Abstract
The circadian regulatory network is organized in a hierarchical fashion, with a central oscillator in the suprachiasmatic nuclei (SCN) orchestrating circadian oscillations in peripheral tissues. The nature of the relationship between central and peripheral oscillators, however, is poorly understood. We used the tetOFF expression system to specifically restore Clock function in the brains of ClockΔ19 mice, which have compromised circadian clocks. Rescued mice showed normal locomotor rhythms in constant darkness, with activity period lengths approximating wildtype controls. We used microarray analysis to assess whether brain-specific rescue of circadian rhythmicity was sufficient to restore circadian transcriptional output in the liver. Compared to Clock mutants, Clock-rescue mice showed significantly larger numbers of cycling transcripts with appropriate phase and period lengths, including many components of the core circadian oscillator. This indicates that the SCN oscillator overcomes local circadian defects and signals directly to the molecular clock. Interestingly, the vast majority of core clock genes in liver were responsive to Clock expression in the SCN, suggesting that core clock genes in peripheral tissues are intrinsically sensitive to SCN cues. Nevertheless, most circadian output in the liver was absent or severely low-amplitude in Clock-rescue animals, demonstrating that the majority of peripheral transcriptional rhythms depend on a fully functional local circadian oscillator. We identified several new system-driven rhythmic genes in the liver, including Alas1 and Mfsd2. Finally, we show that 12-hour transcriptional rhythms (i.e., circadian “harmonics") are disrupted by Clock loss-of-function. Brain-specific rescue of Clock converted 12-hour rhythms into 24-hour rhythms, suggesting that signaling via the central circadian oscillator is required to generate one of the two daily peaks of expression. Based on these data, we conclude that 12-hour rhythms are driven by interactions between central and peripheral circadian oscillators.
Author Summary
Circadian rhythms confer adaptive advantage by allowing organisms to anticipate daily rhythms in their environment, such as light and temperature cycles. In mammals, the central circadian time-keeping mechanism resides within the hypothalamus, integrating information from the retina and synchronizing circadian oscillations in peripheral tissues, such as the liver. Nevertheless, the nature of this regulatory relationship is not completely understood. Here we use mice with disrupted circadian rhythmicity stemming from a mutation in Clock, a key component of the circadian time-keeping mechanism. Using recently developed transgenic technology, we expressed normal CLOCK protein in the brain, resulting in mice with normal brain rhythmicity, but defective circadian rhythmicity everywhere else. Using microarrays to detect transcriptional rhythms in the livers of these mice, we found that 95 genes showed normal oscillations (albeit with generally lower amplitudes), even in the presence of a defective liver circadian clock. This observation suggests that many rhythmic genes in peripheral tissues are driven directly from the brain. Finally, we showed that harmonics of circadian oscillations, e.g. 12-hour rhythms, revert to 24-hour rhythms in these mice, indicating that both liver and brain-derived cues drive different aspects of these rhythms.
Introduction
Circadian rhythms are daily oscillations of behavior and physiology that allow organisms to anticipate and respond to predictable daily changes in their environment [1]–[4]. In animals, these environmental variables include light, temperature, food availability, and predation. As a consequence, circadian rhythms regulate behaviors such as feeding rhythms and sleep/wake cycles [2]. At a tissue and cellular level, circadian rhythms compartmentalize the activity of biochemical pathways to appropriate times of day in tissues throughout the body [1]. Taken as a whole, the circadian regulatory network significantly influences normal organismal physiology, and contributes to the pathogenesis of clinically significant conditions including cancer, heart disease, and metabolic disorders [5]–[8].
Self-sustained circadian oscillations are generated at a molecular level via an elaborate transcriptional/translational feedback loop [9]. The positive arm of this feedback loop is mediated by bHLH-PAS transcription factors, BMAL1 and CLOCK/NPAS2 [10], [11], which heterodimerize and drive the expression of downstream target genes. Among these target genes are Period (Per) and Cryptochrome (Cry), whose protein products accumulate in the cytoplasm, associate with each other, and ultimately translocate to the nucleus. Once in the nucleus, PER and CRY inhibit BMAL1/CLOCK activity, repressing their own transcription and thus forming the negative arm of the circadian oscillator [9]. In parallel, a second feedback loop is generated via RORE binding activators (Rora, Rorb, Rorc) and repressors (Rev-erb-alpha,Rev-erb-beta), whose transcription is driven by BMAL1/CLOCK [12].
In conjunction with accessory genes that regulate the stability and activity of key circadian proteins [9], these feedback loops comprise the circadian clock. Ultimately, these molecular oscillations drive 24-hour rhythms of transcription in downstream target genes. Termed “circadian output genes", these transcriptional rhythms are not necessary for sustaining the core circadian oscillator, but are required for mediating circadian regulation of physiology and behavior [13].
Both core circadian oscillations and rhythmic output genes are found in tissues throughout the body [13], [14]. However, not every tissue is equally important for maintaining proper circadian rhythmicity at an organismal level. Rhythms in peripheral tissues, such as liver and skeletal muscle, are self-sustaining in vitro, but require inputs from the central circadian oscillator in the suprachiasmatic nuclei (SCN) in the hypothalamus for proper coordination in intact animals [15]. The nature of the regulatory signals between the SCN and peripheral tissues (as well as regulatory cues between peripheral tissues) is poorly understood, but thought to involve neuronal circuitry, humoral factors (e.g. glucocorticoids), and cascades of behavior (i.e. the impact of the sleep wake cycle on eating and elimination).
The general mechanism of circadian oscillations and the genes required for their maintenance is largely conserved between different tissues and species [3], [13], [16], [17]. Nevertheless, circadian output genes are tissue specific, as one would expect given the diverse physiologies regulated by the clock [18]–[20]. Consequently, considerable efforts have been made to characterize circadian transcriptional output at a genome level using microarray technologies [18]–[33]. These studies have made considerable progress towards understanding how tissue-level circadian oscillations are translated into rhythms of organismal physiology. At the same time, this systems-level approach can be used in conjunction with tissue-specific manipulation of gene expression to dissect the relationship between central and peripheral oscillators.
For example, a recent study used liver-specific over-expression of REV-ERB-alpha to knock-down BMAL1 expression in the liver, thereby ablating the local circadian oscillator in an otherwise wildtype animal [21]. They examined the impact of this on circadian regulation of liver gene expression by microarray analysis. This analysis showed that the vast majority of circadian output requires a functional circadian oscillator within the liver. Interestingly, there were several exceptions to this rule, indicating that some circadian genes are driven by systemic cues rather than the local circadian clock. Those genes, including Per2, are therefore strong candidates to function as the relay between the SCN and peripheral tissues [21]. Thus, this study established that a functional liver oscillator is required for normal circadian regulation of liver gene expression.
In this manuscript, we seek to extend this research by examining the contribution of central circadian clock function on peripheral physiology. To do this, we employed a tet-OFF expression system to specifically rescue wildtype CLOCK expression in the brains of Clock-mutant mice [21], [34]–[36]. Brain-specific Clock rescue restored normal behavioral rhythmicity in constant conditions with approximately wildtype period lengths. We used genome-wide transcriptional profiling every two hours for two full days followed by JTK_CYCLE analysis to assess transcriptional circadian output in the mouse liver [37]. The majority of rhythmic genes in the liver required a fully functional liver circadian clock; however, 95 genes still oscillate with circadian periods in the livers of brain-rescued mice, albeit with diminished amplitudes in most cases. We observe that 12-hour transcriptional rhythms (i.e., circadian ‘harmonics’ [19]) are entirely lost in Clock-mutant background. Interestingly, brain-specific rescue of Clock restores 24-, but not 12-hour rhythmicity to these genes, suggesting that systemic and locally-derived circadian cues are independently required for different peaks of these 12-hour rhythms.
Results
Inducible, brain-specific expression of wildtype Clock was achieved using the tet-OFF system [34]–[36], [38]. Wildtype Clock was tagged with an HA epitope and linked to a tTA-responsive tetO promoter (tetO::Clock-HA). At the same time, tTA was expressed under the control of the Secretogranin II (Scg2) promoter (Scg2::tTA) which drives expression specifically in the brain, pituitary, and adrenals, with especially high expression in the SCN [39]. When both transgenes were present in the same animal, wildtype Clock was expressed at constitutively high levels in the SCN [36]. When these mice were treated with low-doses of Doxycycline (Dox) via their drinking water, the binding of tTA to tetO promoters was inhibited, and Clock expression was abolished [36].
These mice were then crossed into a ClockΔ19 background. ClockΔ19 is an ENU-generated allele of Clock which results in the loss of exon 19 from mature Clock transcripts [40], [41]. ClockΔ19 acts as a dominant-negative by binding to BMAL1 and inhibiting its activity [41]. Consequently, ClockΔ19 animals have severely disrupted circadian behavioral rhythms, with extremely long period lengths and arrhythmicity in prolonged constant conditions [40], [42]–[44].
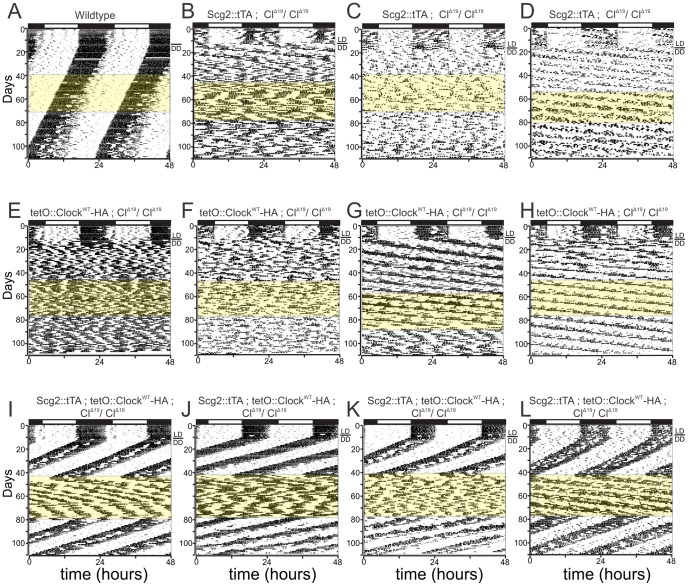
Wildtype animals showed robust circadian oscillations in 12-hour light/12-hour dark (LD) conditions with most locomotor activity restricted to the dark phase. The mice maintained these rhythms in constant darkness (DD), with period lengths slightly shorter than 24-hours, in agreement with previous studies (Figure 1A). When either Scg2::tTA or tetO::Clock-HA were expressed by themselves in a ClockΔ19 background, the mice showed normal LD activity rhythms, but quickly became arrhythmic in DD or had extremely long period lengths (Figure 1B–1H). In all three control genetic backgrounds, the addition of Dox to the drinking water (highlighted in yellow), did not change the circadian behavior of these mice (Figure 1A–1H).
Figure 1. Reversible restoration of circadian rhythm in brain-rescued Clock mutant mice.
Representative actograms of (A) wildtype mice, control littermates (B, C and D) Scg2::tTA; ClΔ19/ClΔ19, (E, F, G and H) tetO::ClockWT; ClΔ19/ClΔ19, and (I, J, K and L) brain-rescued Clock mice (Scg2::tTA;tetO::ClockWT; ClΔ19/ClΔ19) on 10 µg/ml Dox treatment. Activity records were double plotted so that 48 hours is represented horizontally, with each day presented beneath and to the right of the preceding day. Wheel-running activity is indicated by black markings. The initial light cycle is depicted at the top of the record. All animals were maintained on a 12-hour light/12-hour dark cycle (LD) for the first 10 to 15 days shown and then transferred to constant darkness (DD), as indicated by the bar next to each record. The yellow shading denotes the time of Dox administration in DD. Brain-rescued Clock mice (I, J, K, and L) show restoration of free-running period in DD. When the mice are treated with Dox, brain-rescued Clock mice display loss of rhythmicity, similar to that of Clock mutant mice (ClΔ19/ClΔ19). A water washout returns the mice back to their rhythmic state. Control littermates (A–H) display no circadian rhythmicity in DD. Dox treatment has no effect on activity rhythm in control littermates or wildtype mice (A–H).
Combining Scg2:tTA and tetO::Clock-HA in the ClockΔ19 background resulted in mice with normal LD rhythmicity. Unlike their littermate controls (i.e., Figure 1B–1H), however, these mice showed robust circadian rhythmicity in constant conditions (Figure 1I–1L). Previous studies have shown [45] that over-expression of wildtype Clock is sufficient to rescue the behavioral phenotype of ClockΔ19. In agreement with these studies, we detected a modest decrease in the average period length of these animals compared to wildtype (Figure 1A and 1I–1L).
When the rescued mice were treated with Dox (thus inactivating the wildtype Clock transgene expression), normal circadian oscillations were quickly lost (Figure 1I–1L). We observed significant animal-to-animal variability in the severity of the resulting phenotype. Generally, however, these mice were either arrhythmic or showed extremely long period lengths while being treated with Dox, consistent with the expected phenotype of ClockΔ19 mice (Figure 1I–1L). This phenotype was completely reversible; when Dox was removed from the drinking water (thus restoring Clock transgene expression), normal circadian rhythmicity was quickly reestablished (Table 1). Based on these behavioral data and the previously published expression pattern of Scg2::tTA ; tetO::Clock-HA mice [36], we conclude that this system permits the brain-specific rescue of circadian rhythmicity in locomotor activity in a conditional and reversible manner.
Table 1. Clock expression via the tet-OFF system reversibly rescues normal circadian rhythmicity in DD (+/−standard deviation).
| Treatment | Period Length | Number Rhythmic | Number Arrhythmic |
| Before Dox Treatment | 22.6 hours (+/−0.3) | 14 | 0 |
| During Dox Treatment | 27.1 hours (+/−0.8) | 4 | 10 |
| After Dox Treatment | 22.2 hours (+/−0.5) | 14 | 0 |
Although brain-specific rescue of Clock is sufficient to restore normal behavioral rhythms, it was unclear whether transcriptional and metabolic rhythms in peripheral tissues would be similarly rescued. To answer this, we collected liver samples from wildtype animals as well as Scg2:tTA ; tetO::Clock-HA mice that were treated with either normal drinking water or Dox (hereafter referred to as tetO::Clock H2O and tetO::Clock DOX). Based on the behavioral data presented above, we expected the H2O-treated animals (i.e., Clock transgene-expressing) to have normal brain rhythmicity and behavioral rhythms, while the Dox-treated animals (i.e., Clock-defective) would have disrupted rhythms. It is important to note that in both cases the Scg2 promoter does not express in the liver, and thus, this peripheral clock is presumed to be Clock-defective.
Liver samples were collected every two hours for 48 hours in constant darkness (Figure 2A). Total RNA was extracted and global gene expression was profiled using Affymetrix Mouse Exon Arrays. Cycling genes were detected using JTK_Cycle with false-discovery rates (FDRs) based on the Benjamini-Hochberg procedure [37]. We found 576 cycling genes in wildtype mice at a FDR cutoff of q<0.05 (corresponding to a p-value threshold of p<0.0011), which is consistent with expected levels of transcriptional oscillations given the different sampling resolutions of these studies [18], [19], [46]. Over half of the oscillating genes in this study were previously identified as rhythmic [19], which is an encouraging level of overlap given the typically low agreement between circadian microarray studies [47], [48]. Known core clock genes – including Per2, Bmal1, and Rev-erb-alpha – showed high-amplitude oscillations with expected phase differences, as measured by microarray and confirmed by quantitative PCR (qPCR) (Figure S1). Taken together, these data indicate that this microarray study accurately reflects the underlying circadian transcriptome.
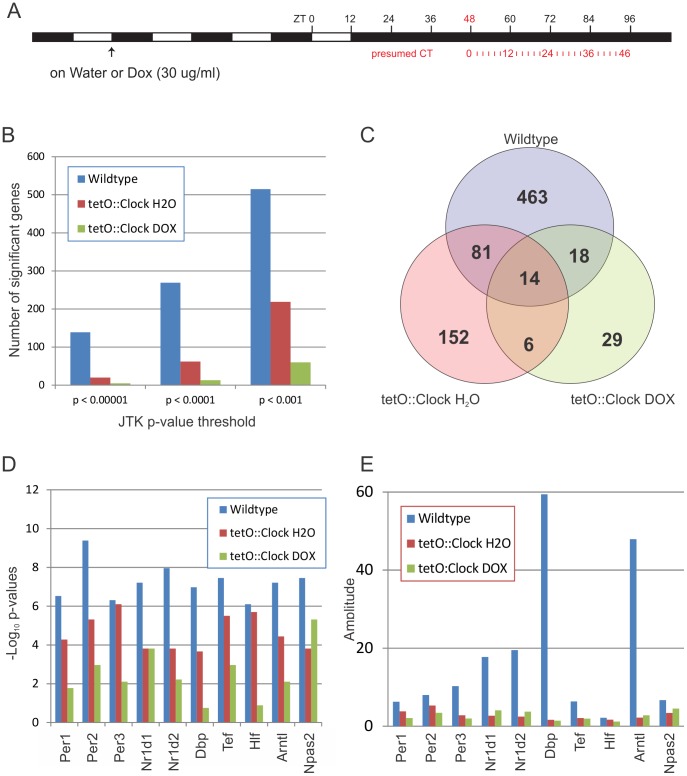
Figure 2. Brain-specific rescue of Clock partially restores transcriptional circadian output in the liver.
(A) Wildtype and Clock-expressing mice (Scg2::tTA;tetO::ClockWT; ClΔ19/ClΔ19) were entrained to a 12 h∶12 h light∶dark (LD) cycle and then treated with either H2O or doxycycline (Dox, 30 µg/ml) 3 days before releasing the mice to constant darkness. Liver samples were collected every two hours for two complete days, starting 48 hours after the final lights-on. (B–E) Cycling genes were detected in microarray datasets using JTK_Cycle. At every p-value threshold tested (B), wildtype livers show considerably more cycling genes than Clock-mutant mice (tetO::Clock DOX), while the Clock-rescue animals (tetO::Clock H2O) have an intermediate number of rhythmic genes. Panel (C) shows the number of cycling genes overlapping between the different genotypes/treatments at a p-value threshold of p<0.0011 (corresponding to q<0.05 in wildtype, all period lengths between 10 and 40 hours). The strength of rhythmicity of core clock genes and key circadian output genes (as measured by −Log10 p-values) is generally higher in Clock-rescue animals (tetO::Clock H2O) versus the Clock-mutants (tetO::Clock DOX). (D) However, Clock-rescue (tetO::Clock H2O) does not restore wildtype-levels of circadian amplitude in most of these genes (E).
At every statistical threshold we examined, wildtype livers showed significantly more rhythmic transcripts than Dox-treated tetO::Clock mice (i.e., Clock-mutant). Heatmaps of all cycling genes detected, as well as histograms of their amplitudes are shown in Figure S2. H2O-treated (i.e., Clock-rescue) mice had an intermediate, partially-rescued phenotype, with considerably more cycling genes detected than Dox-treated animals, though still less than wildtype (Figure 2B and 2C). This pattern was also seen in core clock genes and high-amplitude circadian output genes. The strength of rhythmicity (as measured by the statistical confidence of their detection) consistently demonstrated that Clock-rescue (tetO::Clock H2O) mice had an intermediate phenotype between wildtype and Clock-mutant mice (Figure 2D and Table S1). Brain-specific expression of Clock did not rescue the amplitude of most high-amplitude transcriptional rhythms (Figure 2E and Figures S1 and S2). Even though Clock-rescue restored a considerable portion of the normal circadian output of the liver, these rhythms were frequently low-amplitude relative to wildtype, indicating that the local circadian oscillator – and in particular, wildtype Clock expression – is essential for generating high-amplitude rhythms.
At the statistical threshold we have selected, there are 187 genes that cycle in one or both of the tetO::Clock samples without cycling in wildtype. Of these 187 genes, 151 were analyzed in Hughes et al. 2009 [19], and 102 of them were found to oscillate (Table S2). Based on this, we conclude that the majority of non-wildtype cycling genes in the present study are actually bona fide cyclers that did not meet significance given the relatively stringent statistical threshold used. Consistent with this idea, the median p-value for these 102 genes in wildtype samples in the present study is ∼0.1. Given the false-discovery rates for Clock-rescue and Clock-mutant animals (q<0.10 and q<0.37, respectively at p<0.0011), we conclude that cycling genes specific to non-wildtype backgrounds are rare, in agreement with Kornmann et al. 2007 [21].
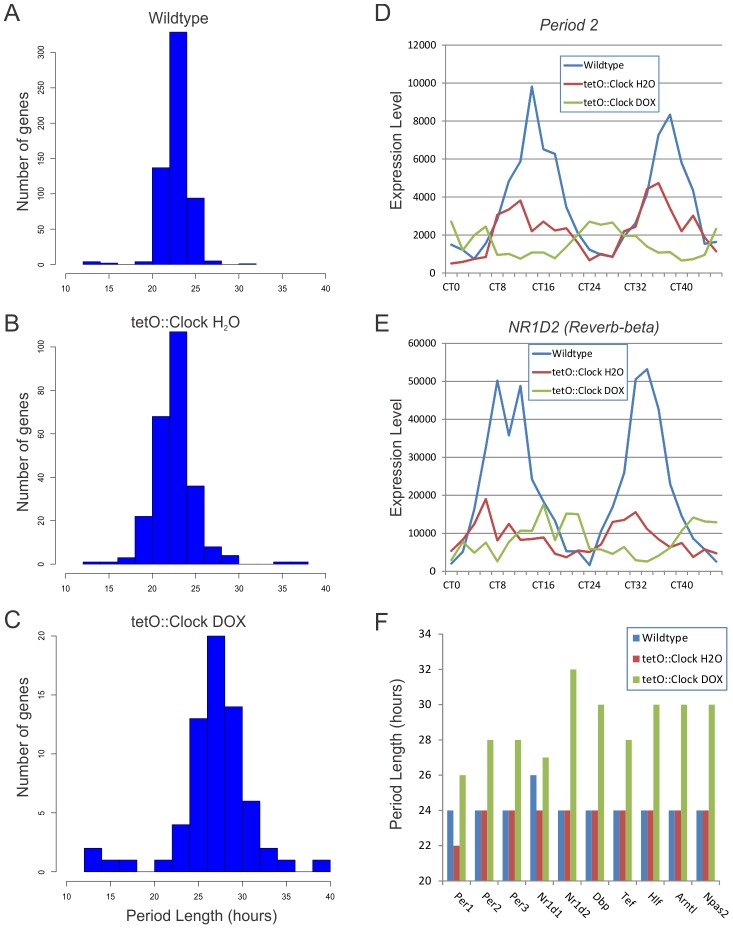
As expected, the average period length of transcriptional rhythms in wildtype mice was ∼24-hours (Figure 3A). Consistent with the behavioral profiles discussed above (Table 1), Clock-rescue mice (tetO::Clock H2O) also showed average period lengths of ∼24-hours, while Clock-defective mice (tetO::Clock DOX) had considerably longer period lengths as would be expected in ClockΔ19 mutant mice (Figure 3B and 3C). This phenotype is illustrated by the profiles of two core clock genes, Per2 and Rev-erb-beta (Figure 3D and 3E, and Figure S1). Both wildtype and Clock-rescue mice showed normal 24-hour rhythms with phases in agreement in both genotypes. In contrast, Per2 and Rev-erb-beta in Clock-defective mice had a longer-period phenotype with peak expression out-of-phase with wildtype. Since these animals were housed in constant darkness for 2 days before sample collection, we believe this apparent phase difference is a consequence of their free-running period length phenotype. These observations were seen in every core clock gene tested, as well as many key circadian output genes (Figure 3F). In each case, these genes had ∼24-hour rhythms in wildtype and Clock-rescue mice, with considerably longer period length in Clock-defective mice.
Figure 3. Clock-rescue restores ∼24-hour transcriptional rhythms.
Panels A–C show histograms of the period lengths of cycling genes in wildtype (A), Clock-rescue (B) and Clock-mutant (C) livers (p<0.0011 for each genotype/treatment, corresponding to a q<0.05 in wildtype). Clock-mutant mice have long transcriptional rhythms with periods approximating their behavioral rhythms (mean period = 27.6+/−4.5 h), while wildtype and Clock-rescue animals have period lengths of approximately 24-hours (23.7+/−1.7 h and 23.6+/−2.6 h, respectively). Note that the difference in y-axis scales reflects the overall level of rhythmic transcriptional output in these mice. Two components of the core circadian clock, Per2 (D) and Nr1d2 (E), illustrate the long-period phenotype of the Clock-mutant mice. This effect is seen in most core clock genes as well as key circadian output genes (F). Additional examples are shown in Figure S1. The apparent phase difference in Clock-mutant mice compared to wildtype/Clock-rescue mice is believed to be a consequence of their free-running period length phenotype.
Overall, 95 genes were found to cycle in both wildtype and Clock-rescue animals (p<0.0011, corresponding to a q<0.05 in wildtype). Three of these genes had period lengths of ∼12-hours in wildtype animals (i.e. circadian harmonics) and are discussed in greater detail below. The remaining 92 genes all showed period lengths of ∼24-hours in both wildtype and rescue animals, and 77 of the 92 (84%) have been previously seen to oscillate in mouse liver (Table 2) [19].
Table 2. Clock-rescue restores 24-hour rhythmicity to 92 circadian genes.
| Wildtype | tetO::Clock H2O (rescue) | 24-hour rhythm in Hughes et al. 2009? | Systemically-driven in Kornmann et al. 2007? | Systemically-driven in JTK_Cycle re-analysis of Kornmann et al. 2007? | Reverb-alpha target in Le Martelot et al.? | Bmal target in Rey et al. 2011? | |||||||||
| Gene | q-value | p-value | Period | Phase (hours) | Amplitude | q-value | p-value | Period | Phase (hours) | Amplitude | |||||
| 0610010D20Rik | 0.043499 | 0.001074 | 26 | 9 | 1.3 | 0.019096 | 3.60E-05 | 26 | 7 | 1.7 | YES | ||||
| 1110067D22Rik | 0.000239 | 7.88E-07 | 24 | 22 | 4.1 | 0.005596 | 4.80E-06 | 24 | 21 | 1.5 | YES | ||||
| 5630401D24Rik | 0.035929 | 0.00079 | 26 | 10 | 1.3 | 0.08455 | 0.00079 | 26 | 11 | 1.4 | |||||
| A630005I04Rik | 0.000239 | 7.88E-07 | 24 | 22 | 10.6 | 0.04346 | 0.000216 | 22 | 21 | 1.8 | YES | ||||
| Abcb11 | 0.017929 | 0.000302 | 24 | 2 | 1.8 | 0.051058 | 0.000302 | 21 | 4.5 | 1.2 | YES | ||||
| Abcg5 | 0.000334 | 1.26E-06 | 22 | 11 | 2.8 | 0.063073 | 0.000419 | 26 | 6 | 1.5 | YES | ||||
| Abcg8 | 0.006556 | 7.56E-05 | 24 | 9 | 4.5 | 0.022227 | 5.24E-05 | 26 | 6 | 1.9 | YES | ||||
| Adarb1 | 0.003737 | 3.03E-05 | 24 | 12.5 | 2.0 | 0.08455 | 0.00079 | 24 | 12 | 1.3 | YES | ||||
| Adora1 | 7.02E-05 | 1.05E-07 | 26 | 9 | 1.8 | 0.08455 | 0.00079 | 26 | 9 | 1.4 | YES | ||||
| Agpat1 | 0.035929 | 0.00079 | 22 | 15 | 1.5 | 0.019096 | 3.60E-05 | 26 | 12 | 1.5 | |||||
| Agtr1a | 0.017929 | 0.000302 | 22 | 6 | 1.9 | 0.037685 | 0.000153 | 24 | 4 | 1.3 | YES | ||||
| AI842396 | 0.006556 | 7.56E-05 | 23 | 6.5 | 1.7 | 0.073204 | 0.000577 | 24 | 6 | 1.5 | YES | ||||
| Alas1 | 0.002233 | 1.66E-05 | 24 | 14 | 3.2 | 0.035038 | 0.000108 | 24 | 12 | 5.6 | YES | YES | |||
| Alpl | 4.93E-05 | 6.13E-08 | 22 | 17 | 1.9 | 0.08455 | 0.00079 | 24 | 15 | 1.5 | |||||
| Ankhd1 | 0.000175 | 4.87E-07 | 26 | 15 | 3.2 | 0.08455 | 0.00079 | 24 | 13.5 | 1.7 | YES | ||||
| Arl4a | 0.010948 | 0.000153 | 24 | 22 | 2.1 | 0.019096 | 3.60E-05 | 22 | 2 | 1.3 | YES | ||||
| Arntl | 4.93E-05 | 6.13E-08 | 24 | 23 | 34.5 | 0.019096 | 3.60E-05 | 24 | 21 | 2.2 | YES | YES | YES | ||
| Atg2a | 0.043499 | 0.001074 | 22 | 9 | 1.6 | 0.011737 | 1.11E-05 | 26 | 5 | 1.4 | |||||
| Atp1a1 | 0.000903 | 4.80E-06 | 24 | 14.5 | 1.5 | 0.019096 | 3.60E-05 | 24 | 13 | 1.3 | YES | ||||
| BC016495 | 0.000133 | 2.96E-07 | 26 | 14 | 3.3 | 0.004223 | 1.99E-06 | 28 | 14 | 2.0 | YES | YES | |||
| Cabc1 | 0.000334 | 1.26E-06 | 24 | 1 | 4.0 | 0.011737 | 1.11E-05 | 22 | 4 | 1.8 | YES | YES | YES | YES | |
| Camk1d | 0.00861 | 0.000108 | 24 | 0 | 2.4 | 0.04346 | 0.000216 | 22 | 2 | 2.3 | YES | ||||
| Ccng2 | 0.010948 | 0.000153 | 24 | 22 | 2.3 | 0.04346 | 0.000216 | 24 | 0 | 1.7 | YES | ||||
| Ccrn4l | 0.00046 | 1.99E-06 | 24 | 14 | 4.1 | 0.08455 | 0.00079 | 22 | 13 | 7.4 | YES | YES | YES | ||
| Celsr1 | 0.00046 | 1.99E-06 | 24 | 21 | 2.3 | 0.04346 | 0.000216 | 24 | 18 | 1.9 | YES | ||||
| Chkb | 0.005073 | 5.24E-05 | 22 | 4 | 1.6 | 0.037685 | 0.000153 | 22 | 3 | 1.4 | YES | ||||
| Clpx | 1.38E-05 | 5.92E-09 | 24 | 23 | 7.4 | 0.001228 | 6.13E-08 | 24 | 23 | 2.0 | YES | YES | |||
| Coq10b | 0.000334 | 1.26E-06 | 24 | 13 | 3.9 | 0.04346 | 0.000216 | 24 | 13 | 2.7 | YES | ||||
| Crot | 0.000175 | 4.87E-07 | 22 | 5 | 2.7 | 0.022227 | 5.24E-05 | 22 | 5 | 1.6 | YES | ||||
| Dapk1 | 0.003837 | 3.60E-05 | 24 | 23 | 2.6 | 0.08455 | 0.00079 | 22 | 21 | 1.5 | YES | YES | |||
| Dbp | 7.02E-05 | 1.05E-07 | 24 | 11 | 10.7 | 0.04346 | 0.000216 | 24 | 9 | 1.6 | YES | YES | YES | ||
| Enpp2 | 0.003061 | 2.45E-05 | 24 | 13 | 1.8 | 0.08455 | 0.00079 | 22 | 11 | 1.4 | YES | ||||
| Etnk2 | 0.000334 | 1.26E-06 | 24 | 12 | 1.5 | 0.099034 | 0.001074 | 24 | 11 | 1.9 | YES | YES | |||
| Fbxo21 | 0.003837 | 3.60E-05 | 22 | 8 | 2.9 | 0.037685 | 0.000153 | 24 | 5 | 1.8 | YES | YES | YES | ||
| Fkbp4 | 0.017929 | 0.000302 | 24 | 19 | 1.7 | 0.012876 | 1.66E-05 | 24 | 16 | 1.7 | YES | ||||
| Fmo5 | 0.000239 | 7.88E-07 | 22 | 17 | 2.1 | 0.04346 | 0.000216 | 24 | 15 | 1.5 | YES | YES | YES | ||
| Gabarapl1 | 0.017929 | 0.000302 | 22 | 7 | 2.0 | 0.037685 | 0.000153 | 24 | 4 | 1.8 | YES | ||||
| Gpt2 | 0.000175 | 4.87E-07 | 24 | 14 | 1.9 | 0.022227 | 5.24E-05 | 22 | 13 | 1.6 | YES | YES | |||
| Hlf | 0.000239 | 7.88E-07 | 24 | 14 | 2.3 | 0.004223 | 1.99E-06 | 24 | 13 | 1.6 | YES | YES | |||
| Ivns1abp | 0.028179 | 0.000577 | 24 | 14.5 | 1.6 | 0.063073 | 0.000419 | 24 | 13 | 1.4 | YES | YES | |||
| Kcnk5 | 0.002233 | 1.66E-05 | 24 | 13 | 3.1 | 0.063073 | 0.000419 | 22 | 11 | 3.2 | YES | ||||
| Klf15 | 0.006556 | 7.56E-05 | 24 | 12 | 1.5 | 0.051058 | 0.000302 | 24 | 12 | 1.6 | YES | YES | |||
| Mad2l2 | 0.000175 | 4.87E-07 | 24 | 2 | 3.0 | 0.035038 | 0.000108 | 22 | 3 | 1.4 | YES | ||||
| Man2a1 | 0.010948 | 0.000153 | 24 | 14 | 1.6 | 0.012876 | 1.66E-05 | 24 | 14 | 1.4 | YES | ||||
| Mapk14 | 0.001229 | 7.32E-06 | 24 | 16 | 1.6 | 0.08455 | 0.00079 | 24 | 14.5 | 1.3 | YES | ||||
| Marveld1 | 1.96E-05 | 1.09E-08 | 24 | 15 | 2.1 | 0.016353 | 2.45E-05 | 24 | 13 | 1.7 | YES | YES | |||
| Mfsd2 | 0.000903 | 4.80E-06 | 24 | 14 | 7.3 | 0.099034 | 0.001074 | 24 | 12.5 | 6.2 | YES | ||||
| Mgrn1 | 0.001229 | 7.32E-06 | 24 | 10 | 1.5 | 0.073204 | 0.000577 | 28 | 5 | 1.3 | YES | YES | |||
| Mov10 | 0.013737 | 0.000216 | 22 | 8 | 1.9 | 0.005596 | 4.80E-06 | 26 | 4 | 1.9 | |||||
| Mpzl1 | 0.001624 | 1.11E-05 | 24 | 0 | 2.6 | 0.073204 | 0.000577 | 22 | 2 | 1.4 | YES | ||||
| Mreg | 0.000133 | 2.96E-07 | 22 | 2 | 2.3 | 0.08455 | 0.00079 | 22 | 2 | 1.4 | |||||
| Mthfr | 1.38E-05 | 5.92E-09 | 24 | 23 | 3.3 | 0.099034 | 0.001074 | 24 | 20 | 1.2 | YES | ||||
| Mtss1 | 0.00861 | 0.000108 | 24 | 12 | 2.1 | 0.001384 | 1.78E-07 | 24 | 13 | 2.6 | YES | YES | |||
| Ndrg1 | 0.000133 | 2.96E-07 | 24 | 2.5 | 2.4 | 0.063073 | 0.000419 | 20 | 5 | 2.3 | YES | YES | |||
| Npas2 | 3.72E-05 | 3.51E-08 | 24 | 1 | 2.7 | 0.037685 | 0.000153 | 24 | 22 | 3.3 | YES | ||||
| Nr1d1 | 4.93E-05 | 6.13E-08 | 26 | 7 | 12.3 | 0.037685 | 0.000153 | 24 | 6 | 1.7 | YES | YES | YES | ||
| Nr1d2 | 1.96E-05 | 1.09E-08 | 24 | 10.5 | 11.3 | 0.037685 | 0.000153 | 24 | 8 | 2.7 | YES | YES | YES | ||
| P2rx4 | 0.013737 | 0.000216 | 23 | 10.5 | 1.7 | 0.063073 | 0.000419 | 28 | 4 | 1.6 | YES | ||||
| Park2 | 0.000631 | 3.11E-06 | 26 | 8 | 1.5 | 0.003676 | 1.26E-06 | 26 | 7 | 1.7 | |||||
| Per1 | 0.000133 | 2.96E-07 | 24 | 13 | 4.3 | 0.022227 | 5.24E-05 | 22 | 12 | 3.7 | YES | YES | |||
| Per2 | 4.78E-06 | 4.10E-10 | 24 | 15 | 5.9 | 0.005596 | 4.80E-06 | 24 | 13 | 5.0 | YES | YES | YES | ||
| Per3 | 0.000175 | 4.87E-07 | 24 | 13 | 11.2 | 0.003676 | 7.88E-07 | 24 | 12.5 | 2.5 | YES | YES | |||
| Pex26 | 0.003061 | 2.45E-05 | 26 | 12 | 1.6 | 0.037685 | 0.000153 | 24 | 12.5 | 1.3 | YES | YES | |||
| Pik3ap1 | 0.010948 | 0.000153 | 24 | 8 | 2.1 | 0.022227 | 5.24E-05 | 24 | 9 | 1.5 | YES | YES | |||
| Pla2g12a | 0.006556 | 7.56E-05 | 22 | 11 | 2.3 | 0.073204 | 0.000577 | 24 | 8 | 1.6 | YES | YES | |||
| Por | 6.35E-06 | 1.63E-09 | 24 | 14 | 3.5 | 0.04346 | 0.000216 | 24 | 13 | 1.6 | YES | YES | |||
| Ppp1r3b | 0.013737 | 0.000216 | 24 | 18.5 | 3.8 | 0.051058 | 0.000302 | 24 | 14 | 1.9 | YES | YES | |||
| Rhbdd2 | 0.000334 | 1.26E-06 | 24 | 14 | 2.3 | 0.016353 | 2.45E-05 | 26 | 11 | 1.4 | YES | ||||
| Rnf125 | 0.010948 | 0.000153 | 22 | 2 | 4.9 | 0.003676 | 1.26E-06 | 22 | 2 | 4.4 | YES | YES | |||
| Scap | 0.00046 | 1.99E-06 | 22 | 8 | 1.6 | 0.08455 | 0.00079 | 26 | 3 | 1.5 | YES | YES | |||
| Serpinf2 | 0.013737 | 0.000216 | 22 | 7 | 1.4 | 0.063073 | 0.000419 | 24 | 5 | 1.4 | YES | ||||
| Sh3bp2 | 0.043499 | 0.001074 | 26 | 8 | 1.2 | 0.099034 | 0.001074 | 28 | 6 | 1.3 | YES | ||||
| Slc17a3 | 0.017929 | 0.000302 | 24 | 21 | 2.1 | 0.037685 | 0.000153 | 26 | 16 | 1.7 | YES | YES | |||
| Slc37a4 | 0.000175 | 4.87E-07 | 24 | 14 | 1.7 | 0.037685 | 0.000153 | 22 | 13 | 2.5 | YES | ||||
| Slc5a6 | 0.00861 | 0.000108 | 24 | 11 | 2.1 | 0.051058 | 0.000302 | 24 | 11 | 1.5 | YES | YES | |||
| Spon2 | 6.48E-05 | 8.33E-08 | 22 | 3 | 2.9 | 0.005578 | 3.11E-06 | 24 | 1 | 2.0 | YES | ||||
| Srm | 0.005073 | 5.24E-05 | 22 | 16 | 1.8 | 0.035038 | 0.000108 | 24 | 13.5 | 2.2 | YES | YES | |||
| St3gal5 | 0.000631 | 3.11E-06 | 24 | 14.5 | 3.2 | 0.028443 | 7.56E-05 | 24 | 11 | 2.4 | YES | YES | |||
| St5 | 0.001624 | 1.11E-05 | 24 | 23 | 5.6 | 0.022227 | 5.24E-05 | 22 | 0 | 1.7 | |||||
| Syt1 | 0.013737 | 0.000216 | 24 | 9 | 3.4 | 0.001728 | 2.96E-07 | 24 | 6 | 1.9 | YES | ||||
| Tef | 3.72E-05 | 3.51E-08 | 24 | 12 | 7.0 | 0.005578 | 3.11E-06 | 24 | 13 | 2.0 | YES | YES | |||
| Tmem218 | 0.001624 | 1.11E-05 | 26 | 1 | 3.1 | 0.001228 | 1.05E-07 | 22 | 2 | 1.8 | YES | ||||
| Tns1 | 0.02232 | 0.000419 | 22 | 15 | 1.8 | 0.012876 | 1.66E-05 | 24 | 12 | 1.5 | YES | ||||
| Tsc22d3 | 0.002233 | 1.66E-05 | 24 | 17 | 2.6 | 0.063073 | 0.000419 | 22 | 13 | 3.1 | YES | YES | YES | ||
| Ubxn1 | 0.000175 | 4.87E-07 | 22 | 5 | 2.0 | 0.012876 | 1.66E-05 | 22 | 5 | 1.6 | YES | ||||
| Ulk1 | 0.010948 | 0.000153 | 22 | 9 | 1.9 | 0.08455 | 0.00079 | 24 | 4 | 1.2 | YES | ||||
| Usp2 | 0.000631 | 3.11E-06 | 24 | 13 | 15.9 | 0.019096 | 3.60E-05 | 24 | 13 | 4.5 | YES | YES | |||
| Wbscr27 | 0.00861 | 0.000108 | 24 | 5 | 1.8 | 0.037685 | 0.000153 | 24 | 4 | 1.5 | YES | ||||
| Wdr45 | 0.010948 | 0.000153 | 22 | 9 | 1.5 | 0.051058 | 0.000302 | 24 | 6 | 1.7 | YES | ||||
| Wrnip1 | 0.006556 | 7.56E-05 | 24 | 0 | 1.8 | 0.005596 | 4.80E-06 | 22 | 3 | 1.6 | YES | YES | |||
| Ypel2 | 0.017929 | 0.000302 | 24 | 22.5 | 3.6 | 0.022227 | 5.24E-05 | 22 | 2 | 2.8 | |||||
| Zfp295 | 9.88E-05 | 1.78E-07 | 24 | 21 | 1.7 | 0.028443 | 7.56E-05 | 24 | 17 | 1.4 | YES | ||||
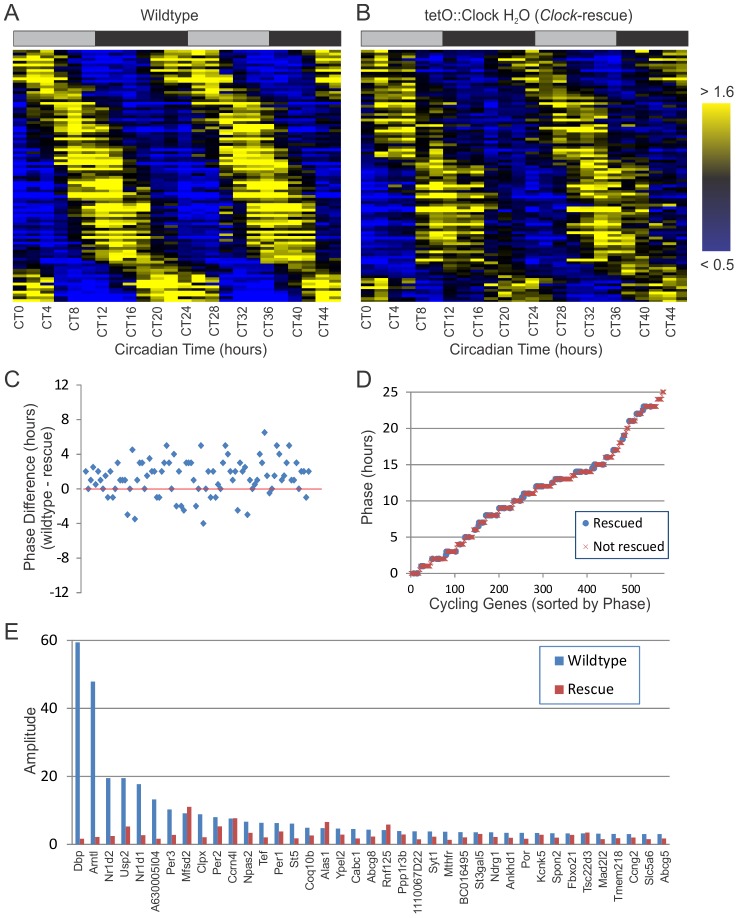
The phase of circadian output rhythms was restored by brain-specific Clock-rescue. The heatmaps in Figure 4A and 4B demonstrate striking similarity between the profiles of the circadian transcriptomes in both sets of animals. Figure 4C shows the phase difference between wildtype and Clock-rescue as a scatter plot for each of the 92 rescued circadian genes. These data points were centered near zero, although there was a modest (∼1.5-hour) phase-advance in Clock-rescue versus wildtype. This phase difference was less than one standard deviation from zero, so we do not consider it to be statistically significant, although we note that the slightly faster behavioral rhythms in Clock-rescue animals may account for this modest phase advance (Figure 1 and Table 1). Figure 4D shows the phases of all rescued (blue circles) and non-rescued genes (red x's). Rescued genes were found with peak expression at every time of day, and there was no significant bias in the phase of rescued versus non-rescued genes (chi-squared test).
Figure 4. Clock-rescue restores appropriate phase to circadian output genes.
24-hour cycling genes (p<0.0011 for both genotypes, period ≥20 hours, N = 92) identified in both wildtype (A) and Clock-rescue animals (B) were median-normalized, sorted by phase, and plotted as a heatmap (yellow = high expression, blue = low expression). Each row represents the temporal expression of one cycling gene common to both datasets. Shaded bars above the heatmaps represent subjective day and night. In (C), the phase difference between wildtype and Clock-rescue is plotted for all 92 rescued genes, ordered by amplitude in wildtype. On average, there is a ∼1.5 h phase advance in the Clock-rescue samples. Panel (D) shows the phases of every cycling gene identified in wildtype, including those rescued in Clock-rescue animals (blue circles) and those not rescued (red x's). There was no bias in the phase of rescued genes. Panel (E) shows the amplitudes of high-amplitude cycling genes (AMP>3.0) in wildtype versus Clock-rescue animals. Several genes have been re-plotted from Figure 2E for the sake of completeness.
Overall, 76 of 92 rescued genes (82%) had lower amplitudes in Clock-rescue animals versus wildtype, and the median amplitude in Clock-rescue was 20% lower than in wildtype. As would be expected given the un-rescued expression of ClockΔ19 in the liver, the most significantly affected genes were high-amplitude cyclers that are direct targets of BMAL1/CLOCK. We also compared this dataset to known Bmal1 and Reverb-alpha target genes (Table 2) [49], [50]. For example, Dbp, Bmal1 (Arntl), Rever-alpha (Nr1d1), Reverb-beta (Nr1d2), Per3, and Tef are among the cycling genes with the most significant amplitude defect in rescue animals (Figure 4E). Likewise, Ubiquitin Specific Protease 2 (Usp2) has significantly diminished amplitude in the rescue, and has also been shown to modify circadian rhythms [51] and is a direct target of CLOCK [52]. Taken as a whole, these data indicate that brain-specific rescue of Clock function is sufficient to restore normal period lengths and phases to a significant fraction of peripheral circadian output. However, the generation of robust circadian output (as measured by the number of cycling genes and their amplitude) depends on an intact peripheral oscillator.
Core clock genes were preferentially rescued by Clock expression in the SCN. We performed DAVID analysis to determine whether rescued genes represented specific pathways or ontologies [53]. We found that core clock genes were the only ontological group significantly enriched in this data set (n = 92, enrichment = 0.79, p<0.0005, q<0.07). Consistent with this observation, we found that core clock genes were preferentially rescued, even compared to other high-amplitude cycling genes (amplitude >5.0 peak∶trough). Of the 11 high-amplitude core clock genes, 9 showed normal rhythms in Clock-rescue mice. In contrast, only 7 of 28 high-amplitude output genes were rescued in these samples. Taken together, we conclude that key components of the circadian clock are sensitive to either direct or indirect signals from the SCN, even in the absence of a functional local circadian clock.
Taking this line of investigation one step further, a direct comparison between our 92 rescued circadian genes and the 31 system-driven genes identified by Kornmann et al. [21] is of obvious importance. Surprisingly, there was very little overlap between these two data sets (Table 2). Only three genes were common to both sets: Per2 (Figure 3D), Nocturnin (Ccrn41) (Figure 5A), and Fbxo21 (Figure 5B). We reasoned that the apparent disagreement between these two data sets may be a consequence of inconsistencies in the underlying statistical analyses. To address this, we re-analyzed Kornmann et al.'s data using JTK_Cycle (Table S3). At the same statistical threshold used in the present study (p<0.0011), we found only three genes that were systemically-driven (Ccdc12, Cry1, and Dbp), none of which were previously identified. In the interest of identifying as many similarities as possible between the present study and Kornmann et al.'s data, we therefore loosened the statistical threshold to p<0.1, which corresponds to a FDR of q<0.41 in the wildtype (non-Reverb-alpha over-expresssing) condition. At this confidence level, we found 47 unique genes that were systemically-driven (Table S3), including five that overlap with Kornmann et al. (1200016E24Rik, 4833417J20Rik, Fus, Hsap1b, Tuba4) and four that overlap with the present study (Alas1, Cabc1, Dbp, and Nr1d2).
Figure 5. Examples of System-driven circadian transcripts.
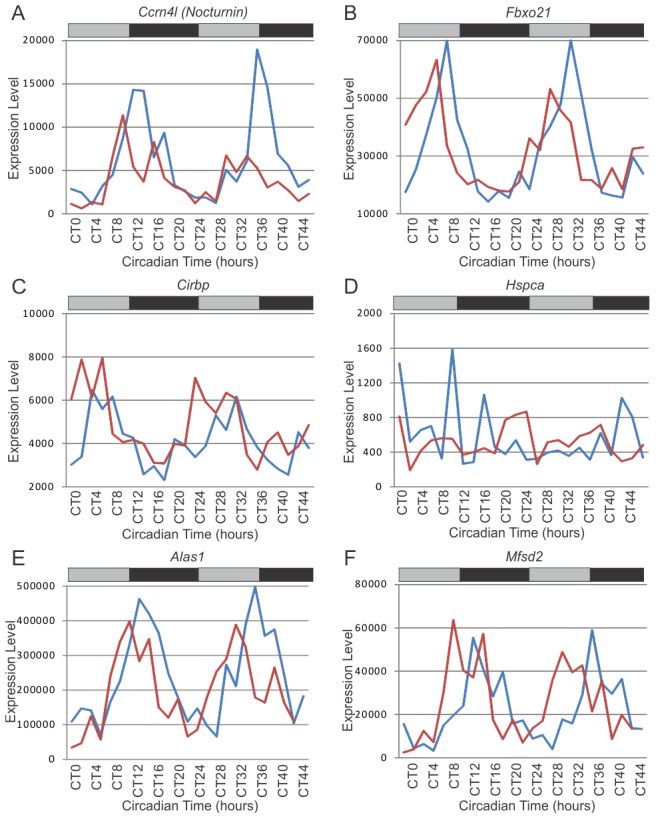
Microarray expression levels in wildtype (blue) and Clock-rescue (red) animals for six notable system-driven genes, including Nocturnin (A), Fbxo21 (B), Cirbp (C), Hspca (D), Alas1 (E), and Mfsd2 (F). Grey and black bars represent subjective day and night, respectively.
Therefore, the apparent disagreement between the present study and Kornmann et al. can be partially reconciled by standardizing the statistical analyses. For example, cold inducible RNA binding protein (Cirbp) was identified and validated by Kornmann et al. as a system-driven gene. Although we do not detect it as being rhythmic in the Clock-rescue condition, if we loosen our statistical threshold, it is clearly rhythmic in both wildtype and Clock-rescue (Figure 5C). Further liberalizing statistical criteria can identify additional similarities between these studies, albeit at the expense of considerably more false-discoveries. Nevertheless, some genes are clearly divergent between these datasets, such as Hspca which is system-driven in Kornmann et al. but arrhythmic in our data (Figure 5D).
We speculate that these differences are due to the different genetic manipulations used in these studies. Kornmann et al. over-expressed Reverb-alpha to systematically inhibit Bmal1-mediated transcription, as well as every other Reverb-alpha target gene. Similarly, our study used ClockΔ19 as a dominant mutant to knock-down Bmal1/Clock activity. However, ClockΔ19 is not expected to dramatically affect Reverb-alpha target genes, which is supported by the presence of 10 Reverb-alpha targets among the 92 rescued circadian genes (Table 2) [50]. Moreover, ClockΔ19 is insufficient to abolish all circadian molecular oscillations, as evidenced by the weak, long-period transcriptional rhythms seen in Clock-mutant mice. For both these reasons, we expected to see rescued transcriptional rhythms not previously seen in Kornmann et al. These rescued transcriptional rhythms could be bona fide system-driven genes, or downstream genes driven by the residual activity of the molecular oscillator in ClockΔ19, or some combination thereof.
Two candidate system-driven genes are shown in Figure 5E and 5F. Aminolevulinic acid synthase 1 (Alas1) was not identified by Kornmann et al., but was detected as system-driven in our re-analysis of their dataset (Table S3). It oscillates with normal period and phase in both wildtype and Clock-rescue, and has an amplitude approximately the same in both genotypes/treatments, as would be expected from a system-driven gene (Figure 4E). Interestingly, Alas1 forms a junction between the circadian clock and heme bioactivity. It is the rate-limiting enzyme in heme biosynthesis and is regulated by Npas2. Additionally, it regulates the activity of Bmal1/Npas2, ultimately affecting the expression of Per1 and Per2 [54], thereby making it a strong candidate for conveying system-driven cues into the peripheral circadian clock.
Similarly, major facilitator superfamily domain containing 2A (Mfsd2) is rhythmic in Clock-rescue animals (Figure 5F). Like other potential system-driven genes (e.g. Per2 and Nocturnin), Mfsd2 expression is largely unchanged between wildtype and Clock-rescue animals (Figure 4E). Interestingly, Mfsd2 is highly induced in liver and brown fat by fasting and cold-induced thermogenesis [55], consistent with (and a possible molecular mechanism for) Kornmann et al.'s hypothesis that temperature is a major entrainer of peripheral circadian clocks. At the least, Mfsd2 is an excellent candidate for conveying nutritional signals to the liver clock.
In addition to 24-hour transcriptional rhythms, the liver and other tissues express ultradian rhythms with period lengths of 12- and 8- hours [19]. A recent study has demonstrated that at least some of these circadian ‘harmonics’ are disorganized in mice with genetically-disrupted circadian oscillations [56]. However, the extent to which these rhythms are driven by central versus peripheral oscillators is unclear.
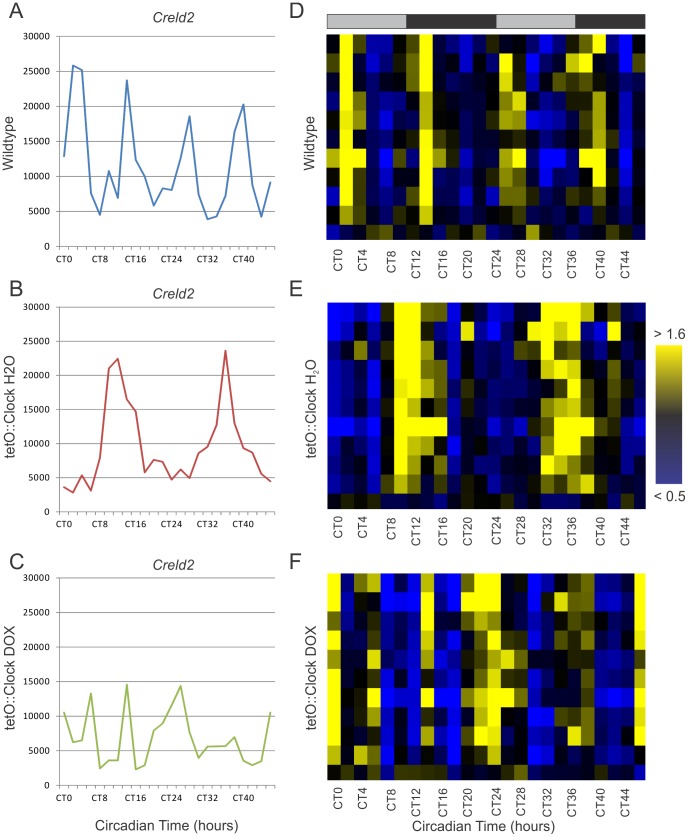
To address this, we examined the transcriptional profiles of circadian harmonics in wildtype, Clock-rescue, and Clock-defective mice. For example, Creld2 was previously identified as a 12-hour oscillator [19], and re-confirmed by this study (Figure 6A). This ultradian oscillator reverted to a 24-hour period length in the Clock-rescue mice (Figure 6B), and became disorganized with lower overall expression in Clock-defective mice (Figure 6C). This phenotype is consistent with the other 12-hour rhythms detected in these data (N = 3, Table 3), as well as previously identified 12-hour cyclers [19] with some evidence of oscillatory behavior (period = ∼12-hours AND p<0.1) in the present data set. In the every case, 12-hour oscillations revert to 24-hour period lengths in Clock-rescue mice (Figure 6D and 6E, and Table 3). Interestingly, the disorganization of these rhythms in Clock-defective mice is consistent between genes (Figure 6F), strongly suggesting that their promoters share common transcriptional inputs. Moreover, the amplitudes of normal 12-hour rhythms and rescued 24-hour rhythms are largely indistinguishable (Figure 7A), indicating that at least one of the two daily peaks of expression in wildtype is largely driven by systemic cues and not the local, peripheral oscillator. Likewise, the phases of the normal and rescued rhythms largely fall into a single cluster (Figure 7B), consistent with the idea that they are responding to identical circulating cues. Given the unambiguous defect in generating 12-hour rhythms in Clock-rescue animals, we conclude that the local, peripheral oscillator is absolutely required for generating 12-hour transcriptional rhythms. However, the appearance of 24-hours in the absence of an intact liver clock suggests that half the peaks of these oscillations are derived from circulating cues downstream of the central oscillator in the SCN.
Figure 6. Clock-rescue restores 24- but not 12-hour rhythms to circadian harmonics.
Panels A–C show the time course of Creld2 expression (a known 12-hour cycling gene) in wildtype (A), Clock-rescue (B) and Clock-mutant (C) livers. Panels D–F show a subset (N = 11) of wildtype 12-hour rhythms median-normalized and plotted as a heatmap (yellow = high expression, blue = low expression). Every line represents the temporal expression of one wildtype 12-hour cycling gene. Shaded bars above the heatmaps represent subjective day and night.
Table 3. 12-hour transcriptional rhythms revert to 24-hour rhythms when Clock is driven in the SCN.
| Wildtype | tetO::Clock H2O | tetO::Clock DOX | ||||||||
| Probe Set ID | Gene | p-value (FDR) | Period (hours) | Phase (hours) | p-value (FDR) | Period (hours) | Phase (hours) | p-value (FDR) | Period (hours) | Phase (hours) |
| 6837437 | Arfgap3 | 0.027 (0.45) | 12 | 2.0 | 0.034 (0.61) | 22 | 11.0 | 1.00 (1.0) | 28.4 | 16.9 |
| 6800427 | Dnajb9 | 0.003 (0.08) | 12 | 2.0 | 0.286 (1.0) | 20 | 12.0 | 1.00 (1.0) | 28.4 | 17.5 |
| 6992306 | Gmppb | 3.02 E-04 (0.02) | 12 | 2.0 | 7.56 E-05 (0.03) | 24 | 13.0 | 1.00 (1.0) | 28.4 | 15.1 |
| 6997671 | Tmed3 | 0.043 (0.61) | 12 | 3.0 | 0.003 (0.18) | 24 | 12.0 | 0.193 (1.0) | 12 | 1.0 |
| 6992409 | Nme6 | 0.054 (0.71) | 12 | 3.0 | 0.001 (0.10) | 24 | 12.0 | 1.00 (1.0) | 28.4 | 15.5 |
| 6998396 | Srprb | 0.003 (0.10) | 12 | 3.0 | 2.16 E-04 (0.04) | 24 | 12.5 | 1.00 (1.0) | 28.4 | 16.4 |
| 6832530 | Creld2 | 0.010 (0.22) | 12 | 3.0 | 1.26 E-06 (0.004) | 24 | 13.0 | 1.00 (1.0) | 28.4 | 16.1 |
| 6876342 | Hspa5 | 0.001 (0.04) | 12 | 3.0 | 1.53 E-04 (0.38) | 24 | 13.0 | 0.235 (1.0) | 30 | 27.0 |
| 6807251 | Tmed9 | 0.068 (0.82) | 12 | 3.0 | 0.006 (0.24) | 24 | 13.0 | 1.00 (1.0) | 28.4 | 15.1 |
| 6881837 | Sec23b | 2.45 E-05 (0.003) | 12 | 3.0 | 0.001 (0.10) | 24 | 13.0 | 0.416 (1.0) | 12 | 0.0 |
| 6817412 | Samd8 | 0.005 (0.12) | 12 | 7.0 | 0.104 (1.0) | 20 | 5.0 | 1.00 (1.0) | 28.4 | 9.4 |
Figure 7. Clock-rescue restores normal amplitude and phase to circadian harmonics.
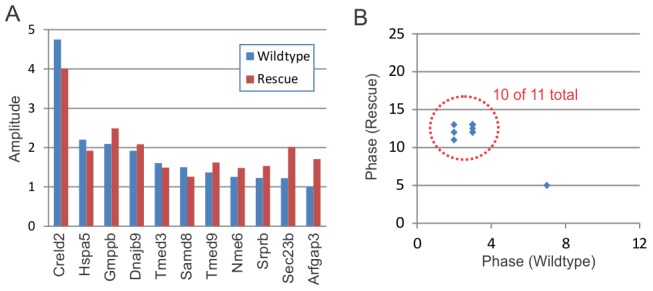
Panel (A) shows the amplitude of circadian harmonics in wildtype (blue) and Clock-rescue (red) animals. In every case, there was no obvious diminishment of amplitude in the rescue animals. Panel (B) plots the phase of these rhythms in wildtype (x-axis) versus Clock-rescue (y-axis). 10 of the 11 total genes had similar phases in both genotypes.
Discussion
The circadian regulatory network is organized in a hierarchical fashion with signals originating in the SCN orchestrating rhythms in peripheral tissues. Understanding the nature of these signals and how they are communicated to peripheral tissues is a significant challenge for the field [57]. Here, we have used the tet-OFF expression system to rescue normal circadian function in the brains of otherwise circadian-defective mice. This approach allowed us to dissect aspects of peripheral rhythms that depend on central oscillator function from those that depend on local circadian oscillators.
Using microarray analysis to characterize the circadian transcriptome of wildtype, Clock-rescue, and Clock-mutant mice, we found that brain-specific rescue of Clock partially restores circadian transcriptional output in the liver. The number of cycling genes was significantly higher in Clock-rescue mice compared to Clock-defective mice (Figure 2B). Likewise, the confidence with which core clock genes were identified as cycling was greater in the rescue animals (Figure 2D). Both period length (Figure 3) and phase (Figure 4) of transcriptional rhythms in the liver of Clock-rescue mice were consistent with their wildtype counterparts. In contrast, rhythms in Clock-defective mice were out-of-phase with wildtype and had significantly longer period lengths. These results agree with studies of transplanted fibroblasts that have been shown to synchronize their local circadian oscillators to the SCN of their host [58]. Taken as whole, these data indicate that signaling downstream of the central circadian oscillator is sufficient to reset the period and phase of many peripheral transcriptional rhythms in tissues that otherwise have disrupted circadian oscillations.
Nevertheless, the total amount of circadian transcriptional output (as measured by the number of cycling genes and their amplitudes) was still significantly less than wildtype (see Figure 2B and 2E, Figure 3D and 3E, and Figure 4). This observation is consistent with a recent study by Kornmann et al. that profiled the liver circadian transcriptome in ad libitum fed mice with disrupted circadian oscillations in the liver due to the over-expression of Rev-erb-alpha [21]. In that study, ∼10% of cycling transcripts (31 of 351) continued to oscillate with normal periods and phases independently of the genetic manipulation to their peripheral clock, although the amplitudes of these rhythms were generally diminished. Similarly, we saw 16.5% (952 of 576) of genes had restored oscillations in Clock-rescue animals at a threshold of p<0.0011, corresponding to a FDR of q<0.05 (Figure 2C).
Unlike the Kornmann et al. study, however, we detected 24-hour oscillations of many key circadian genes in Clock-rescue animals, including Nr1d1, Arntl, Per1/Per2/Per3, and Dbp/Tef/Hlf (Table 2). This disagreement between the present study and Kornmann et al. may not be as dramatic as it initially appears. When re-analzying the Kornmann et al. dataset with JTK_Cycle, we found Cry1, Dbp, and Nr1d2 to all be system-driven. We therefore conclude that 24-hour periodicity in many components of the core circadian clock can be driven by oscillations in the SCN. The extent to which this rescue of peripheral clock periodicity is due to restored behavioral rhythmicity, humoral cues, or their interaction is unknown.
We noticed that core clock genes were preferentially rescued by restoring CNS clock function. This held true even when compared with other high amplitude oscillating genes in the liver. We speculate that the promoters of clock genes may have evolved configurations of response elements that are particularly sensitive to humoral and behavioral cues. One can imagine that this property – sensitivity to the CNS clock – would be particularly advantageous for resetting to different light schedules. Core clock gene action, subsequently, would then synchronize tissue specific peripheral gene expression appropriately.
At a broader level, we discovered 92 circadian genes (including clock factors) that have normal phases and periodicity in Clock-rescue animals. Since the rescue of Clock expression is largely confined to the brain, this restoration of normal phase and period is may be due to direct signaling from humoral cues emerging from the SCN, or alternatively due to indirect signaling, via the SCN's regulation of locomotor and feeding rhythms. In either case, a subset of rescued transcriptional rhythms is expected to be system-driven.
Kornmann et al. have previously identified 31 system-driven circadian genes by over-expressing Reverb-alpha in the liver [21]. Surprisingly, there were only three genes in common between their dataset and the present study. Even when using standardized statistical methods, only 7 of the 92 rescued genes identified herein were found in common with Kornmann et al. This is not entirely unexpected given the difference in the genetic manipulations used. For example, over-expressing Reverb-alpha would be expected to inhibit the expression of any genes under the control of ROR elements. Consistent with this idea, we found 10 rescued genes not previously identified by Kornmann et al. that are known targets of Reverb-alpha [50].
The genetic lesion used in this study, ClockΔ19, significantly diminishes the number of rhythmic genes as well as their amplitudes. However, some rhythmicity still persists, as evidenced by the weak, long-period rhythms still seen in Clock-mutant animals (Figure 2). At a mechanistic level, this implies that rescued genes in our dataset may be bona fide system-driven genes or peripheral clock-driven genes that are insensitive to CLOCK signaling. Consequently, strong candidates for system-driven genes such as Per2 and Nocturnin (identified by both Kornmann et al. and the present study) have amplitudes that are largely independent of genetic manipulation (Figure 4E). Based on this criteria, we identified two novel system-driven candidates: Alas1 and Mfsd2. Alas1 regulates and is regulated in turn by clock factors, while providing a link between the clock and heme biosynthesis [54]. Similarly, Mfsd2 is known to be driven by the clock, and is upregulated by fasting and cold-induced thermogenesis [55]. Both genes are thus potential nodes through which systemic cues synchronize and drive peripheral circadian rhythms.
Both our study and Kornmann et al. agree that the number of system-driven circadian genes may be relatively high. Even accounting for the measurable false discovery rate of both studies, there are likely several dozen and potentially as many as a hundred cycling genes in the liver than can be driven in part by systemic cues. We speculate that this may be an evolutionary strategy to tightly link peripheral clocks with the physiological status of the animal. Unlike the SCN, peripheral clocks (especially the liver) are sensitive to a wide array of behavioral, environmental, and physiological stimuli. By having many different input pathways to synchronize liver rhythms, evolution may have built considerable redundancy into the system. This is analogous to the coupling of SCN neurons, which renders the entire timekeeping mechanism considerably more resistant to perturbation [59], [60].
Similarly to 24-hour rhythms, circadian harmonics (i.e., 12- and 8-hour rhythms), are transcriptional oscillations found in tissues throughout the body that persist in constant darkness [19], [56]. These rhythms have been confirmed to exist at the protein level, and may be a consequence of rhythms of lipid metabolism and ER stress [56]. Consistent with this observation, 12-hour transcriptional rhythms are disrupted in mice subjected to restricted feeding [19]. Nevertheless, the extent to which central and peripheral oscillators contribute to circadian harmonics is an open question.
We found that disrupting Clock function throughout the body disorganized and diminished 12-hour rhythms, indicating that these rhythms derive (at least in part) from the conventional 24-hour circadian clock (Figure 5 and Table 3). Interestingly, brain-specific rescue of Clock function restored 24- but not 12-hour rhythmicity to these genes, with no discernible loss of amplitude (Figure 6, Figure 7A, and Table 3). This observation is reminiscent of previous studies that demonstrated that both restricted feeding and Hlf/Dbp/Tef loss-of-function convert 12-hour rhythms to 24-hour period lengths [19], [56]. Moreover, the phase of 12-hour transcription is precisely the same regardless of which tissues are examined, suggesting a common signaling origin [19].
Since 24-hour periodicity of most clock genes is restored in the livers of Clock-rescue mice (albeit at consistently low amplitudes), we acknowledge that the rescue of 12-hour rhythms may be downstream of local oscillations in the liver. Nevertheless, we favor the explanation that circulating, tissue-non-autonomous signaling cues drive one of the two daily peaks of 12-hour transcriptional rhythms. Two observations support this interpretation. First, the amplitude of rescued circadian harmonics is largely the same as in wildtype (Figure 7B). In contrast, most core clock genes and key circadian outputs have low-amplitude oscillations in the rescued livers (Figure 4E and Table 2). We consider it more likely that systemic cues downstream of fully rescued SCN and the resulting behavioral rhythms drive these rescued harmonic oscillations rather than low-amplitude oscillations of clock genes in the liver. Second, previous studies in dissociated cells with normal molecular oscillations have consistently failed to detect harmonic transcriptional rhythms [19], [46]. Although these studies do not speak to the necessity of peripheral circadian clocks for generating 12-hour rhythms, they do indicate that cell-autonomous circadian oscillations are not sufficient for generating circadian harmonics. Regardless, based on the present study we can conclude for the first time that the conventional 24-hour circadian oscillator (whether central or peripheral) is the ultimate origin of harmonic transcriptional rhythms, and circulating cues downstream of the SCN are sufficient to restore one of the two daily peaks of expression.
Materials and Methods
Circadian tissue collection
Mice were housed in light-tight boxes and entrained to a 12 hour light, 12 hour dark schedule for one week before being switched to complete darkness. Wildtype mice (C57BL/6J) were acquired from Jackson Labs; experimental animals (Scg2::tTA; tetO::Clockwt; ClockΔ19/Δ19) were generated as previously described [36], [40]. The Scg2::tTA mice were congenic on a C57BL/6J background, and the tetO::Clockwt transgenic mice were co-isogenic on a C57BL/6J background. Mice were supplied with regular food and water ad libitum. Three days before the final lights off (at ZT12), experimental animals were treated with either water or doxycycline (30 ug/ml; Sigma-Aldrich) supplied in the drinking water. Starting two days after the first day in DD (i.e., at CT48), four wildtype mice (two females and two males) and Clock-rescue mice (one female and one male) were sacrificed in the dark every two hours. Liver samples were quickly excised and snap-frozen in liquid nitrogen. Liver samples were homogenized in Trizol (Invitrogen) and total RNA was purified using the manufacturer's protocol. All animal experiments were performed with the approval of the Committee on Animal Care and Use at Northwestern University.
Circadian activity analysis
Mice were placed in individual running wheel cages and activity was recorded and analyzed using the ClockLab Data Collection System (Actimetrics, Wilmette, IL). Mice were entrained to a 12-hour light/12-hour dark cycle (LD) for a minimum of 10 days before they were released into constant darkness (DD). Doxycycline was supplied in the drinking water at a concentration of 10 µg/ml for the behavioral analysis as described previously [36], [40]. Mice were supplied with regular food and water with or without doxycycline ad libitum. Dox-containing water was renewed every 2–3 days.
Quantitative PCR
RNA isolated from the liver were reverse transcribed with SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). cDNA (1.25 µl) was pre-amplified for specific target amplification (STA) using pooled 0.2× TaqMan Gene Expression Assays mix (Applied Biosystems). The thermal cycling conditions used for STA were 95°C hold for 10 minutes followed by 14 cycles of 95°C for 15 seconds and 60°C for 4 minutes. TaqMan Gene Expression Assays (Applied Biosystems) used in this study are listed in the supplemental data (Table S4). Preamplified cDNA was diluted with TE buffer (1∶5) and qPCR assays were performed using the BioMark 48.48 Dynamic Array as specified (Fluidigm). Assays were run in triplicate on each array and data were analyzed by the use of BioMark Real-Time PCR Analysis Software Version 2.0 (Fluidigm) to obtain Ct and Δ ΔCt values. Quantitative PCR analysis (Figure S1) was performed from the same RNA samples as those used for the microarray analysis (see below).
Microarray analysis
5 µg total RNA per time point was submitted to the University of Pennsylvania School of Medicine Microarray Facility for labeling and hybridization to Affymetrix Mouse Exon 1.0 ST Arrays. Expression values were extracted using RMA implemented in Expression Console (Affymetrix) at the core gene level. JTK_Cycle implemented in R (version 2.12.1, 64-bit) was used to detect cycling genes as previously described [37], using a period length window of 10–40 hours. Due to the lower sampling resolution, re-analysis of the Kornmann et al. 2009 dataset was performed using a 24 hour period length window. Raw data and statistics were compiled into an Access database (Microsoft). Heatmaps were generated using custom scripts implemented in MATLAB (Mathworks, version R2010b). DAVID analysis was performed as previously described [53], using all rhythmic genes in wildtype (N = 576, q<0.05, p<0.0011) as a background list, and all 24-hour rescued genes (n = 92) as the principal gene list. Amplitude estimates were made using JTK_Cycle, modified as previously described ((2*JTK.AMP)+(percentile(array, 0.1)/(percentile(array, 0.1)) [61] All .CEL files are available from GEO (accession number: GSE30411); custom scripts for MATLAB and R are available on demand.
Supporting Information
qPCR validation of microarray data in core clock genes. Wildtype rhythms (top panels, solid blue lines, right axis) for Per1 (A), Per2 (B), Per3 (C), Arntl (D), Npas2 (E), and Nr1d1 (F) show robust circadian oscillations with phases in close agreement with microarray data (grey dashed lines, left axis). Although lower amplitude in Clock-rescue animals (middle panels, red solid lines, right axis), these samples show appropriate period lengths and phases, in agreement with microarray data (grey dashed lines, left axis). In contrast, Clock-mutant mice (lower panels, green solid lines, right axis) show long-period rhythms out of phase with the wildtype samples and in agreement with microarray data (grey dashed lines, left axis). This apparent phase difference is believed to be a consequence of the free-running period length phenotype of Clock-mutant mice. Input RNA samples were the same for qPCR and microarray measurements and should therefore be considered technical replicates.
(TIF)
Cycling genes and their amplitudes. Microarray expression levels for circadian genes (p<0.0011, period ≥20 hours) in wildtype (A), Clock-rescue (C), and Clock-mutant (E) animals were median-normalized, sorted by phase, and plotted as a heatmap (yellow = high expression, blue = low expression). Shaded bars above the heatmaps represent subjective day and night. The amplitudes of these cycling genes are shown for wildtype (B), Clock-rescue (D), and Clock-mutant (F) animals (wildtype N = 570, Clock-rescue N = 248, Clock-mutant N = 63).
(TIF)
JTK_Cycle statistics of the top cycling genes in wildtype, tetO::Clock H2O, and tetO::Clock DOX datasets (JTK_Cycle p-value<0.1; key circadian genes shown in bold).
(XLSX)
Non-wildtype cycling genes. There are 187 genes that cycle (p<0.0011) in Clock-rescue and Clock-mutant animals but do not cycle in wildtype. Of these, 102 were previously found to oscillate in wildtype animals [19].
(XLSX)
Acknowledgments
We thank Jeanne Geskes, Vinhfield Ta, the Northwestern Animal Facility, and the University of Pennsylvania School of Medicine Microarray facility for excellent technical assistance. We also thank Julie Baggs, Jack Qian, and members of the Takahashi and Hogenesch lab for helpful discussions during the course of these studies.
Footnotes
The authors have declared that no competing interests exist.
MEH is supported by NIH grant F32GM096577-01. This work was supported by the National Institute of Mental Health (NIMH) P50 Conte Center grant MH074924 (Research Center Grant awarded to Center Director JST, Project Principal Investigator JBH); the National Heart, Lung, and Blood Institute (NHBLI, 1R01HL097800); and Pennsylvania Commonwealth Health Research Formula Funds (JBH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 2.Andretic R, Franken P, Tafti M. Genetics of sleep. Annu Rev Genet. 2008;42:361–388. doi: 10.1146/annurev.genet.42.110807.091541. doi: 10.1146/annurev.genet.42.110807.091541. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 4.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 5.Klerman EB. Clinical aspects of human circadian rhythms. J Biol Rhythms. 2005;20:375–386. doi: 10.1177/0748730405278353. doi: 10.1177/0748730405278353. [DOI] [PubMed] [Google Scholar]
- 6.Curtis AM, Fitzgerald GA. Central and peripheral clocks in cardiovascular and metabolic function. Ann Med. 2006;38:552–559. doi: 10.1080/07853890600995010. [DOI] [PubMed] [Google Scholar]
- 7.Halberg F, Cornélissen G, Ulmer W, Blank M, Hrushesky W, et al. Cancer chronomics III. Chronomics for cancer, aging, melatonin and experimental therapeutics researchers. J Exp Ther Oncol. 2006;6:73–84. [PMC free article] [PubMed] [Google Scholar]
- 8.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 9.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 Spec No 2:R271–277. doi: 10.1093/hmg/ddl207. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 10.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debruyne JP. Oscillating perceptions: the ups and downs of the CLOCK protein in the mouse circadian system. J Genet. 2008;87:437–446. doi: 10.1007/s12041-008-0066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 13.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 14.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 15.Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 16.Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 17.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes M, Deharo L, Pulivarthy SR, Gu J, Hayes K, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol. 2007;72:381–386. doi: 10.1101/sqb.2007.72.011. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storch K-F, Lipan O, Leykin I, Viswanathan N, Davis FC, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 21.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K, Diangelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13:639–654. doi: 10.1016/j.cmet.2011.05.001. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y, Han M, Shimada B, Wang L, Gibler TM, et al. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proceedings of the National Academy of Sciences. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, et al. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 26.Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, et al. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 28.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 29.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 31.Storch K-F, Paz C, Signorovitch J, Raviola E, Pawlyk B, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 33.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 34.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, et al. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 35.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong H-K, Chong JL, Song W, Song EJ, Jyawook AA, et al. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genet. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schönig K, Bujard H. Generating conditional mouse mutants via tetracycline-controlled gene expression. Methods Mol Biol. 2003;209:69–104. doi: 10.1385/1-59259-340-2:69. [DOI] [PubMed] [Google Scholar]
- 39.Mahata SK, Mahata M, Marksteiner J, Sperk G, Fischer-Colbrie R, et al. Distribution of mRNAs for Chromogranins A and B and Secretogranin II in Rat Brain. Eur J Neurosci. 1991;3:895–904. doi: 10.1111/j.1460-9568.1991.tb00101.x. [DOI] [PubMed] [Google Scholar]
- 40.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oishi K, Miyazaki K, Ishida N. Functional CLOCK is not involved in the entrainment of peripheral clocks to the restricted feeding: entrainable expression of mPer2 and BMAL1 mRNAs in the heart of Clock mutant mice on Jcl:ICR background. Biochem Biophys Res Commun. 2002;298:198–202. doi: 10.1016/s0006-291x(02)02427-0. [DOI] [PubMed] [Google Scholar]
- 43.Ochi M, Sono S, Sei H, Oishi K, Kobayashi H, et al. Sex difference in circadian period of body temperature in Clock mutant mice with Jcl/ICR background. Neurosci Lett. 2003;347:163–166. doi: 10.1016/s0304-3940(03)00688-8. [DOI] [PubMed] [Google Scholar]
- 44.Kennaway DJ, Varcoe TJ, Mau VJ. Rhythmic expression of clock and clock-controlled genes in the rat oviduct. Mol Hum Reprod. 2003;9:503–507. doi: 10.1093/molehr/gag067. [DOI] [PubMed] [Google Scholar]
- 45.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atwood A, DeConde R, Wang SS, Mockler TC, Sabir JSM, et al. Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis. Proc Natl Acad Sci U S A. 2011;108:18560–18565. doi: 10.1073/pnas.1115753108. doi: 10.1073/pnas.1115753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keegan KP, Pradhan S, Wang J-P, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scoma HD, Humby M, Yadav G, Zhang Q, Fogerty J, et al. The de-ubiquitinylating enzyme, USP2, is associated with the circadian clockwork and regulates its sensitivity to light. PLoS ONE. 2011;6:e25382. doi: 10.1371/journal.pone.0025382. doi: 10.1371/journal.pone.0025382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 53.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 55.Angers M, Uldry M, Kong D, Gimble JM, Jetten AM. Mfsd2a encodes a novel major facilitator superfamily domain-containing protein highly induced in brown adipose tissue during fasting and adaptive thermogenesis. Biochem J. 2008;416:347–355. doi: 10.1042/BJ20080165. doi: 10.1042/BJ20080165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47–57. doi: 10.1016/j.cmet.2009.11.002. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Kennedy D, Norman C. What Don't We Know? Science. 2005;309:75. doi: 10.1126/science.309.5731.75. doi: 10.1126/science.309.5731.75. [DOI] [PubMed] [Google Scholar]
- 58.Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–117. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- 59.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyazaki M, Schroder E, Edelmann SE, Hughes ME, Kornacker K, et al. Age-Associated Disruption of Molecular Clock Expression in Skeletal Muscle of the Spontaneously Hypertensive Rat. PLoS ONE. 2011;6:e27168. doi: 10.1371/journal.pone.0027168. doi: 10.1371/journal.pone.0027168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qPCR validation of microarray data in core clock genes. Wildtype rhythms (top panels, solid blue lines, right axis) for Per1 (A), Per2 (B), Per3 (C), Arntl (D), Npas2 (E), and Nr1d1 (F) show robust circadian oscillations with phases in close agreement with microarray data (grey dashed lines, left axis). Although lower amplitude in Clock-rescue animals (middle panels, red solid lines, right axis), these samples show appropriate period lengths and phases, in agreement with microarray data (grey dashed lines, left axis). In contrast, Clock-mutant mice (lower panels, green solid lines, right axis) show long-period rhythms out of phase with the wildtype samples and in agreement with microarray data (grey dashed lines, left axis). This apparent phase difference is believed to be a consequence of the free-running period length phenotype of Clock-mutant mice. Input RNA samples were the same for qPCR and microarray measurements and should therefore be considered technical replicates.
(TIF)
Cycling genes and their amplitudes. Microarray expression levels for circadian genes (p<0.0011, period ≥20 hours) in wildtype (A), Clock-rescue (C), and Clock-mutant (E) animals were median-normalized, sorted by phase, and plotted as a heatmap (yellow = high expression, blue = low expression). Shaded bars above the heatmaps represent subjective day and night. The amplitudes of these cycling genes are shown for wildtype (B), Clock-rescue (D), and Clock-mutant (F) animals (wildtype N = 570, Clock-rescue N = 248, Clock-mutant N = 63).
(TIF)
JTK_Cycle statistics of the top cycling genes in wildtype, tetO::Clock H2O, and tetO::Clock DOX datasets (JTK_Cycle p-value<0.1; key circadian genes shown in bold).
(XLSX)
Non-wildtype cycling genes. There are 187 genes that cycle (p<0.0011) in Clock-rescue and Clock-mutant animals but do not cycle in wildtype. Of these, 102 were previously found to oscillate in wildtype animals [19].
(XLSX)