective way to repress deterioration of postharvest fruit is to control the balance of ethylene-mediated regulation in fruit ripening. Along with this viewpoint, we have been focusing on the availability of the Sletr1 alleles as a potential breeding material for increasing shelf life of post-harvest tomato fruit. To determine the causal gene, genetic complementation analysis is required since EMS randomly induces point mutations throughout the whole genome in addition to the responsible gene. Confirming the association between identified mutation and expected phenotype is essential before the practical use of Sletr1 alleles in the tomato breeding.
In this study, we generated and characterized transgenic tomato lines over-expressing mutant versions of a tomato ethylene receptor gene SlETR1, Sletr1-1 and Sletr1-2, to confirm whether Sletr1-1 and Sletr1-2 actually confer reduced ethylene sensitivity. In addition, the availability of Micro-Tom mutant library with use of TILLING in mutation breeding is discussed by comparing the transgenic tomato lines and Sletr1 mutant alleles with the objective of controlling fruit ripening.
Materials and Methods
Plant material and transformation
Tomato (Solanum lycopersicum) cv. Micro-Tom was used for transformation. Full-length coding region of Sletr1-1 and Sletr1-2 were cloned into the pENTR vector (Invitrogen), each has an amino acid substitution in the predicted transmembrane region of SlETR1, c152t (P51L) for Sletr1-1 and t206a (V69D) for Sletr1-2. The each coding region was introduced into the binary vector pBI-OX-GW (Inplanta Innovations Inc.) for subsequent tomato transformation by Agrobacterium tumefaciens GV2260. Tomato transformation was performed using the highly efficient protocol established by Sun et al. (2006). Sletr1-1 and Sletr1-2 mutant alleles were used for characterizing fruit ripening phenotype in transgenic lines as comparisons.
Seedling ethylene triple response assay
Seeds were sterilized with 10% commercial bleach including a detergent (Kitchen Haiter, Kao, Tokyo, Japan) for 20 min and then rinsed with sterilized water three times for 5 min each. The seeds were germinated in a 50-ml glass bottle containing 10 ml of 1/2 MS medium (Murashige and Skoog 1962). Ethylene was added to the bottles sealed with silicon rubber at designed concentrations (10 ppm) and seedlings were grown for 5 days in the dark at 25°C.
Characterization of phenotypes in transgenic lines
The date of initiation of fruit coloration (breaker stage) was tagged for the evaluation of fruit ripening phenotype in wild type, Sletr1-1, Sletr1-2 and T0 transgenic lines. Seeds of wild-type Micro-Tom and T1 seeds were sown on a wet filter paper and placed for two to three days at 25°C to stimulate seed germination and then germinated seeds were transplanted into soil and grown under a photoperiod of 16 h light at 25°C. The date of flowering was tagged for the time course observation of fruit ripening and evaluation of fruit shelf life.
Southern blotting of T0 generation of transgenic tomato lines
To confirm the copy number of transgene in transgenic tomato plants expressing Sletr1-1 and Sletr1-2, leaves from each transgenic line were collected in 2-ml micro test tubes, frozen in liquid nitrogen, and homogenized with a pestle. Genomic DNA was extracted using a Maxwell16 Tissue DNA Purification Kit (Promega, USA). Extracted genomic DNA (10 μg) was digested with Hind III, electrophoresed on an 0.8% agarose gel at 50 V for 3 hr and transferred to a Hybond-N+ nylon membrane (GE Healthcare, UK). The membrane was hybridized overnight at 60°C in high-SDS buffer [50% deionized formamide (v/v), 5× SSC, 7% SDS, 2% blocking reagent (Roche Diagnostics, Mannheim, Germany)], 50 mM sodium phosphate (pH 7.0) and 0.1% N-lauroylsarcosine sodium salt (w/v) containing a NPT II-specific DIG-labeled probe at 45°C. The hybridization signals were detected using an LAS4000 mini Image Analyzer (Fujifilm Co. Ltd., Tokyo, Japan).
RNA extraction and RT-PCR
Total RNA was extracted from leaves using RNeasy plant mini kit (Qiagen). cDNA was synthesized from 1 μg of total RNA using Superscript VILO cDNA Synthesis Kit (Invitrogen). The cDNA was ten times-diluted with sterile water and 1 μl of diluted cDNA was used for RT-PCR analysis in a 20 μl reaction volume. cDNA was mixed with 2 μl 10 × Ex-taq buffer, 1.6 μl 2.5 mM dNTP, 0.4 μl 10 μM primers and 0.1 μl Ex-taq Hotstart version (5 U/μl, Takara) using SlETR1-speicific primers and Actin primers as the internal control. PCR amplification for the detection of each gene was performed with following primer pairs; SlETR1 forward (5′-ATGGGATCTCTTCTCCGGATG-3′) and SlETR1 reverse (5′-CACCAGTGCAGTCAAGGC-3′), Actin forward (5′-GATGGATCCTCCAATCCAGACACT GTA-3′) and Actin reverse (5′-GTATTGTGTTGGACTCT GGTGATGGTGT-3′). The PCR program was consisted of first denature step for 2 min at 95°C; followed by 26 cycles of 30 seconds at 95°C, 30 second at 57°C and 40 seconds at 72°C; with final extension for 5 min at 72°C. Then, 10 μμl of PCR products were subjected to agarose gel electrophoresis and visualized by 1.5% and 2.0% agaroge gels containing SYBR Safe (Invitrogen) for Actin and SlETR1, respectively.
Results
Generation of transgenic Micro-Tom lines over-expressing mutant versions of a ethylene receptor gene SlETR1, Sletr1-1 and Sletr1-2
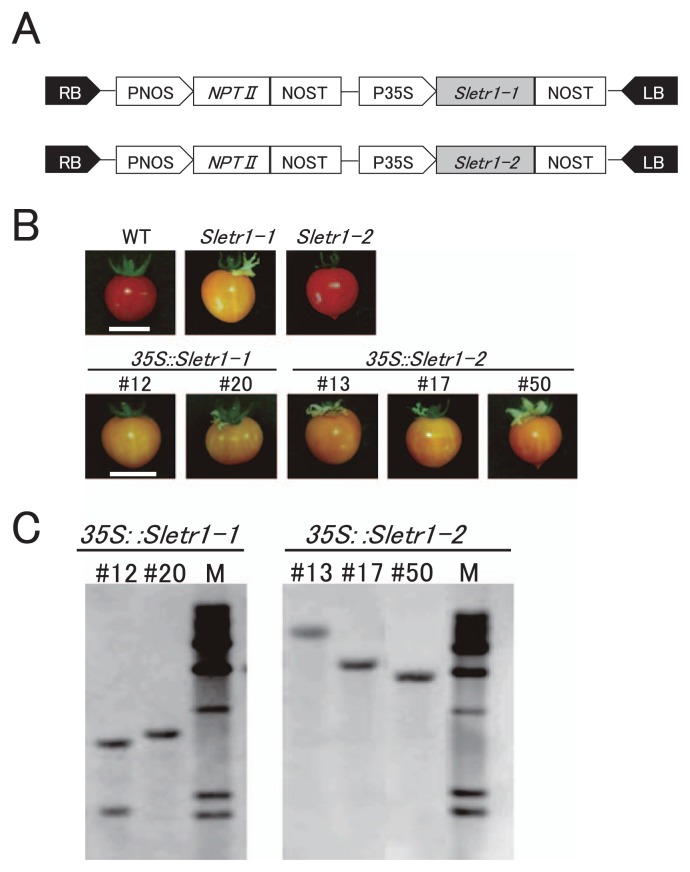
Mutant versions of SlETR1, full-length coding sequence of Sletr1-1 and Sletr1-2 were transformed into tomato cv. Micro-Tom via Agrobacterium-mediated transformation (Fig. 1A). Diploid T0 transgenic plants were subjected to evaluation of fruit ripening phenotype and genomic Sourthern blot analysis. Finally, two lines (#12 and #20) for the 35S::Sletr1-1 construct and 3 lines (#13, #17 and #50) for the 35S::Sletr1-2 construct showing delayed fruit ripening phenotype and carrying two copies or single copy of transgene(s) were selected (Fig. 1B, 1C). The seeds from T0 transgenic lines were collected for subsequent seedling triple response assay and phenotype characterization in the T1 generation.
Fig. 1.
Southern blot analysis and comparison of fruit-ripening phenotype in the T0 generation. (A) T-DNA region of pBI-OX-GW used for the transformation. RB and LB, right and left borders of T-DNA; PNOS, nopaline synthase gene promoter; NPT II, neomycin phospho-transferase gene; NOST, nopaline synthase gene terminator; P35S, cauliflower mosaic virus 35S promoter; Sletr1-1 and Sletr1-2, coding region of mutated tomato ethylene receptor gene SlETR1/P51L and SlETR1/V69D. (B) Fruit ripening phenotype of T0 transgenic lines. Fruits were harvested at breaker plus 14 days. Fruits of wild type and Sletr1-1, Sletr1-2 mutants are shown as comparisons. (C) The result of Southern blot analysis in T0 transgenic lines. Transgenes were detected by the NPT II probe.
Ethylene response of seedlings in the T1 generation of transgenic lines
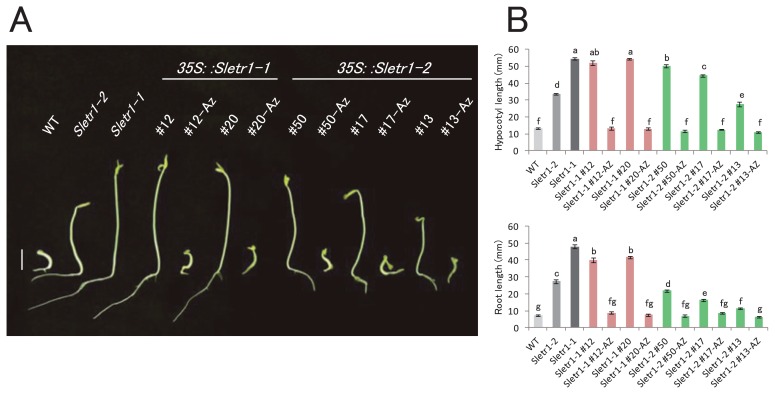
Ethylene response of seedlings was tested in the T1 transgenic lines over-expressing Sletr1-1 and Sletr1-2, to confirm whether they show expected ethylene insensitivity. T1 transgenic seedlings of Sletr1-1 and Sletr1-2 and mutants carrying either of the Sletr1 alleles exhibited ethylene insensitive phenotypes, whereas wild-type and azygous seedlings became swollen, and hypocotyl and root elongation were inhibited in response to the ethylene treatment (Fig. 2A). With respect to the insensitivity level, hypocotyl and root length of Sletr1-1 transgenic lines (#12, #20) and the Sletr1-1 mutant was similar. In contrast, each Sletr1-2 transgenic line (#13, #17, #50) showed various hypocotyl and root length (Fig. 2B). Compared to Sletr1-1 transgenic lines, all the Sletr1-2 transgenic lines exhibited shorter root length (Fig. 2). These tendencies of hypocotyls and root length in the transgenic lines appear to correlate with the level of ethylene sensitivity in the Sletr1-2 allele. These results indicate that transgenes of mutated ethylene receptor confer ethylene insensitivity in the seedlings.
Fig. 2.
The ethylene response of transgenic lines in the T1 generation. (A) Triple response phenotype of the seedlings of wild type (WT), Sletr1-1, Sletr1-2 and transgenic lines. Seeds were surface sterilized and sown on 1/2 MS medium in the presence of 10 ppm C2H4 and incubated at 25°C in the dark for five days. Bar = 1 cm. (B) Quantification of ethylene-induced inhibition of root and hypocotyl growth. At least twenty seedlings of wild type, Sletr1-1, Sletr1-2 and T1 transgenic lines were measured, except for each azygous line (n > 3). Az represents the azygous plant of each transgenic line. Different letters between lines represent significant difference at P < 0.05, as determined by the Tukey-Kramer test. Vertical bars represent SE.
Over-expression of mutated ethylene receptor genes confers delayed fruit ripening and prolonged fruit shelf-life phenotype
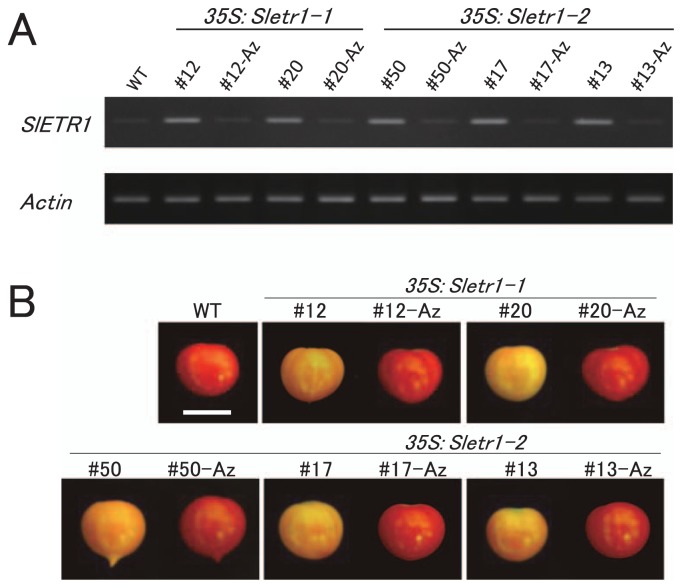
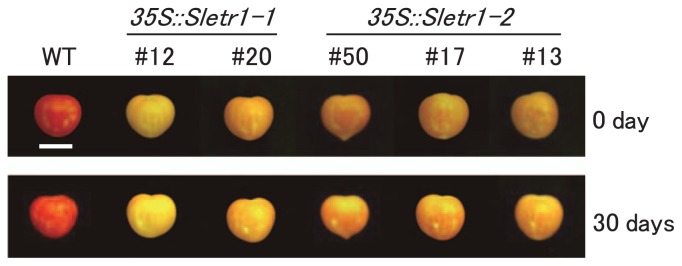
To confirm the expression level of mutated ethylene receptor genes, mRNA expression was investigated in each T1 transgenic line and its azygous line and wild type. Presence of the transgene in the T1 plants was confirmed by genomic PCR prior to this experiment. SlETR1 expression in the leaves was detected by semi-quantitative RT-PCR. SlETR1 expression was increased in all transgenic lines compared to the wild type and each azygous line. Similar level of increased expression was observed in all transgenic lines. The expression levels in wild type and azygous lines were similarly low (Fig. 3A). T1 progenies derived from two or three independent transgenic lines showed the association of the transgene with Sletr1-1 or Sletr1-2 over-expression, thus a link between the transgene and the delayed ripening phenotype was clearly demonstrated (Fig. 3A, 3B). We have previously documented about delayed fruit ripening associated with the Sletr1 mutant alleles (Okabe et al. 2011). In support of this observation, several transgenic lines displayed delayed fruit ripening phenotype (Fig. 3B). Contrary to our expectation, although the Sletr1-2 allele showed nearly normal ripening in our previous work as shown in Fig. 1C, Sletr1-2 transgenic lines exhibited delayed fruit ripening (Fig. 3B). To better evaluate the fruit phenotype, phenotype of post-harvest fruits was investigated in the transgenic lines. The wild-type fruits wilted at 30 days after harvest, whereas the fruit surface of transgenic lines remained intact (Fig. 4). Consistent with the phenotype of fruit ripening, the fruit shelf life in transgenic lines was apparently prolonged compared to wild type. These results suggest that the weaker type of mutated ethylene receptor is sufficient for conferring reduced ethylene sensitivity in fruits.
Fig. 3.
Expression analysis of SlETR1 gene and comparison of fruit ripening phenotype in T1 transgenic lines. (A) SlETR1 expression in leaves was investigated using semi-quantitative RT-PCR. Common region in the native SlETR1 and transgenes were amplified. Actin was used as an internal standard. Az represents the azygous plant of each transgenic line. (B) Fruit ripening phenotype of T1 transgenic lines. Fruits were harvested at 50 days after flowering. At least 3 fruits were evaluated in each line. Bar = 2 cm.
Fig. 4.
Shelf life of postharvest fruits in T1 transgenic lines. Fruits were harvested at 50 days after flowering and stored at 25°C, 55–70% humidity for 30 days in the growth chamber. The appearance of T1 transgenic fruits was compared with that of wild type at 30 days after harvest. At least 3 fruits were evaluated in each line. Bar = 2 cm.
Discussion
We previously reported about the identification of novel Sletr1 alleles (Sletr1-1 and Sletr1-2) showing ethylene insensitivity and impaired fruit ripening phenotype at different levels. Segregation analysis in the F2 populations suggested an association between these alleles and ethylene insensitive phenotype (Okabe et al. 2011). In this study, to confirm whether Sletr1 mutations confer reduced ethylene sensitivity and to discuss the availability of Micro-Tom mutant library combined with TILLING in mutation breeding of tomato, we characterized transgenic tomato lines over-expressing mutant versions of SlETR1, Sletr1-1 and Sletr1-2.
In the seedling ethylene triple response analysis, both of T1 transgenic lines showed different levels of ethylene insensitive phenotype depending on the transgene type. No significant difference in the hypocotyl length was observed in the Sletr1-1 transgenic lines compared to the Sletr1-1 mutant line. Although the root length of the Sletr1-1 transgenic lines showed significant difference, the values were close to that of the Sletr1-1 mutant line. This phenotype is similar to what had been observed in transgenic tomato plants expressing Nr (Wilkinson et al. 1995). In contrast to the Sletr1-1 transgenic lines, various levels of reduced ethylene sensitivity were observed in the Sletr1-2 transgenic lines (Fig. 2). This observation is likely to correspond to the ethylene insensitivity level of the two Sletr1 alleles, since the expression level of the transgene was similar in all tested transgenic lines as shown in Fig. 3A. These results indicate that Sletr1-1 confers nearly complete ethylene insensitive phenotype, whereas Sletr1-2 confers reduced ethylene sensitive phenotype.
Regarding fruit phenotypes of the transgenic lines, although we initially expected that the Sletr1-2 transgenic lines would display weaker ripening impairment compared to the Sletr1-1 transgenic lines, Sletr1-2 transgenic lines were similarly impaired in fruit ripening and shelf life (Fig. 3B, Fig. 4). These phenotypes resembled that of transgenic tomato plants expressing Arabidopsis etr1-1 (Wilkinson et al. 1997). It is suggested that the enhancement of ethylene insensitivity affected differently to various organs (i.e. fruit, hypocotyl and root), thus resulted in nonuniform ethylene sensitivity. It is also speculated that the fruit ripening process is more sensitive to ethylene, since the enhanced expression of the weaker allele (Sletr1-2) resulted in the fully inhibition of ripening. Furthermore, one possible explanation for the difference of fruit ripening phenotype between Sletr1-2 mutant and Sletr1-2 transgenic lines is that fruit ripening inhibition in transgenic tomato fruits may depend on the Sletr1 expression level, because SlETR1 is expressed in all tissues including fruit at a constant level (Kevany et al. 2007, Lashbrook et al. 1998).
So far, it has been reported that genetic engineering of ethylene-mediated biological regulation including ethylene biosynthesis, perception and signaling is available for manipulating fruit ripening. Suppression of ethylene biosynthesis genes in previous studies have revealed that antisense transgenic lines of tomato ACC gene had resulted in decreased ethylene production in fruits and impaired fruit ripening (Oeller et al. 1991), RNAi transgenic lines of ACC oxidase (ACO) gene, which is a rate-limiting enzyme of ethylene biosynthesis had resulted in decreased ethylene production, delayed fruit ripening, and prolonged fruit shelf life (Xiong et al. 2005). Similarly, with respect to ethylene perception and signaling, transgenic tomato lines constitutively expressing Arabidopsis etr1-1 exhibited reduced ethylene sensitivity and delayed fruit ripening (Wilkinson et al. 1997), fruit specific suppression of SlETR4 gene using the E8-promoter resulted in early ripening phenotype (Kevany et al. 2008), antisense transgenic lines of ethylene response factor 1 (ERF1) gene showed longer fruit shelf life (Li et al. 2007). However in many cases, these transgenic plants displayed excessive ripening inhibition or undesired developmental effect by the ectopic expression or global suppression of transgenes under the CaMV 35S promoter. Our data also showed that ripening inhibition occurred even in the Sletr1-2 transgenic lines (Fig. 3B). From the standpoint of breeding, a material showing severe ripening inhibition is not suited for practical use. A case study was successful in generating transgenic tomato plants with altered ripening but not an agronomic penalty by using a fruit-specific E8-promoter (Kevany et al. 2008). Another study demonstrated that transgenic tomato plants expressing Arabidopsis mutant ethylene receptor etr1-1 using an inducible promoter conferred reduced ethylene sensitivity depending on the concentration of inducer (Gallie 2010). Utilization of such approach is not widespread due to the limited availability of appropriate or flexible promoters that can induce the expression of transgene at expected level. Therefore, production of the numerous variations in target traits by transgenic approach could be realistically difficult.
Contrary to genetic engineering of agronomic traits by transgenic approach, TILLING allows obtaining a broad range of variants from EMS mutagenized population. As a beneficial property of EMS mutagenesis, the resulting population theoretically includes various levels of mutants, such as null and leaky mutant of a target gene. It is also considered that TILLING approach in mutation breeding is more efficient as a strategy compared to the transgenic approach in which selecting different kinds of promoter is necessary to create variations in targeted agronomic trait. We previously identified the Sletr1-2 allele by TILLING as a potential breeding material for improving the fruit shelf life without agronomic penalties such as excessive ripening inhibition and apparent loss of color in ripen fruit (Okabe et al. 2011). We consider that if we did not perform TILLING, the Sletr1-2 allele probably would not have been selected from the Micro-Tom mutant population, since the fruit ripening phenotype of Sletr1-2 plant is not easily distinguished from wild type when the mutant populations are grown in large scale.
Furthermore, to manage the field trials, the phenotype screening of 10,000 tomato M2 families consisting of twenty individuals per family in common tomato cultivar would require approximately 20 hectares of field space, i.e., one individual/m2 (Giovannoni 2007). A mutant screen in such scale would not be realistic for majority of laboratories. In contrast, the plant size of Micro-Tom enables growing in limited spaces. Meissner et al. (1997) showed that Micro-Tom can be grown at high density, up to 1,357 individuals/m2. Thus, the combination of Micro-Tom mutant library and TILLING permitted to efficiently isolate a valuable material, such Sletr1-2. This approach could be applied to other important agronomic traits.
Acknowledgments
The Micro-Tom seeds used in this research were provided by the National BioResource Project (NBRP), MEXT, JAPAN. This work was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Research for the Future Program of the Japan Society for the Promotion of Science (JSPS, No. 21580004) to H. E. and Adaptable and Seamless Technology Transfer Program through target-driven R&D (A-STEP) from the Japan Science and Technology Agency (No. AS231Z04705E).
Literature Cited
- Aoki K., Yano K., Suzuki A., Kawamura S., Sakurai N., Suda K., Kurabayashi A., Suzuki T., Tsugane T., Watanabe M., et al. (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat V.A., Trivedi P.K., Ghosh A, Sane V.A., Ganapathi T.R., Nath P. (2010) Ripening of fleshy fruit: molecular insight and the role of ethylene. Biotechnol. Adv. 28: 94–107 [DOI] [PubMed] [Google Scholar]
- Barry C.S., Mcquinn R.P., Thompson A.J., Seymour G.B., Grierson D, Giovannoni J.J. (2005) Ethylene insensitivity conferred by the Green-ripe and Never-ripe 2 ripening mutants of tomato. Plant Physiol. 138: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., Giovannoni J.J. (2007) Ethylene and fruit ripening. J. Plant Growth Regul. 26: 143–159 [Google Scholar]
- Brummell D.A., Harpster M.H. (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol. Biol. 47: 311–340 [PubMed] [Google Scholar]
- Carrari F, Fernie A.R. (2006) Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Colbert T., Till B.J., Tompa R, Reynolds S, Steine M.N., Yeung A.T., McCallum C.M., Comai L, Henikoff S. (2001) High-throughput screening for induced point mutations. Plant Physiol. 126: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Henikoff S. (2006) TILLING: practical single-nucleotide mutation discovery. Plant J. 45: 684–694 [DOI] [PubMed] [Google Scholar]
- Dahmani-Mardas F., Troadec C., Boualem A., Lévêque S., Alsadon A.A., Aldoss A.A., Dogimont C., Bendahmane A. (2010) Engineering melon plants with improved fruit shelf life using the TILLING approach. PloS One 5: e15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D.R. (2010) Regulated ethylene insensitivity through the inducible expression of the Arabidopsis etr1-1 mutant ethylene receptor in tomato. Plant Physiol. 152: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. (2001) MOLECULAR BIOLOGY OF FRUIT MATURATION AND RIPENING. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Giovannoni J.J. (2004) Genetic regulation of fruit development and ripening. Plant Cell 16: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J.J. (2007) Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Hoshino T., Takagi Y, Anai T. (2010) Novel GmFAD2-1b mutant alleles created by reverse genetics induce marked elevation of oleic acid content in soybean seeds in combination with GmFAD2-1a mutant alleles. Breed. Sci. 60: 419–425 [Google Scholar]
- Kevany B.M., Tieman D.M., Taylor M.G., Cin V.D., Klee H.J. (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51: 458–467 [DOI] [PubMed] [Google Scholar]
- Kevany B.M., Taylor M.G., Klee H.J. (2008) Fruit-specific suppression of the ethylene receptor LeETR4 results in early-ripening tomato fruit. Plant Biotechnol. J. 6: 295–300 [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Ito H, Shiina T, Nakamura N, Inakuma T, Kasumi T, Ishiguro Y, Yabe K, Ito Y. (2005) Characterization of tomato fruit ripening and analysis of gene expression in F1 hybrids of the ripening inhibitor (rin) mutant. Physiol. Plant. 123: 331–338 [Google Scholar]
- Kurowska M., Daszkowska-Golec A, Gruszka D, Marzec M, Szurman M, Szarejko I, Maluszynski M. (2011) TILLING: a shortcut in functional genomics. J. Appl. Genet. 52: 371–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook C.C., Tieman D.M., Klee H.J. (1998) Differential regulation of the tomato ETR gene family throughout plant development. Plant J. 15: 243–252 [DOI] [PubMed] [Google Scholar]
- Li Y., Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y. (2007) LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 26: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Manning K., Tör M, Poole M, Hong Y, Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Matsukura C., Aoki K., Fukuda N., Mizoguchi T., Asamizu E., Saito T., Shibata D., Ezura H. (2008) Comprehensive resources for tomato functional genomics based on the miniature model tomato Micro-Tom. Curr. Genomics 9: 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum C.M., Comai L, Greene E.A., Henikoff S. (2000a) Targeted screening for induced mutations. Nat. Biotechnol. 18: 455–457 [DOI] [PubMed] [Google Scholar]
- McCallum C.M., Comai L, Greene E.A., Henikoff S. (2000b) Targeting induced local lesions in genomes (TILLING) for plant functional genomes. Plant Physiol. 123: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R., Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A.A. (1997) A new model system for tomato genetics. Plant J. 12: 1465–1472 [Google Scholar]
- Mueller L.A., Solow T.H., Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright M.H., Ahrens R, Wang Y, et al. (2005a) The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol. 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L.A., Tanksley S.D., Giovannoni J.J., van Eck J., Stack S., Choi D., Kim B.D., Chen M., Cheng Z., Li C., et al. (2005b) The Tomato Sequencing Project, the first cornerstone of the International Solanaceae Project (SOL). Comp. Funct. Genomics 6: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L.A., Lankhorst R.K., Tanksley S.D., Giovannoni J.J., White R., Vrebalov J. (2009) A snapshot of the emerging tomato genome sequence. Plant Genome 2: 78–92 [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Oeller P.W., Lu M.W., Taylor L.P., Pike D.A., Theologis A. (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254: 437–439 [DOI] [PubMed] [Google Scholar]
- Okabe Y., Asamizu E, Saito T, Matsukura C, Ariizumi T, Brès C, Rothan C, Mizoguchi T, Ezura H. (2011) Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 52: 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron F., Nicolaï M., Minoïa S., Piednoir E., Moretti A., Salgues A., Zamir D., Caranta C., Bendahmane A. (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PloS One 5: e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, Mizoguchi T, Yamazaki Y, Aoki K, Ezura H. (2011) TOMATOMA: a novel tomato mutant database distributing MicroTom mutant collections. Plant Cell Physiol. 52: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrani J.C., Ruiz-Rivero O, Fos M, García-Martínez J.L. (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 56: 922–934 [DOI] [PubMed] [Google Scholar]
- Shirasawa K., Isobe S, Hirakawa H, Asamizu E, Fukuoka H, Just D, Rothan C, Sasamoto S, Fujishiro T, Kishida Y, et al. (2010) SNP discovery and linkage map construction in cultivated tomato. DNA Res. 17: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade A.J., Fuerstenberg S.I., Loeffler D, Steine M.N., Facciotti D. (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 23: 75–81 [DOI] [PubMed] [Google Scholar]
- Sun H.J., Uchii S, Watanabe S, Ezura H. (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wilkinson J.Q., Lanahan M.B., Yen H.C., Giovannoni J.J., Klee H.J. (1995) An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Wilkinson J.Q., Lanahan M.B., Clark D.G., Bleecker A.B., Chang C, Meyerowitz E.M., Klee H.J. (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat. Biotechnol. 15: 444–447 [DOI] [PubMed] [Google Scholar]
- Xiong A.S., Yao Q.H., Peng R.H., Li X, Han P.L., Fan H.Q. (2005) Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant Cell Rep. 23: 639–646 [DOI] [PubMed] [Google Scholar]
- Yin Y.G., Kobayashi Y, Sanuki A, Kondo S, Fukuda N, Ezura H, Sugaya S, Matsukura C. (2010) Salinity induces carbohydrate accumulation and sugar-regulated starch biosynthetic genes in tomato (Solanum lycopersicum L. cv. ‘Micro-Tom’) fruits in an ABA- and osmotic stress-independent manner. J. Exp. Bot. 61: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]