Abstract
The emerging discipline of systems pharmacology aims to combine analysis and computational modeling of cellular regulatory networks with quantitative pharmacology approaches to drive the drug discovery processes, predict rare adverse events, and catalyze the practice of personalized precision medicine. Here, we introduce the concept of enhanced pharmacodynamic (ePD) models, which synergistically combine the desirable features of systems biology and current PD models within the framework of ordinary or partial differential equations. ePD models that analyze regulatory networks involved in drug action can account for a drug’s multiple targets and for the effects of genomic, epigenomic, and posttranslational changes on the drug efficacy. This new knowledge can drive drug discovery and shape precision medicine.
Over the last decade, it has become clear that the responses of individuals to drugs can vary. Often, divergent responses can be attributed in part to differences in the genomic makeup of the individual patient. Variations in genomic characteristics, such as polymorphisms in drug metabolizing enzymes and drug targets, have led to the understanding that drug dosage needs to be matched to the genomic makeup of the individual patient. This has been best shown in anticoagulation therapy with warfarin, for which single-nucleotide polymorphism (SNP) arrays (“chips”) that permit identification of the isoforms of cytochrome P450 involved in warfarin metabolism and of the drug’s target vitamin K epoxide reductase can aid the clinician in determining the most beneficial dose for an individual patient (1). For many other diseases and drugs the relationship is not so straight forward. The patient may have multiple genomic and epigenomic characteristics that determine the efficacy of the drug response. How can we incorporate this genomic information into predictive models of drug action? Systems biology approaches could be the answer.
Systems biology provides a broad perspective of cellular- and tissue-level events that requires the application of quantitative tools and computational models (2) to develop and study the functional capabilities of molecular networks (3, 4). Systems biologists use many data-mining and statistical tools to identify trends in large genomic data sets (5). Such trends enable network analyses to map the topology of biological systems (6) and to construct dynamic ordinary differential equation (ODE) models that represent biochemical reaction mechanisms from ligand-receptor binding to cell outputs (2, 7). These are plausible consensus models that form one class within multiple classes of models (table S1).
Pharmacokinetic (PK) and pharmacodynamic (PD) researchers take a different approach to building integrated PK/PD models of drug action in biological systems (8–10). With their focus on drug disposition and dynamics, these models place greater reliance on obtaining experimental data on drug concentrations and associated biological responses collected over time and at various drug doses in specific systems of interest. In contrast to systems biology ODE models, PK/PD models are often “hardwired” to the data (unless they are being used as simulation tools). This enables the implementation of model-fitting algorithms (weighted least squares, maximum likelihood, and Bayesian) that contain weighting and statistical functions that help select the best-fit model.
BIRTH OF AN INTEGRATED DISCIPLINE
Systems pharmacology is an emerging integrated discipline (11–18) that combines many features of systems biology and pharmacology. Such integration is feasible in many facets of pharmacology, including pharmacodynamics. Although systems modelers and PK/PD modelers have distinct goals and contrasting approaches to model development, there are also convergences that are catalyzing the development of systems pharmacology. Both groups of scientists use ODE-based models of biological systems. Cell signaling networks, a focus of systems modelers, are highly pertinent to drug action and are the PD maps that provide the basic architecture of networks that describe the mechanisms of action of drugs.
We propose that such maps of cellular regulatory networks could be cast as enhanced PD (ePD) models, which would be related to traditional PK/PD models in that they are data-driven and be similar to systems biology models in their mechanism-based representation of cellular processes affected by drugs. ePD models would seek to specify a detailed topology of the cellular networks in order to identify regulatory motifs, such as feed-forward and feedback loops. A defining characteristic of ePD models is the ability to explicitly account for how genomic, epigenomic, and posttranslational regulatory characteristics in individual patients can alter the response to drugs. These ODE models would be developed from measurements in cells or tissue systems of interest with the goal of using model-fitting algorithms that result in identifiable models (that is, models with experimentally validated parameters) (Table 1). The actual formulation of ePD models in terms of data collection and model construction requires much further study.
Table 1.
Features of ePD models
| • Incorporate the mechanistic details of systems biology models and the identifiable characteristics of current PD models |
| • Link to PK models |
| • Contain representations of all pertinent regulatory networks and motifs, such as feedback loops and drug targets in the specific biological system of interest |
| • Incorporate the effects of genomic, epigenomic, and posttranslational characteristics on model parameters |
| • Data-driven to minimize degrees of freedom of model parameters and obtain a locally identifiable model through model-fitting optimization methods |
| • Serve as a platform for scale-up to explain response to drug therapy in individual patients with differing genomic and epigenomic characteristics (personalized medicine) |
| • Enable binning of individual patient’s drug response into tranches to develop a limited set of appropriate treatment regimens (precision medicine) |
The ePD modeling approach we propose draws on advances in physiologically-based (9, 19) and genomics-based (20) PK and PD models that provide a fuller understanding of drug action within the broader cellular and physiological context of the whole system. ePD models build on this approach. In ePD models, networks wherein regulatory motifs, such as feedback and feed-forward loops, are clearly identified form the base to explicitly represent the effects of genomic and epigenomic alterations on drug action. This type of representation will allow us to determine how genomic and epigenomic effects are manifested at the tissue/organ and organ-systems levels. As vital components of systems pharmacology, ePD models emphasize the importance of examining drug effects on a multiscale network, from biochemical cellular events to organ pathophysiology, for treatment of an individual patient.
Standard PD models often rely on a single end point (that is, a biomarker) as a measure of drug activity. This practice is a disadvantage, because drugs often have multiple targets that are parts of networks that comprise multiple components that determine drug activity. ePD models should provide a network view of drug action and consider both on- and off-targets that likely will affect multiple interconnected pathways. Although some off-targets may be considered minor, their importance may be revealed under different conditions, such as when combined with other drugs that affect targets and pathways within the same regulatory network. A central goal of an ePD model is to provide a comprehensive picture of drug action—both therapeutic and adverse—on cell-, tissue-, and organ-level physiological functions.
AN ePD MODEL
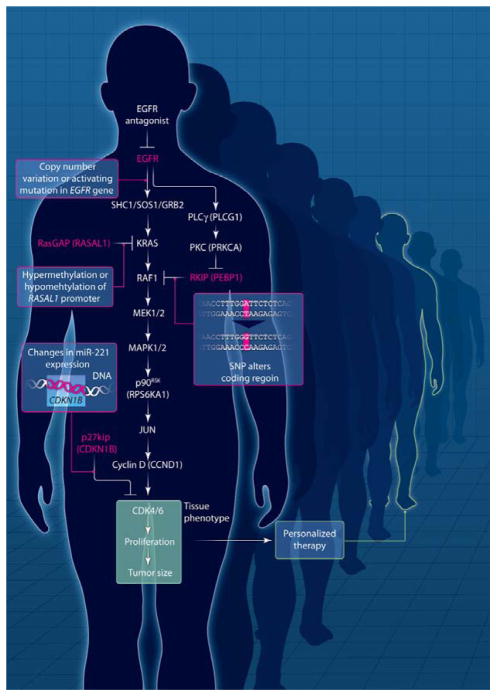
We have constructed an operational ePD model to illustrate its salient features. This model tracks the effect of an epidermal growth factor receptor (EGFR) inhibitor (such as gefitinib) on tumor growth and accounts for multiple genomic variations within the cellular regulatory network that controls tumor response (Fig. 1). The simplified regulatory network in this model is a linear pathway with a single additional coherent feed-forward motif from EGFR to RAF. In this ePD model, the link to a PK model is simplified by using the extent of fractional inhibition of an activated receptor as the input.
Fig. 1. Operational ePD model for EGFR inhibitor therapeutics.
A simplified EGFR signaling network that contains a single coherent feed-forward loop. The model is multiscale in that it includes cell- and tissue-level phenomena. Cell-level signaling reactions feed into tissue-level reactions (in green box), resulting in tissue phenotype. The ODE model incorporated biochemical reactions and linked them to cell proliferation and tumor size. Simulations were run for different combinations of genomic and epigenomic changes, highlighted in red and as specified in Fig. 2. See Supplementary Materials for details.
A number of simplified patient scenarios demonstrate how genomic and epigenomic variations in individual cancer patients can be modeled within the dynamics of anti-EGFR therapy to demonstrate the potential power of an ePD model. In this model, all hypothetical patients have driver mutations that activate the EGFR or copy number variations that increase amounts of EGFR, either of which results in increased proliferation and tumor formation. All patients are being treated with an EGFR tyrosine kinase inhibitor such that there is 80% inhibition of receptor activity. Details of the model and simulations can be found in the Supplementary Materials, and the results of the simulations at the tissue/organ level (that is, tumor size) are shown in Fig. 2.
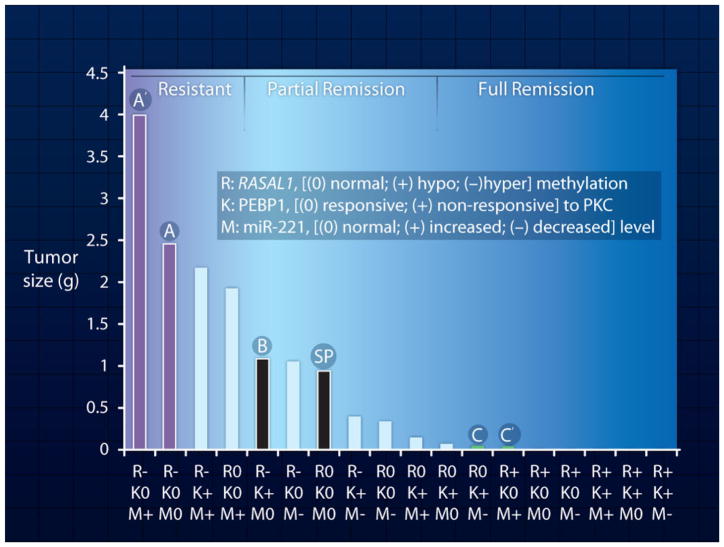
Fig. 2. Enhancing prediction of drug efficacy.
ePD model prediction of drug efficacy based on genomic and epigenomic status of the individual patient. Each bar represents tumor size calculated from simulations. Tumor size is calculated after drug treatment for a patient with the indicated genomic or epigenomic statuses. All patients are assumed to have the same oncogenic mutation in EGFR or increase in EGFR gene copy number. Drug treatment is assumed to block receptor activity by 80% for all patients. The other genomic and epigenomic changes for each patient are specified under the bars. SP represents a standard patient with no changes other than an oncogenic EGFR mutation. The relationship between drug responsiveness and genomic and epigenomic statuses of patients A, A′, B, C, C′, and SP are described in the text. For details on simulations see Supplementary Materials.
These simulations show how the response to a drug in each patient can vary on the basis of the number and type of genomic or epigenomic alterations. Consider a hypothetical standard patient (SP) who has the driver EGFR alterations but no additional changes in the genome and epigenome related to this tumorigenetic network. This patient would show decreased tumor growth upon drug treatment [Fig. 2, black bar (SP)]. The profiles of activated RAS, RAF, and MEK1/2 amounts and levels of the cell cycle activator cyclin D are shown in fig. S1.
Now consider patient B (Fig. 2), who displays the following features: (i) hypermethylation of RASAL1, which results in lowered amounts RasGAP (table S3) and (ii) a single-nucleotide polymorphism (SNP) (such as rs55716409) in the RKIP/PEBP gene (21). The RKIP/PEBP gene encodes the protein RKIP, an inhibitor of RAF1 (Fig. 1). We assume that this SNP results in an RKIP protein with greatly lowered affinity for protein kinase C (PKC) and that RKIP is not phosphorylated by PKC. Thus the increased signal from KRAS that results from the reduced amount of RasGAP is effectively suppressed at the level of RAF by the increased amounts of active RKIP, resulting in levels of active MEK1/2 that are similar to what is seen in the standard patient. Therefore, in patient B, the SNP in RKIP/PEBP effectively cancels out the epigenetic (DNA methylation) change in the RASAL1 gene and results in the same partial-remission response to the drug as the SP [Fig. 2, black bar (B)]. The profiles of activated RAS, RAF, and MEK1/2 amounts and cyclin D levels for patient B are shown in fig. S2.
In contrast, patient A has (i) a hypermethylated RASAL1 gene, but (ii) no other changes in RKIP/PEBP or alterations in the amount of microRNA-221 (miR-221), which is known to modulate the cell cycle regulatory protein p27kip (22). In this patient, the decrease in RasGAP protein levels leads to an uncompensated increase in RAS activity, which eventually results in over-expression of the cell cycle activator cyclin D in response to EGFR stimulation, even in the presence of the EGFR inhibitor. As a result, patient A has a tumor variant that is resistant to drug therapy and proliferates even when treated with a drug dose that blocks 80% of EGFR activity [Fig. 2, purple bar (A)]. The profiles of activated RAS, RAF, and MEK1/2 amounts and cyclin D levels of patient A are shown in fig. S3. If patient A accumulates additional deleterious changes, such as increased amounts of miR-221, which lead to decreased amounts of p27kip, then such a patient (A′) would show even greater increases in cyclin-dependent kinase 4/6 (CDK 4/6) activity (Fig. 1); this change leads to increased tumor growth even when the patient is treated with the drug [Fig. 2, left-most bar, purple (A′)].
Now consider patient C, who is represented in Fig. 2 as a green bar. This patient has (i) a normal RASAL1 gene, (ii) a SNP in the RKIP/PEBP gene that produces a hyperactive RKIP because of its lack of responsiveness to PKC regulation, and (iii) decreased amounts of miR-221. The increased amounts of active RKIP inhibit RAF activity, and the decreased miR-221 amounts lead to an increase in p27kip; together, these changes yield greatly decreased amounts of functional cyclin D and CDK4/6, which result in cancer cell proliferation and tumor growth (Fig. 1). The profiles of activated RAS, RAF, and MEK1/2 amounts and cyclin D levels for patient C are shown in fig. S4. Patient C′, who has (i) hypomethylated RAS-AL1 (which leads to increased amounts of RasGAP), (ii) a normal RKIP/PEBP1 gene, and (iii) increased amounts of miR-221 (which leads to decreased amounts of p27 kip), is also highly responsive to drug therapy [Fig. 2, green bar (C′)]. For patient C′, the epigenetic changes in the RASAL1 gene compensate for increased miR-221 amounts and the subsequent decrease in the cell cycle inhibitor p27 kip.
These hypothetical cases clearly indicate how multiple genomic and epigenomic changes can produce a wide range of responses to drug therapy. Even when patients have the same initial oncogenic driver mutation, distinct genomic and epigenomic changes can profoundly affect drug response. As shown in Fig. 2, operational ePD models, such as the one described here, provide a mechanistic understanding of why such variability occurs and can handle a range of genomic and epigenomic variations and predict drug response.
It is likely that the complexity of patient responses will be far more diverse than the simple example depicted in Fig. 2, because many drugs have multiple targets and are used in combination therapies. ePD models can make the problem of deciphering and predicting patient responses in these complex cases explicitly tractable when such computational models are developed in a data-driven manner and computational analyses become an integral part of clinical decision-making. Although we are not there yet, current progess is slow but steady.
At a conceptual level, the myriad genomic, epigenomic, translational, and posttranslational changes that are possible appear to be very complicated. However, all alterations at the various levels of biological regulation fall into only two types of changes in an ePD model. Mutations, SNPs, and posttranslational modifications such as protein phosphorylation can change biochemical reaction rates. Missense mutations and changes in DNA methylation, histone modifications, microRNA expression, ubiquitination, and protein turnover all alter the concentrations of the reactants. With the appropriate data, a range of genomic, epigenomic, and posttranslational changes can be readily represented with precision in ePD models. Hence, the complexity of ePD models is operational rather than conceptual.
FROM ePD MODELS TO CLINICAL DECISIONS
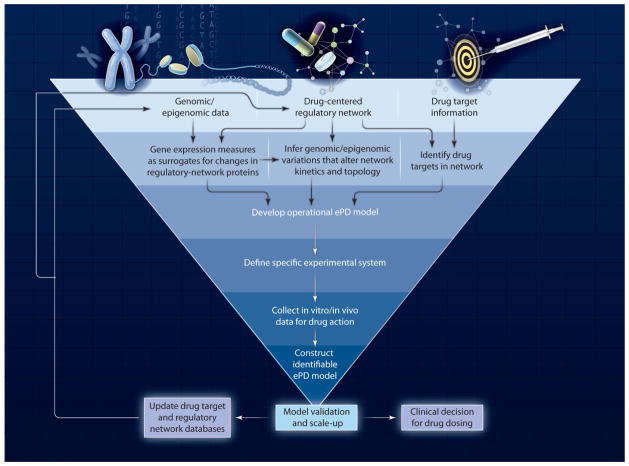
Our examples depicted in Fig. 1 and Fig. 2 highlight the network-centric nature of ePD models and the need to obtain target cell–dependent and drug-dependent measurements of drug responses. How data from in vitro animal and human cell systems or target tissues in animal models may be merged into reliable ePD models remains to be determined; however, such a merging will require genomic, epigenomic, and functional protein measurements collected under a range of drug exposures. The use of animal models provides a means by which to obtain drug concentration measurements and thus the ability to build PK models that are the logical input of ePD models. With appropriate genomic and epigenomic constraints, model organisms could be a practical approach for building PK-ePD models that, can not only can be used to probe drug efficacy in preclinical disease models, but further provide a basis for predicting drug action in patients. The construction of ePD models is not likely to be a linear path from data collection to final model but may use an iterative process (Fig. 3) of hypothesis-generating ePD models and associated data collection to fine-tune the model. There are multiple challenges in implementing the process we propose in Fig. 3.
Fig. 3.
Process diagram for building ePD models.
Unless patient cell and tissue samples are available in sufficient amounts and from diverse times in the pathophysiological process, ePD and PK/ePD models will rely on preclinical systems. Species-to-species extrapolations have been considered in the PK domain (23, 24) but much less so in the PD domain (25). ePD models rooted in the genome of the species being studied should be able to account for species differences by taking into account isoform diversity and the resultant changes in macromolecular interactions and functions. Physiologically-based PK-PD models are useful for pre-clinical-to-clinical scale-up (26). However, explicit experimental interspecies scaling of PD models has only begun to be addressed by researchers. For example, experimental challenges associated with obtaining data for ePD models will include identification and characterization of the relationships between genomic and epigenomic changes and alterations in corresponding cellular proteins and their functional interactions in different species.
Integration of drug-target networks with target cells and tissue-specific regulatory networks may streamline model development and lead to libraries of dynamic ePD models. However, building such libraries of ePD models will depend on broad and easy access to the systems biology and pharmacology datasets. As such, three types of databases will be needed: (i) Databases that provide detailed descriptions of genomic and epigenomic changes associated with clinical and pathophysiological characteristics for individual patients within a sufficiently large cohort of patients. The glioblastoma data set within The Cancer Genome Atlas (TCGA) provides us with a view of how such databases may be developed (27). (ii) Databases that relate genomic and epigenomic changes to changes in protein concentrations. For example, such a database would provide numbers that correlate changes in levels of DNA methylation to changes in levels of messenger RNA transcripts and proteins. (iii) Databases of kinetic constants and concentrations of cellular components—the Quantitative Human Interactome. The latter two types of databases currently do not exist but need to be developed.
The accuracy and benefits of clinical decision-making with the use of ePD models will need to be demonstrated. We predict that multiple cycles of iteration will lead to progressively more accurate clinical decisions regarding drug selections and designing of dosage regimens for optimal efficacy. A key issue that must be addressed is how ePD model implementation and possible refinement for individual patients can be synchronized with the clinical treatment timeline.
Further investigations are needed to extend ePD models across patient populations. Lemaire and colleagues developed a bone molecular and cellular biological homeostasis model (28), which has been modified to describe the central tendencies of metabolic and clinical biomarkers in several patient populations (29). A natural progression would include patient-specific genomic, epigenomic, and proteomic characteristics to understand intersubject variability in drug response. This approach would contrast with traditional methods of population-based PK-PD systems analyses that have complex structural models, which often start with applying standard model-reduction strategies to arrive at a minimal number of identifiable drug- and system-specific terms, followed by nonlinear mixed-effects modeling to estimate mean parameters and the magnitude of inter- and intrasubject and residual variability. Schmidt et al. (30) derived such a reduced model of bone remodeling that might be extended to fitting population-level responses to therapy. This methodology assumes that inter- and intrasubject variability and residual error are random effects attributed to model parameter values and error variance functions. With our increasing knowledge, we can now hypothesize that such variations may originate in the mixing and matching of genomic and epigenomic changes that lead to different drug responses (Fig. 2). Identifying clinical covariates continues to be a data-driven endeavor, requiring relatively large patient populations with appropriately distributed demographics and an unbiased covariate model-building technique. Such clinical covariates will now have to be matched with genomic and epigenomic changes to develop population-level models with variances in parameters. Although not all parameters will be uniquely identifiable, PK-ePD models should provide a platform for integrating sources of variability in exposure-response relationships, which in turn should enable the prospective prediction of the probability of clinical outcomes for specific genotypes and phenotypes and thus eliminate the practice of unguided clinical decisions for pharmacotherapy.
One clear outcome from the development of the library of ePD models summarized in Fig. 2 is the conceptual transition from personalized to precision medicine. It can readily be seen in Fig. 2 that, even though each of the 18 patients has a unique set of genomic and epigenomic characteristics, their tumor-growth response to drug therapy can be binned into three categories to facilitate implementation of drug therapy. Knowing which tranche a patient falls in provides a precise basis for selecting the most appropriate drugs and treatment regimen. Thus, ePD models can be viewed as a useful tool for precision medicine and used for personalized chemotherapy.
BENEFITS OF INTEGRATION
The fields of systems biology and PK-PD thus far have evolved on different tracks yet have significant interrelationships that can enhance drug discovery and enable optimized therapy for each patient. Progress in comprehensive PK assessments of drug candidates in the early phases of drug discovery has substantially reduced the number of PK failures later in the drug-development process (31). Similarly, establishment of ePD models early in drug development could aid in the selection of the most efficacious agents for the appropriate subpopulations of varying genomic and epigenomic signatures. This should reduce failures during phase II and phase III clinical trials and so aid in reducing the cost of drug development. The quantitative nature of ePD models will allow systems biology and PK-PD researchers to collaborate in the design of experimental protocols that advance large-scale data collection (biochemical parameters and physiological outputs) and of mathematical tools. Together, these endeavors should yield a predictive understanding of drug pharmacology and form the basis for integrating genomics and drug action for personalized precision medicine.
Supplementary Material
Footnotes
http://stm.sciencemag.org/content/4/126/126ps7/suppl/DC1
Model description
Figures and tables
Fig. S1. Temporal profiles of active signaling components of the EGFR network in the standard patient (SP)
Fig. S2. Temporal profiles of active signaling components of the EGFR network in patient B
Fig. S3. Temporal profiles of active signaling components of the EGFR network in patient A
Fig. S4. Temporal profiles of active signaling components of the EGFR network in patient C
Table S1. Characteristics of different types of computational model
Table S2. Equations for the multiscale ODE model
Table S3. ODE model parameters for the EGFR network
Table S4. Relationship between free cyclin d, mass doubling given growth fraction, and tumor size
Table S5. Simulation conditions for multiple genomic/epigenomic alterations in the EGFR network References
REFERENCES AND NOTES
- 1.Jonas DE, McLeod HL. Genetic and clinical factors relating to warfarin dosing. Trends Pharmacol Sci. 2009;30:375–386. doi: 10.1016/j.tips.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Ma’ayan A, Blitzer RD, Iyengar R. Toward predictive models of mammalian cells. Annu Rev Biophys Biomol Struct. 2005;34:319–349. doi: 10.1146/annurev.biophys.34.040204.144415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 4.Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 7.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 8.Sheiner LB, Steimer JL. Pharmacokinetic/pharmaco-dynamic modeling in drug development. Annu Rev Pharmacol Toxicol. 2000;40:67–95. doi: 10.1146/annurev.pharmtox.40.1.67. [DOI] [PubMed] [Google Scholar]
- 9.Xu L, Eiseman JL, Egorin MJ, D’Argenio DZ. Physiologically-based pharmacokinetics and molecular pharmacodynamics of 17-(allylamino)-17-demethoxygel-danamycin and its active metabolite in tumor-bearing mice. J Pharmacokinet Pharmacodyn. 2003;30:185–219. doi: 10.1023/a:1025542026488. [DOI] [PubMed] [Google Scholar]
- 10.Mager DE, Jusko WJ. Development of translational pharmacokinetic-pharmacodynamic models. Clin Pharmacol Ther. 2008;83:909–912. doi: 10.1038/clpt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma’ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mt Sinai J Med. 2007;74:27–32. doi: 10.1002/msj.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 13.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 14.Berger SI, Ma’ayan A, Iyengar R. Systems pharmacology of arrhythmias. Sci Signal. 2010;3:ra30. doi: 10.1126/scisignal.2000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie L, Xie L, Bourne PE. Structure-based systems biology for analyzing off-target binding. Curr Opin Struct Biol. 2011;21:189–199. doi: 10.1016/j.sbi.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorger PK, co-chair, Allerheiligen SRB., co-chair Quantitative and Systems Pharmacology in the Post-genomic Era: New Approaches to Discovering Drugs and Understanding Therapeutic Mechanisms. An NIH White Paper by the QSP Workshop Group; October 2011; http://www.nigms.nih.gov/NR/rdonlyres/8ECB1F7C-BE3B-431F-89E6-A43411811AB1/0/SystemsPharmaWPSorger2011.pdf. [Google Scholar]
- 17.van der Graaf PH, Benson N. Systems pharmacology: bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharm Res. 2011;28:1460–1464. doi: 10.1007/s11095-011-0467-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S, Iyengar R. Systems pharmacology: Network analysis to identify multiscale mechanisms of drug action. Annu Rev Pharmacol Toxicol. 2012;52:505–521. doi: 10.1146/annurev-pharmtox-010611-134520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowland M, Peck C, Tucker G. Physiologically-based pharmacokinetics in drug development and regulatory science. Annu Rev Pharmacol Toxicol. 2011;51:45–73. doi: 10.1146/annurev-pharmtox-010510-100540. [DOI] [PubMed] [Google Scholar]
- 20.Panetta JC, Sparreboom A, Pui CH, Relling MV, Evans WE. Modeling mechanisms of in vivo variability in methotrexate accumulation and folate pathway inhibition in acute lymphoblastic leukemia cells. PLOS Comput Biol. 2010;6:e1001019. doi: 10.1371/journal.pcbi.1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Database of Single Nucleotide Polymorphisms (dbSNP Build ID: 135); dbSNP accession: rs55716409. National Center for Biotechnology Information, National Library of Medicine; Bethesda, MD: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- 22.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boxenbaum H. Interspecies pharmacokinetic scaling and the evolutionary-comparative paradigm. Drug Metab Rev. 1984;15:1071–1121. doi: 10.3109/03602538409033558. [DOI] [PubMed] [Google Scholar]
- 24.Mahmood I. Application of allometric principles for the prediction of pharmacokinetics in human and veterinary drug development. Adv Drug Deliv Rev. 2007;59:1177–1192. doi: 10.1016/j.addr.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Mager DE, Woo S, Jusko WJ. Scaling pharmacodynamics from in vitro and preclinical animal studies to humans. Drug Metab Pharmacokinet. 2009;24:16–24. doi: 10.2133/dmpk.24.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Q, Guo P, Kruh GD, Vicini P, Wang X, Gallo JM. Predicting human tumor drug concentrations from a preclinical pharmacokinetic model of temozolomide brain disposition. Clin Cancer Res. 2007;13:4271–4279. doi: 10.1158/1078-0432.CCR-07-0658. [DOI] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas: Glioblastoma multiforme. http://tcga-data.nci.nih.gov/tcga/tcgaCancerDetails.jsp?diseaseType=GBM&diseaseName=Glioblastomamultiforme.
- 28.Lemaire V, Tobin FL, Greller LD, Cho CR, Suva LJ. Modeling the interactions between osteoblast and osteoclast activities in bone remodeling. J Theor Biol. 2004;229:293–309. doi: 10.1016/j.jtbi.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 29.Marathe A, Peterson MC, Mager DE. Integrated cellular bone homeostasis model for denosumab pharmacodynamics in multiple myeloma patients. J Pharmacol Exp Ther. 2008;326:555–562. doi: 10.1124/jpet.108.137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt S, Post TM, Peletier LA, Boroujerdi MA, Danhof M. Coping with time scales in disease systems analysis: Application to bone remodeling. J Pharmacokinet Pharmacodyn. 2011;38:873–900. doi: 10.1007/s10928-011-9224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.