Abstract
Amino acids can induce yeast cell adhesion but how amino acids are sensed and signal the modulation of the FLO adhesion genes is not clear. We discovered that the budding yeast Saccharomyces cerevisiae CEN.PK evolved invasive growth ability under prolonged nitrogen limitation. Such invasive mutants were used to identify amino acid transporters as regulators of FLO11 and invasive growth. One invasive mutant had elevated levels of FLO11 mRNA and a Q320STOP mutation in the SFL1 gene that encodes a protein kinase A pathway regulated repressor of FLO11. Glutamine-transporter genes DIP5 and GNP1 were essential for FLO11 expression, invasive growth and biofilm formation in this mutant. Invasive growth relied on known regulators of FLO11 and the Ssy1-Ptr3-Ssy5 complex that controls DIP5 and GNP1, suggesting that Dip5 and Gnp1 operates downstream of the Ssy1-Ptr3-Ssy5 complex for regulation of FLO11 expression in a protein kinase A dependent manner. The role of Dip5 and Gnp1 appears to be conserved in the S. cerevisiae strain ∑1278b since the dip5 gnp1 ∑1278b mutant showed no invasive phenotype.
Secondly, the amino acid transporter gene GAP1 was found to influence invasive growth through FLO11 as well as other FLO genes. Cells carrying a dominant loss-of-function PTR3647::CWNKNPLSSIN allele had increased transcription of the adhesion genes FLO1, 5, 9, 10, 11 and the amino acid transporter gene GAP1. Deletion of GAP1 caused loss of FLO11 expression and invasive growth. However, deletions of FLO11 and genes encoding components of the mitogen-activated protein kinase pathway or the protein kinase A pathway were not sufficient to abolish invasive growth, suggesting involvement of other FLO genes and alternative pathways. Increased intracellular amino acid pools in the PTR3647::CWNKNPLSSIN-containing strain opens the possibility that Gap1 regulates the FLO genes through alteration of the amino acid pool sizes.
Introduction
Nitrogen is a vital metabolite in living organisms and nitrogen metabolism governs major developmental decisions in Saccharomyces cerevisiae. For example, nitrogen starvation triggers diploid cells to undergo meiosis [1] and can lead to quiescence of both haploid and diploid cells [2]. Low concentrations of nitrogen induce pseudohyphal growth through elongated growth and polar budding [3] while propagation of haploid cells on rich complex medium leads to biofilm formation and invasive growth [4], [5].
Biofilm, pseudohyphal growth and haploid invasive growth are dependent on the cell wall adhesin gene FLO11 (MUC1), which confers cell-surface adhesion [6], [7]. Cellular access to nitrogen is linked to FLO11 expression through the cyclic AMP-protein kinase A (cAMP-PKA) pathway and the general transcription factor Gcn4 [8], [9]. However, a central question is how the yeast cell senses extracellular nitrogen in the form of amino acids and the signal is transmitted to activation of FLO11.
Nitrogen in the form of ammonium induces transcription of FLO11 via the high-affinity ammonium transporter Mep2 and several observations support the hypothesis that Mep2 is an ammonium receptor [8], [10]. However, the ability to transport ammonium appears to be essential for the signaling function of Mep2 [11] suggesting that Mep2 is a transceptor or transports ammonium to an intracellular receptor. Mep2 is proposed to induce expression of FLO11 via the cAMP-PKA pathway, based on the findings that exogenous cAMP, dominant active RAS2 or GPA2 alleles are able to restore pseudohyphal differentiation in a Δmep2/Δmep2 strain [8].
Lorenz and Heitman observed that pseudohyphal growth on proline and glutamine-based medium was not dependent on MEP2 [8] and hypothesized that amino acid transporters could have a function similar to Mep2. In the current work we have explicitly tested the role of amino acid transporters in the regulation of FLO11, since these transporters have not yet been shown to play a role in biofilm formation, pseudohyphal or invasive growth.
Glutamine transport is mediated by Gnp1, Agp1, Gap1 and Dip5 [12]–[15] while proline is transported by Put4 and the general amino acid transporter Gap1 [16]. Transcription of GNP1, AGP1 and DIP5 is induced by extracellular amino acids through the Ssy1-Ptr3-Ssy5 (SPS) complex and deletion of SSY1 leads to a 5–15 fold reduction in DIP5 and GNP1 expression [13], [17], [18]. Counter intuitively, deletion of SSY1 or PTR3 lead to increased invasive growth in the Σ1278b strain background [17] while the opposite would be expected if invasive growth only depended on GNP1, AGP1 or DIP5. A cause of the invasive growth in the ssy1 and ptr3 mutants may be increased expression of GAP1 and higher levels of intracellular amino acids.
Here we investigated the importance of the amino acid transporters Gap1, Dip5 and Gnp1 in regulation of FLO11-dependent invasive growth. We hypothesize that (i) the general amino acid permease Gap1 is essential for invasive growth and FLO11 expression under conditions where GAP1 is highly expressed and (ii) that Gnp1 and Dip5 are essential for FLO11 dependent invasive growth in conditions where GAP1 is not expressed.
Results
Nitrogen-limited S. cerevisiae Populations Evolve Invasive Growth
To investigate the role of amino acids transporters for invasive growth and FLO11 regulation, we initially tested the phenotype of gnp1, dip5 and gap1 mutants in the Σ1278b strain background but neither of these single gene deletions led to the loss of the invasive growth phenotype (Fig. S1). In an alternative approach we selected invasive mutants and used these as background for further analysis of amino acid regulation of FLO11. Invasive mutants appeared in populations of the normally noninvasive S. cerevisiae (CEN.PK113-7D) when propagated under nitrogen limitation (Fig. 1A–D). Clones were isolated from two haploid CEN.PK113-7D populations of 1010 cells after 250 generations in continuous bioreactors, limited for ammonium or glutamine respectively.
Figure 1. Invasive growth of haploid descendants after prolonged nitrogen limitation.
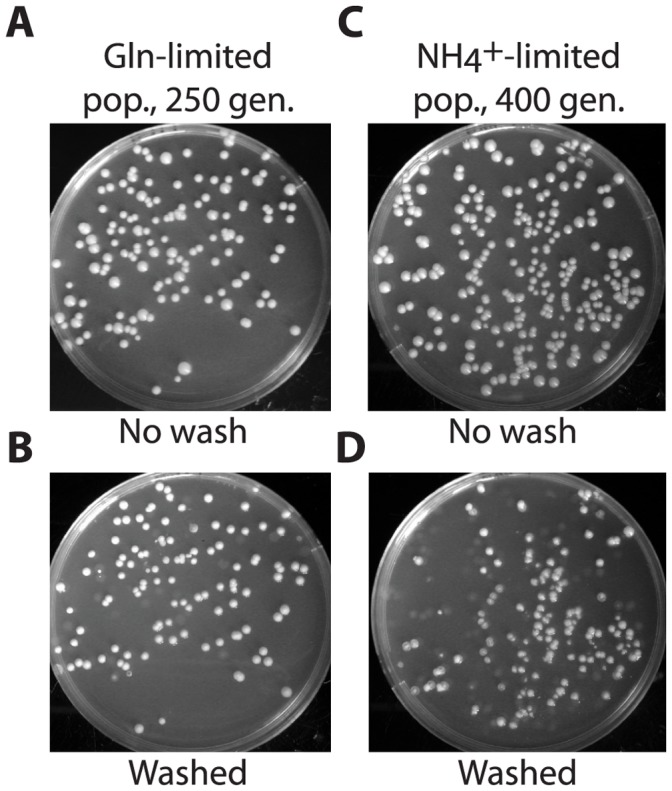
Descendants of CEN.PK113-7D in A, glutamine-limited culture, and C, ammonium-limited culture, grown on rich complex medium (YPD) for 3 days at 30°C and flushed with water to determine the proportion of adhering clones (B and D). Of descendants from the glutamine-limited population, 80% were invasive on YPD, while in the two ammonium-limited populations, 40% and 26% percent had become invasive (n>200).
Clones Sfl1Q320STOP and Ptr3647::C…N Induce Invasive Growth
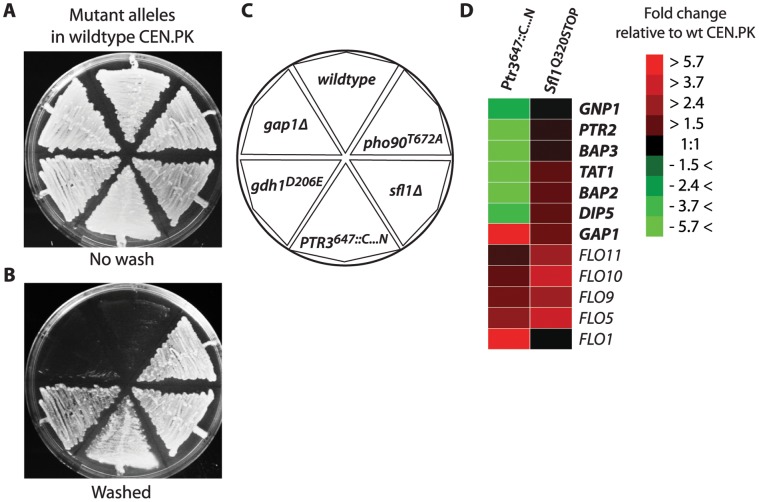
To characterize clones from nitrogen-limited populations, we compared the genomes of six clones to the wildtype using DNA tiling arrays, revealing 12 to 88 single nucleotide polymorphisms (SNPs) per genome. We characterized the SNPs that affected ORFs by PCR and Sanger sequencing and verified up to two mutations in ORFs per clone (Table 1). Mutant alleles identified in the descendants were subsequently introduced into the wildtype strain to test if these caused the invasive growth phenotype. All gave rise to invasive phenotypes, except for a deletion of the general amino acid permease gene GAP1 (Fig. 2A–C).
Table 1. Peptide modifications in six clonal isolates from prolonged nitrogen-limited populations.
| Gene | Glutamine-limited clones | Ammonium-limited clones | ||||
| I | II | III | IV | V | VI | |
| Invasive | Invasive | Non-invasive | Non-invasive | Invasive | Invasive | |
| GAP1 | Δgap1 | S388P | Δgap1 | |||
| MEP2 | G352S | |||||
| PHO90 | T672A | |||||
| SFL1 | Q320 STOP | |||||
| PTR3 | 647::CWNKNPLSSIN | |||||
| GDH1 | D206E | |||||
| SAM4 | Synonymous | |||||
Denoted are the protein modifications resulting from mutations in the clonal isolates. The corresponding mutations are described in Material and Methods.
Figure 2. Mutant alleles restore invasive growth in wildtype strain and invasiveness correlates with increased FLO11 mRNA.
Mutations from invasive descendants of glutamine- and ammonium-limited populations were reconstituted in the wildtype strain (A) and tested for invasive growth (B). Distribution of strains with phoT672A (RB389), sfl1Δ (RB595), PTR3647::C…N (RB567), gdhD206E (RB484), gap1Δ (RB318) (C). mRNA levels for FLO adhesion genes (italic) and amino acid permease genes (italic bold) are shown for glutamine-limited mutants Ptr3647::C…N and Sfl1Q320STOP after growth to mid-exponential phase in liquid YPD (D). Presented are log2-transformed ratios of transcripts. The color code bar illustrates fold-change in mRNA in mutants relative to the wildtype strain CEN.PK113-7D. For the entire clustering of genes with more than 2-fold altered mRNA levels see Fig. S2.
We focused our work on two of the invasive clones from the glutamine-limited population. The clone Sfl1Q320STOP (Table 1) carries two mutant alleles: the nonsense mutation sfl1C958T that leads to a truncation of Sfl1 at residue 320 and the pho90A2014G mutation that substitutes threonine with alanine at residue 672 of Pho90. Clone Ptr3647::C…N (Table 1) carries a partial substitution of the PTR3 ORF with the long terminal repeat (LTR) of a Ty1 element from residue 1941 of PTR3. This leads to substitution of the 31 C-terminal amino acids of Ptr3, starting from residue 647, with 11 novel residues CWNKNPLSSIN encoded by the Ty1 element.
GAP1 Transcription is Induced in Ptr3647::C…N
Ptr3 is part of the SPS complex that responds to the presence of amino acids in the environment and induces transcription of amino acid transporter genes including AGP1, GNP1 and DIP5 [13], [17], [18], which encode glutamine transporters. Transcript profiling of the Ptr3647::C…N clone revealed that DIP5 and other Ptr3-regulated transporter genes (GNP1, PTR2, BAP2, BAP3, TAT1) had strongly reduced mRNA levels relative to the wildtype (Fig. 2D). The general amino acid permease gene GAP1 on the other hand, had a 10-fold increase in transcription level. The Ptr3647::C…N clone also had increased transcript levels of FLO genes, which are involved in cell-surface and cell-cell adhesion and could potentially explain the adhesive phenotype. FLO5 and FLO9 were mildly induced while FLO1 was upregulated 10-fold (Fig. 2D). Northern blot analysis showed that FLO11 had increased mRNA levels in the Ptr3647::C…N mutant showing that also FLO11 had been induced (Fig. 3B). We found a clear discrepancy between FLO11 mRNA levels found in the Northern blot and the array analysis (Fig. 2D). This difference could be caused by binding of unspecific mRNA species to the 60-mer probe used in the array analysis. The 621 bp long probe used for Northern blot analysis was specific for FLO11, and hence, we made the assumption that Northern blot analysis was more representative for the FLO11 mRNA levels.
Figure 3. Gap1-dependent FLO11 expression and invasive growth in Ptr3647::C…N.
Amino acid permease gene product in invasive growth ability in Ptr3647::C…N was tested in a ura3 PTR3647::C…N background (RB428) by introducing gene deletions dip5 (RB468), gnp1 (RB469) or gap1 (RB547). Cells were grown on rich complex medium (YPD) for 2 day at 30°C (A, upper panel) and flushed with water to expose invasive growth (A, lower panel). Northern blot of FLO11 mRNA for strains in A are shown in B (upper panel). RNA for ribosomal subunits 18S and 28S served as loading controls (B, lower panel). Genes of modulating proteins in invasive growth were deleted in the RB428 mutant: flo11 (RB457), tec1 (RB460), tpk2 (RB461), cyr1 (RB465), gcn4 (RB472), mss11 (RB474), ssy1 (RB477) and ptr3 (RB492). Cells were grown on YPD for 3 days at 30°C (C). The plate was flushed with water to test for invasive growth (D). Distribution of deletion strains in C and D are in E.
GAP1 is Essential for FLO11 Expression and Invasive Growth in Ptr3647::C…N
To test if increased GAP1 expression was causative for FLO expression and invasive growth we deleted GAP1. The gap1 Ptr3647::C…N mutant lost the ability to grow invasively while deletion of the glutamine transporter genes DIP5 or GNP1 in Ptr3647::C…N did not affect invasiveness (Fig. 3A). Northern blot analysis showed that FLO11 mRNA could not be detected in the gap1 Ptr3647::C…N mutant (Fig. 3B), suggesting that GAP1 was essential for FLO11 expression and invasive growth in this mutant.
GAP1 also Induce Invasive Growth Independent of FLO11
We next tested if the regulation of invasive growth in the Ptr3647::C…N clone was dependent on FLO11 and known regulators of FLO11 [19]. Invasive growth was still maintained after gene-deletion of either FLO11, the mitogen-activated protein kinase (MAPK) transcription factor gene TEC1, or the PKA-pathway genes TPK2 and CYR1. The same result was obtained when we deleted the transcription factor genes GCN4 and MSS11 as well as the SPS-component gene SSY1 (Fig. 3C–E). Only deletion of the PTR3647::C…N allele with a ptr3::KanMX cassette led to loss of invasive growth. Hence, besides FLO11, a second response was responsible for invasive growth in Ptr3647::C…N. The response was dependent on high GAP1 expression and not dependent on the MAPK-, PKA- and SPS-pathways.
Intracellular Amino Acid Pools are Increased in PTR3647::C…N
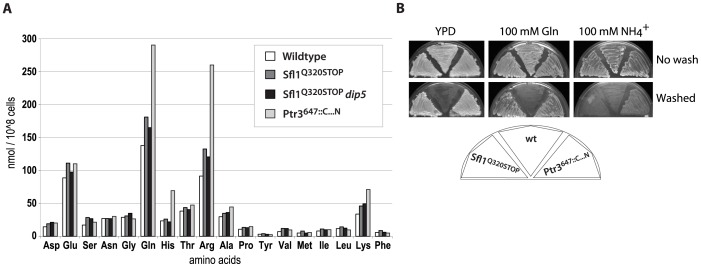
To explain the role of GAP1 for invasive growth, we hypothesized that Gap1 could either i) change the balance in the intracellular pools of certain amino acids, ii) affect the activity of invasive growth regulators, and/or iii) act as receptors, eliciting a signal transduction pathway causing invasiveness. To test the first hypothesis, amino acid pool sizes were determined for clones Ptr3647::C…N and Sfl1Q320STOP (Fig. 4A). Compared to the wildtype strain, glutamine pools were almost two-fold higher in the Ptr3647::C…N clone while the pool of the nitrogen-rich vacuolar amino acids lysine, histidine and arginine was as high as 2.5-fold higher than in the wildtype. We hypothesized that high levels of glutamine induced invasive growth. Both Ptr3647::C…N and Sfl1Q320STOP were invasive when grown on medium with 100 mM glutamine as the only nitrogen source, while the wildtype strain remained noninvasive. The strains were not invasive on ammonium (Fig. 4B) suggesting that the effect was glutamine specific.
Figure 4. Increased Gln, His, Arg and Lys pools in Ptr3647::C…N and invasive growth of mutants on glutamine minimal medium.
(A) shows amino acid concentrations (nmol 108 cells−1) in whole cells of ura3 strains isogenic to the wildtype (CEN.PK113-5D), Sfl1Q320STOP (RB20), Sfl1Q320STOP dip5 (RB317) and Ptr3647::C…N (RB428) after growth to 1–2×107 cells ml−1 in liquid YPD. Values shown are representative of three independent experiments. Descendants from glutamine-limited cultures of Sfl1Q320STOP and Ptr3647::C…N were grown as indicated (B) on complex YPD, synthetic minimal medium with 100 mM glutamine or 100 mM ammonium as the sole nitrogen source for 5 days at 30°C. Plates were washed to reveal adhesion.
PTR3647::C…N is a Dominant Loss-of-function Allele
The transcript profile of Ptr3647::C…N was similar to transcript profiles of ptr3 deletion mutants, with severe reductions in transcript levels of SPS-regulated amino acid and peptide transporters GNP1, PTR2 and DIP5 (Fig 2D; [17]). Hence, PTR3647::C…N behaves as a loss-of-function allele when expressed on YPD. Reconstitution of the PTR3647::C…N allele in the wildtype strain (Fig. 2A–C) and mating of the resultant invasive clone with a wildtype PTR3 strain, CEN.PK110-16D, revealed that the PTR3647::C…N allele was dominant for invasive growth (data not shown). Moreover, the finding that SSY1 deletion did not suppress the invasive phenotype of Ptr3647::C…N (Fig. 3C–E) suggested that the PTR3647::C…N allele is epistatic to ssy1 and supported that it is dominant. Taken together, these data suggest that the PTR3647::C…N allele must be a dominant loss-of-function allele.
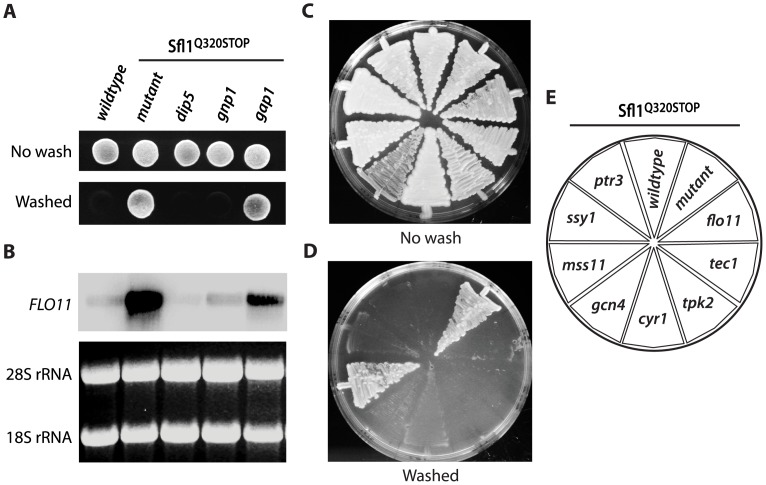
GNP1 and DIP5 are Essential for FLO11 Expression and Invasive Growth in Sfl1Q320STOP
The transcript profile of the invasive mutant Sfl1Q320STOP revealed increased transcript levels of the adhesion genes FLO5, FLO9, FLO10 (Fig. 2D) and FLO11 (Fig. 5B). SFL1 encodes a PKA-regulated suppressor of FLO11 [20]–[22] and the sfl1Q320STOP mutation causes a truncation of the C-terminal part of Sfl1 that normally interacts with the co-suppressor Ssn6 [23]. Because the Ssn6-Sfl1 complex is essential for Sfl1-mediated FLO11 repression, the sfl1Q320STOP allele could be responsible for derepression of FLO11. The Sfl1Q320STOP clone also carried a SNP in the phosphate transporter gene PHO90 (Table 1), however the role of the pho90T672A allele in invasive growth regulation was unclear.
Figure 5. Amino acid permease genes DIP5 and GNP1 are essential for invasive growth in Sfl1Q320STOP.
Amino acid permease genes were deleted in ura3 Sfl1Q320STOP (RB20) to obtain the mutants dip5 (RB317), gnp1 (RB197) and gap1 (RB531). Strains were grown one day on YPD at 30°C (A, upper panel) and washed to expose invasive growth (A, lower panel). FLO11 mRNA from individual mutants was examined by Northern blotting (B, upper panel) with 18S and 28S rRNA as loading controls (B, lower panel). Regulators of invasive growth were deleted in the RB20 background: flo11 (RB184), tec1 (RB540); tpk2 (RB538), cyr1 (RB201), gcn4 (RB475), mss11 (RB488), ssy1 (RB199) and ptr3 (RB486). After 3 days of growth on YPD (C), plates were flushed with water to reveal invasive growth (D). Mutant Sfl1Q320STOP strains in C and D are shown in E.
We next tested the role of glutamine transporter genes for invasive growth of Sfl1Q320STOP. Both a gnp1 Sfl1Q320STOP mutant and a dip5 Sfl1Q320STOP mutant lost their adhesion properties (Fig. 5A) and Northern blot analysis supported these data, showing that FLO11 mRNA levels were reduced in these strains (Fig. 5B). Thus, GNP1 and DIP5 were essential for expression of FLO11 and invasive growth in a PKA pathway dependent manner. Deletion of GAP1 in the Sfl1Q320STOP strain had no effect on invasive growth and only led to a partial reduction of FLO11 expression (Fig. 5B), presumably because (i) GAP1 was not highly expressed as in Ptr3647::C…N and (ii) GNP1 and DIP5 were expressed and responsible for invasive growth.
We also found that GNP1 and DIP5 were essential for invasive growth in the S. cerevisiae strain ∑1278b. In ∑1278b, deletion of both GNP1 and DIP5 was required for loss of invasive growth (Fig. S1). Based on these results, the glutamine transporter genes appear to be a general requirement for invasive growth in the S. cervisiae strains ∑1278b and CEN.PK.
Mutations in FLO11, MAPK, SPS and PKA Pathway Genes Suppress Sfl1Q320STOP Invasiveness
Expression of FLO11 in the Sfl1Q320STOP clone indicated that the FLO11 promoter was in a PKA pathway regulated derepressed state. Northern blot analysis revealed that FLO11 was in fact transcribed in the Sfl1Q320STOP clone and deletion of FLO11 showed that it was responsible for invasive growth (Fig. 5). The absence of Sfl1 repression is known to allow access of transcriptional activators to the FLO11 promoter [20]–[22]. Individual deletions of the genes TEC1, TPK2, CYR1, GCN4 and MSS11, which encode known regulators of FLO11 expression [17], [19], led to the loss of invasive growth in all cases, except for the deletion of MSS11 (Fig. 5C–E). Hence, regulation of invasive growth in the Sfl1Q320STOP clone, CEN.PK background, was similar to regulation in ∑1278b. This result suggests that GNP1 and DIP5 induce FLO11 and invasive growth through identical pathways in the two strain backgrounds involving Gcn4, the MAPK- and PKA-pathways. We tested if regulators of GNP1 and DIP5 were also essential for invasive growth. Ssy1-Ptr3-Ssy5 induces the expression of GNP1 and DIP5 in response to amino acids [13], [17]. We found that both PTR3 and SSY1 were essential for invasive growth in the Sfl1Q320STOP clone (Fig. 5), suggesting that the SPS complex regulates invasive growth through GNP1 and DIP5.
Amino acid pools in the Sfl1Q320STOP mutant and in the Sfl1Q320STOP dip5 mutant were almost identical to the wildtype strain with a slight increase in the glutamine pool. Thus, Sfl1 and Dip5 did not appear to influence invasive growth through pool sizes (Fig. 4A).
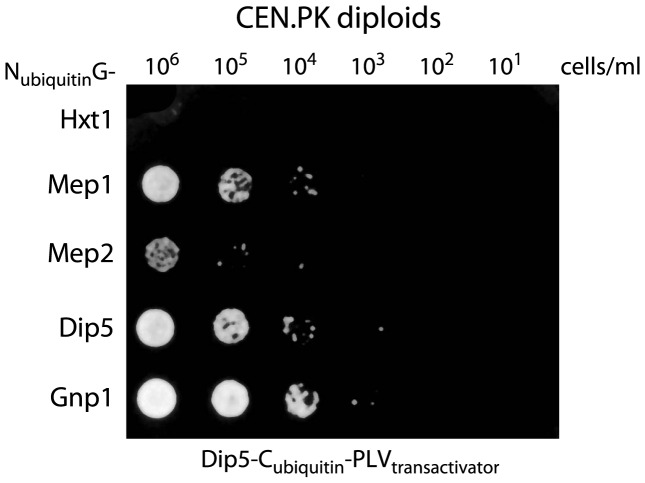
Dip5 Interacts with Gnp1 and Mep2
Interestingly, individual deletions of DIP5 and GNP1 resulted in complete loss of FLO11 expression and invasive growth in Sfl1Q320STOP (Fig. 5A). The finding that two amino acid transporter genes are essential for invasive growth under the same condition indicated corporative behavior between the transporters in the CEN.PK background. We tested the physical interaction between Dip5 and Gnp1, using the split-ubiquitin assay. In accordance with previous findings [24], we found that Dip5 interacts with itself (Fig. 6). Dip5 also interacted with the glutamine transporter Gnp1 and the ammonium permeases Mep1 and Mep2 but did not interact with the glucose transporter Hxt1 (Fig. 6). Physical interaction between Dip5 and Gnp1 might cause a functional dependence for activity of both transporters and explain why deletion of either DIP5 or GNP1 led to complete loss of FLO11 expression in the CEN.PK background.
Figure 6. Dip5 interacts with Gnp1 and Mep2.
Physical interaction between Dip5 and transporters Hxt1, Mep1, Mep2, Dip5, Gnp1 was tested with the membrane protein-based split-ubiquitin system in the CEN.PK background. Ade- His- cells (THY.AP4) expressing the C-terminal part of ubiquitin fused to Dip5 and the transcriptional activator PLV were mated to Ade- His- cells (THY.AP5) expressing the N-terminal part of ubiquitin fused to either Hxt1, Mep1, Mep2, Dip5 or Gnp1. Interacting protein-pairs formed a functional ubiquitin that released PLV for induction of the HIS3 and ADE2 genes, enabling growth on synthetic complete medium without adenine and histidine.
DIP5 is Essential for Biofilm Formation and Pseudohyphal Growth
Invasive growth shares many regulative constituents with biofilm and pseudohyphal growth [19]. We found that the Sfl1Q320STOP mutant formed biofilm on a polystyrene surface and that this was dependent on DIP5 and GNP1 (Fig. 7A). A diploid form of Sfl1Q320STOP, homozygous for the sfl1 mutation, formed pseudohyphal growth on synthetic low ammonium medium (SLAD); however, a sfl1Q320STOP/sfl1Q320STOP dip5/dip5 mutant lost the ability to form pseudohyphae (Fig. 7B). This showed that DIP5 was essential for the pseudohyphal phenotypes. We attempted to construct sfl1Q320STOP/sfl1Q320STOP gnp1/gnp1 mutants to test if GNP1 was required for pseudohyphal growth but were unsuccessful in mating sfl1Q320STOP gnp1 haploids.
Figure 7. DIP5 and GNP1 are essential for biofilm formation and DIP5 depletion abolishes pseudohyphal differentiation of Sfl1Q320STOP.
Biofilm formation on a polystyrene surface was tested in the Sfl1Q320STOP trp1 background (RB52) of Sfl1Q320STOP dip5 (RB317), Sfl1Q320STOP gnp1 (RB196) and compared to ∑1278b strain 10560-2B, ∑1278b dip5 gnp1 (RB157) and the wildtype strain CEN.PK110-16D. After 24 hours growth, in either synthetic complete 0.1% glucose or synthetic complete 2.0% glucose, biofilms were visualized by crystal violet staining and then washed three times (A). Homozygous diploid strains in the Sfl1Q320STOP background were incubated on low-ammonium SLAD medium for 3 days at 30°C to test for pseudohyphal growth (B).
Discussion
The ammonium transporter Mep2 has been shown to be essential for expression of FLO11 and filamentous growth of S. cerevisiae under nitrogen limitation [8]. Our results suggest that amino acid transporters can have a similar function as Mep2, since they are essential for transcription of the cell surface glycoprotein gene FLO11, for biofilm as well as invasive growth in S. cerevisiae. We show that GAP1 is essential for invasive growth and that invasiveness can be maintained even after deletion of FLO11. The highly expressed GAP1 phenotype correlates with an increased amino acid pool and an increased expression of FLO11 and other FLO genes. In cases where GAP1 is not expressed, we find that GNP1 and DIP5 are essential for invasive growth and that they probably exhibit this role through PKA-pathway components and FLO11 expression.
GNP1 and DIP5 induction of FLO11 is likely to be dependent on an active PKA-pathway, since this regulation is observed in a Sfl1Q320STOP mutant. The transcription factor Sfl1 represses transcription of FLO11 when activity of the PKA pathway is low [20], [22]. In a ∑1278b background, Sfl1 is removed from the promoter when phosphorylated by an active PKA subunit, Tpk2, leading to FLO11 transcription [20]–[22]. The PKA pathway is partially inactive in the CEN.PK strain background due to a point mutation in the adenylate cyclase gene, cyr1K1876M [25]. Therefore, the wildtype CEN.PK strain is likely to have permanent low PKA activity that keeps the FLO11 promoter in the repressed state. Mutant Sfl1Q320STOP lost the Sfl1 domain that interacts with the co-repressor Ssn6 [23] and the FLO11 promoter was likely converted to a state amenable to transcription similar to the state found when the PKA pathway is active.
Invasive growth of the Sfl1Q320STOP mutant appeared to be regulated by the same pathways described for the ∑1278b background, including the essential constituents of the PKA-, MAPK- and general amino acid control (GCN) pathways (Fig. 5). Hence, GNP1 and DIP5 are likely to be involved in the regulation of one or more of these signaling pathways. The cAMP-PKA and MAPK pathways crosstalk and both are regulated by the G-protein, Ras2 [26] but how this complex signal network is wired in response to GNP1 and DIP5 could not be determined by conventional epistasis analysis.
The role of the SPS complex (Ptr3, Ssy1 and Ssy5) in regulating invasive growth is so far unclear [17]. Our results suggest that the SPS complex acts on FLO11 expression by inducing GNP1, DIP5 and potentially other amino acid transporter genes (Fig. 8A model). The model is supported by the fact that the function of GNP1 and DIP5 in FLO11 regulation appears to be conserved between CEN.PK and ∑1278b (Fig. S1).
Figure 8. Models of FLO gene regulation by amino-acid transporters.
Extracellular amino acids induce the SPS complex at the plasma membrane (PM) and the activated complex elicits gene expression of amino acid transporters DIP5 and GNP1. Dip5 and Gnp1 activate FLO11 transcription in a manner dependent on activity of the PKA-pathway (A). Inactive SPS signaling indirectly leads to increased transcript levels of GAP1. Gap1 increases the amino acid pool concentration, which in turn triggers a PKA independent signal for induction of FLO genes (B). PM, plasma membrane. NM, nuclear membrane.
In the Ptr3647::C…N strain, GAP1 activity was essential for FLO11 expression (Fig. 3).
The phenotype of this strain appeared to be similar to previously reported recessive ptr3 and ssy1 mutants [17]. Ljungdahl and coworkers found that loss of Ptr3 activity leads to increased GAP1 transcript levels, reduced levels of GNP1 and PTR2 mRNA, invasive growth and increased pools of glutamate, glutamine, histidine, arginine and several other amino acids when grown on YPD [17]. However, in contrast to the recessive ptr3 alleles, the dominant loss-of-function PTR3647::C…N allele complemented an ssy1 deletion, revealing that it was epistatic to SSY1 (Fig. 3C–E). We propose that Ptr3647::C…N leads to an inactive SPS complex, under the tested conditions, causing reduced transcription of GNP1 and DIP5 (Fig. 8B model). GAP1 transcription is not under direct control of the SPS complex but might be induced in Ptr3647::C…N as a result of altered activity of the TOR-regulated nitrogen catabolite repression pathway [27], giving rise to a higher intracellular amino acid pool. The glutamine transporter genes GNP1 and DIP5 are not essential for invasive growth in Ptr3647::C…N probably because these genes have very low expression levels (Fig. 2D).
In addition to Gap1’s role as an amino acid transporter, Gap1 has been reported to induce the PKA pathway, leading to rapid degradation of trehalose and glycogen in nitrogen-starved cells [28]. Hence, Gap1 could be acting on the FLO11 promoter via the PKA pathway. However, deletion of the PKA pathway genes CYR1 and TPK2 in Ptr3647::C…N were not sufficient for loss of invasive growth (Fig. 3C–E). Deletion of other regulatory factors essential for invasive growth in ∑1278b was also not sufficient to eliminate invasive growth by Ptr3647::C…N, suggesting a novel GAP1-dependent regulatory mechanism. This mechanism acts independent of the PKA-pathway and could regulate transcription of FLO genes other than FLO11 (Fig. 8B model). We found that deletion of FLO11 was not sufficient to abolish invasive growth, so alternative adhesion proteins must be involved. S. cerevisiae contains a number of FLO genes that confer cell-cell and cell-surface adhesion [29], [30]. The FLO1, 5, 9 and 10 genes are normally inactive in laboratory strains though FLO1, FLO5 and FLO10 are known to have an impact on S. cerevisiae adhesion [29], [31]–[33]. A likely candidate is FLO1, which was 10-fold induced in the Ptr3647::C…N mutant relative to the wildtype and correlated with a high GAP1 and FLO11 expression (Fig. 2D).
In summary, our data demonstrate that amino acid transporters are involved in biofilm development and invasive growth. We propose that at least two modes of action exist on invasive growth regulation; a FLO11-dependent modulated by Dip5, Gnp1 and Gap1 and a FLO11-independent, regulated by Gap1. The former, Dip5 and Gnp1, are likely amino acid transporters or transceptors that directly activate a cytosolic signaling pathway linked to PKA-pathway components, which in turn induce FLO11. Our observations support that both permeases operate downstream of amino acid sensing complex, SPS, for regulation of FLO11. The latter, Gap1, may potentially induce invasive growth through increased amino acid pool levels that elicit a novel signal-cascade for gene expression of other FLO genes.
Materials and Methods
Strains
Strains used are listed in Table 2. The wildtype strain CEN.PK113-7D and the isogenic strains CEN.PK113-5D (ura3) and CEN.PK110-16D (trp1) were kind gifts from Peter Kötter, Frankfurt am Main, Germany. The two populations of CEN.PK113-7D and two populations of S288c evolved in glucose limitation were kind gifts from Maitreya Dunham, Seattle, USA. The 10560-2B parental strain (∑1276b) and dip5 (RB598), gnp1 (RB599) and gap1 (RB600) mutants in the ∑1276b background were kind gifts from Owen Ryan and Charles Boone, Toronto, Canada [34]. Descendants of CEN.PK113-7D exposed to glutamine limitation for 250 generations were isolated and three clones were chosen for analysis (Table 1). Descendants of CEN.PK113-7D exposed to ammonium limitation for 400 generations were isolated and three clones were chosen for analysis (Table 1).
Table 2. S. cerevisiae strains and populations used in this study.
| Strain | Genotype | Reference/source |
| Wildtype CEN.PK113-7D | MAT a | [47] |
| CEN.PK113-5D | MAT a ura3-52 (generated from progenitor CEN.PK113-7D) | [47] |
| CEN.PK110-16D | MATα trp1 | [47] |
| 10560-2B (Σ1276b) | MAT a ura3-52 leu2::hisG his3::hisG | [48] |
| THY.AP4 | MAT a ura3 leu2 lexA::lacZ::trp1 lexA::HIS3 lexA::ADE2 | [46] |
| THY.AP5 | MATα leu2 trp1 his3 loxP::ade2 | [46] |
| RB1 | MAT a sfl1Q320STOP pho90T672A | this study |
| RB3 | MAT a PTR3647::CWNKNPLSSIN | this study |
| RB12 | MAT a Δgap1 | this study |
| RB16 | MAT a gap1S388P mep2G352S | this study |
| RB17 | MAT a Δgap1 gdh1D206E | this study |
| RB18 | MAT a | this study |
| RB20 | MAT a sfl1Q320STOP pho90T672A ura3 | this study |
| RB52 | MATα sfl1Q320STOP trp1 | this study |
| RB157 | ∑1276b MAT a ura3-52 leu2::hisG his3::hisG dip5:::KanMX gnp1::loxP | this study |
| RB184 | MAT a sfl1Q320STOP pho90T672A ura3 flo11::KanMX | this study |
| RB196 | MATα sfl1Q320STOP trp1 gnp1:: KanMX | this study |
| RB197 | MAT a sfl1Q320STOP pho90T672A ura3 gnp1::KanMX | this study |
| RB199 | MAT a sfl1Q320STOP pho90T672A ura3 ssy1::KanMX | this study |
| RB201 | MAT a sfl1Q320STOP pho90T672A ura3 cyr1::KanMX | this study |
| RB209 | MATα sfl1Q320STOP trp1 dip5::KanMX | this study |
| RB210 | MATα sfl1Q320STOP ura3 mep2::KanMX | this study |
| RB317 | MAT a sfl1Q320STOP pho90T672A ura3 dip5::KanMX | this study |
| RB318 | MAT a ura3-52 gap1 | this study |
| RB389 | MAT a ura3-52 pho90T672A | this study |
| RB428 | MAT a PTR3647::CWNKNPLSSIN ura3 | this study |
| RB457 | MAT a PTR3647::CWNKNPLSSIN ura3 flo11::KanMX | this study |
| RB460 | MAT a PTR3647::CWNKNPLSSIN ura3 tec1::KanMX | this study |
| RB461 | MAT a PTR3647::CWNKNPLSSIN ura3 tpk2::KanMX | this study |
| RB465 | MAT a PTR3647::CWNKNPLSSIN ura3 cyr1::KanMX | this study |
| RB468 | MAT a PTR3647::CWNKNPLSSIN ura3 dip5::KanMX | this study |
| RB469 | MAT a PTR3647::CWNKNPLSSIN ura3 gnp1::KanMX | this study |
| RB472 | MAT a PTR3647::CWNKNPLSSIN ura3 gcn4::KanMX | this study |
| RB474 | MAT a PTR3647::CWNKNPLSSIN ura3 mss11::KanMX | this study |
| RB475 | MAT a sfl1Q320STOP pho90T672A ura3 gcn4::KanMX | this study |
| RB477 | MAT a PTR3647::CWNKNPLSSIN ura3 ssy1::KanMX | this study |
| RB484 | MAT a ura3-52 gdh1D206E | this study |
| RB486 | MAT a sfl1Q320STOP pho90T672A ura3 ptr3::KanMX | this study |
| RB488 | MAT a sfl1Q320STOP pho90T672A ura3 mss11::KanMX | this study |
| RB492 | MAT a PTR3647::CWNKNPLSSIN ura3 ptr3::KanMX | this study |
| RB531 | MAT a sfl1Q320STOP pho90T672A ura3 gap1 | this study |
| RB538 | MAT a sfl1Q320STOP pho90T672A ura3 tpk2::KanMX | this study |
| RB540 | MAT a sfl1Q320STOP pho90T672A ura3 tec1::KanMX | this study |
| RB547 | MAT a PTR3647::CWNKNPLSSIN ura3 gap1 | this study |
| RB567 | MAT a ura3-52 PTR3647::CWNKNPLSSIN | this study |
| RB595 | MAT a ura3-52 sfl1Δ | this study |
| RB598 | ∑1276b MAT a dip5::KanMX can1::STE2p-SpHIS5 lyp1::STE3p-LEU2 his3::HisG leu2 ura3 | [34] |
| RB599 | ∑1276b MAT a gnp1::KanMX can1::STE2p-SpHIS5 lyp1::STE3p-LEU2 his3::HisG leu2 ura3 | [34] |
| RB600 | ∑1276b MAT a gap1::KanMX can1::STE2p-SpHIS5 lyp1::STE3p-LEU2 his3::HisG leu2 ura3 | [34] |
Tiling array analysis of descendants revealed potential SNPs and deletions. SNPs in ORFs were identified by PCR amplification of 500 bp around the potential SNP and Sanger sequencing of the product, while deletions were identified as described previously [35]. Mutations in the three clones from the glutamine limited culture were i) sfl1C958T and pho90A2014G (clone Sfl1Q320STOP); ii) and PTR31941::Ty1 (clone Ptr3647::C…N, allele PTR3647::C…N) and iii) YKRCδ11-Δgap1-YKRCδ12. The three clones from ammonium limitation carried the mutations: i) gap1T1162C and mep2G1054A; ii) YKRCδ11-Δgap1-YKRCδ12 and gdh1C618G and iii) no SNPs were detected in the last clone. The resulting protein modifications are described in Table 1. The PTR3647::C…N insertion initially appeared as a SNP in PTR3 in the tiling array analysis but several primer combinations around residue 1941 of PTR3 failed to give a PCR product. The mutation was identified by digestion of genomic DNA from the Ptr3647::C…N clone with XhoI followed by ligation with T4 ligase and inverted primers identical to regions just upstream of residue 1941 for inverse PCR. Sanger sequencing of the PCR product revealed insertion of a long terminal repeat (LTR) identical to YMRCTy1-4, YJRWTy1-2, YGRWTy1-1 and YOLWTy1-1 (http://www.yeastgenome.org/), given that PTR3647::C…N had an insertion through transposition of one of these Ty1s.
The PTR3647::C…N allele was reconstituted into the wildtype strain background (CEN.PK113-5D isogenic to CEN.PK113-7D except for URA3) by transformation with a chimeric PTR31941::Ty1 PCR product, using the primers 5′-CATGATACTAGTTGAGGAG-3′ (identical to nucleotides 983–1001 of the PTR3 ORF) and 5′-tcgaagatgactatgggtttccgacgattcatgaatgagtcGATTCCATTTTGAGGATTCC-3′ (identical to nucleotides 215–234 of the Ty1 LTR and 213143–213173 of chromosome XI downstream of the stop codon of wild type PTR3). The PCR product was co-transformed into CEN.PK113-5D with a URA3 plasmid. A prototrophic transformant able to adhere and invade agar had the correct insertion of the PTR3647::C…N mutation, identified by PCR and Sanger sequencing. The resultant strain was named RB567. gdh1C618G and pho90A2014G were reconstituted into CEN.PK113-5D by co-transformation of a URA3 plasmid with PCR products carrying gdh1C618G or pho90A2014G. Correct clones were identified through prototrophic, invasive growth and Sanger sequencing and the resultant mutants denoted RB484 and RB389. sfl1C958T was not successfully introduced into CEN.PK113-5D; therefore a full sfl1 deletion was used as substitute (RB595). YKRCδ11-gap1-YKRCδ12 mutants (RB318, RB531, RB547) were selected on D-histidine [36] and YKRCδ11-gap1-YKRCδ12 deletions were confirmed by PCR as described [35].
Auxotrophic ura3 Sfl1Q320STOP and Ptr3647::C…N strains were selected on medium with 5-FOA [37] and the genotype confirmed by complementation with URA3-based plasmids to generate strains RB20 and RB428. PCR-based gene deletions were made as described [38], using primers to the loxP::KanMX::loxP cassette in pUG6 with 5'-flanking sequences of the 40 bp immediately upstream and downstream of the targeted ORF. Deletion mutants were selected on YPD with 200 mg L−1 geneticin. Correct insertion of the pUG6-based PCR fragment was confirmed by diagnostic PCR on genomic DNA from geneticin-resistant clones using primers for the KanMX cassette and the promoter sequence of the targeted gene (see Table 2 for deletion strains).
A MATα Sfl1Q320STOP trp1 strain (RB52) was obtained by mating a MATa sfl1Q320STOP pho90T672A ura3 strain, RB20, and CEN.PK110-16D and sporulating the resultant diploid. Diploid homozygous mutants were obtained by crossing the haploid mutants RB20 (MAT a sfl1Q320STOP pho90T672A ura3) and RB52 (MATα Sfl1Q320STOP trp1). Standard yeast genetics and molecular biology methods were used [37].
Chemostat Cultivation
CEN.PK113-7D MATa was grown in modified minimal medium at dilution rates of 0.20 h−1 as described [39]. Cultures were fed with modified minimal medium containing ammonium (in duplicated culture experiments) or glutamine at a concentration corresponding to 6 mM nitrogen and 250 mM glucose (calculated on nitrogen and carbon atom bases, respectively), which ensured that growth was limited by nitrogen source with all other nutrients in excess. After steady state was reached, cellular dry weight, metabolite concentrations and gas profiles were monitored as described [39] every thirteenth generation. All parameters were constant throughout cultivation including biomass yield, glutamine limitation: 0.22±0.01 g dry weight/g glucose consumed and ammonium limitation: 0.22±0.01 and 0.21±0.01 g dry weight/g glucose consumed. Dry weight was determined in the efflux medium to test if cells accumulated because of flocculation in the bioreactor. Residual concentrations of glutamine were below detection level (<30 µM). In the ammonium-limited chemostat, the residual ammonium concentration was less than 50 µM throughout fermentation. Glucose and nitrogen consumption and metabolites (biomass, ethanol and glycerol) throughout chemostat cultivations showed no changes and the C:N ratio was thus constant over time.
Culture Media
Media for the chemostat cultivations was as described [39]. Standard yeast media, YPD and SC were as described [37]. All standard media were variations of synthetic dextrose medium with 1.7 g L−1 yeast nitrogen base without amino acids and ammonium sulphate, 20 g L−1 glucose, and 18 g L−1 agar. Low ammonium SLAD contained 0.05 mM ammonium sulphate as sole nitrogen source, while synthetic glutamine dextrose medium was supplemented with 100 mM glutamine as the sole nitrogen source.
Gene Expression Analysis
Cells grown in liquid YPD were harvested in mid-exponential phase by addition of an equal volume crushed ice and centrifugation. RNA was prepared by acid phenol extraction and hybridized to Agilent 60-mer yeast ORF expression microarrays as described [40]. Data were acquired using an Agilent scanner and extracted with Agilent software using standard settings. Ratios of mRNA levels in the descendants to the progenitor were sorted using a 3-fold cutoff (Fig. S2). The 106 resultant Log2 transformed transcript ratios were clustered by centered Pearson correlation. All raw microarray data are available from the Princeton Microarray Database (http://puma.princeton.edu).
Tiling Arrays and SNP Identification
Affymetrix Yeast Tiling arrays 1.0 were used. Total genomic DNA was hybridized to probes spaced every 5 bases and the data were analyzed as described [41], [49].
Northern Blots
Northern blot analysis was performed with RNA from cells in YPD in mid-exponential phase. Total RNA was isolated using an acid-phenol extraction protocol [42] and 10 µg separated by electrophoresis, blotted and hybridized as described [43] with the following modifications: a probe was prepared from a PCR-amplified 621 bp FLO11 DNA fragment from the 3′-end of FLO11 (≤60% homologous to other regions in the S. cerevisiae genome). The probe was radioactively labeled with [Υ-P32]-CTP using a Prime-It II Random Primer Labeling Kit (Agilent Technologies, USA) and purified from unincorporated nucleotides using Illustra ProbeQuan G-50 Micro Columns (GE Healthcare, UK). The membrane was pre-hybridized with Ambion ULTRAhyb (Life Technologies Ltd., UK) for 2 hours at 42°C before addition of labeled probe.
Intracellular Amino Acid Pools
Intracellular amino acids were extracted with trichloracidic acid as described [44] and quantified by reverse-phase HPLC using an LKB-Alpha Plus amino-acid analyser [45].
Split-ubiquitin Interaction Assay
The split-ubiquitin interaction assay was essentially as described [46]. S. cerevisiae genes were amplified from genomic DNA [S288c] using gene-specific primers acaagtttgtacaaaaaagcaggctctccaaccaccATGX19–25-5` ORF and tccgccaccaccaaccactttgtacaagaaagctgggtaX19–25-3` strand ORF without stop. The DIP5 PCR product was cloned in vivo by homologous recombination in the THY.AP4 strain, carrying the plasmid pMetYCgate, to form the plasmid pMet-DIP5-Cgate. The THY.AP5 strain, carrying the plasmid pNXgate33-3HA, was transformed with PCR products of either HXT1, MEP1, MEP2, DIP5 or GNP1 to form strains with the plasmids pN-HXT1-gate33-3HA, pN-MEP1-gate33-3HA, pN-MEP2-gate33-3HA, pN-DIP5-gate33-3HA and pN-GNP1-gate33-3HA. THY.AP4 and THY.AP5 are CEN.PK derivatives and transformants of THY.AP4 were selected on SC -leucine and transformants of THY.AP5 were selected on SC -tryptophan. Plasmids were isolated and amplified in Escherichia coli by standard methods and DNA insertion confirmed by PCR. Protein-protein interactions between Dip5, fused to the C-terminal half of ubiquitin (Dip5-CubPLV), and Hxt1, Mep1, Mep2, Dip5, or Gnp1, fused to the N-terminal half of ubiquitin (NubG-X), were determined by mating strain THY.AP4 (Dip5-CubPLV) to strain THY.AP5 (NubG-X). Diploids were selected on SC-leucine and -tryptophan and interaction between Dip5-CubPLV and each NubG-fusion was determined by growth after 3 days on reporter plates (SC+500 µM methionine and without leucine, tryptophan, adenine and histidine).
Pseudohyphal Transition, Invasive Growth and Biofilm Formation
Pseudohyphal growth assays were as described [3] and performed using diploid mutants. Invasive and adhesive growth was tested by incubating haploid cells on solid medium at 30°C for 2 days. Plates were photographed and a gentle stream of water used to rinse non-invasive cells from the plate. Plates were dried for 30 minutes and photographed again. Biofilm formation in polystyrene 96-wells plates was tested as described [5]. Biofilm formation assay was conducted with trp1 auxotrophs (CEN.PK110-16D, RB52, RB157, RB196, RB209, 10560-2B; Table 2).
Supporting Information
Amino acid permeases are essential for invasive growth in Sfl1Q320STOP, Ptr3647::C…N and ∑1276b. The role of amino acid permeases on invasive growth was tested in several strains: a ura3 auxotroph version of the CEN.PK wildtype (CEN.PK113-5D), the mutants Sfl1Q320STOP ura3 (RB20) and Ptr3647::C…N ura3 (RB428) and the Σ1276b strain (10560-2B). In the ura3 Sfl1Q320STOP background we deleted dip5 (RB317), gnp1 (RB197) and gap1 (RB531). In the ura3 Ptr3647::C…N background we deleted dip5 (RB468), gnp1 (RB469) and gap1 (RB547). In the ∑1278b background, we tested invasive growth ability of dip5 (RB598), gnp1 (RB599), gap1 (RB600) and gnp1 dip5 (RB157). All strains were grown as dot spots on rich complex medium (YPD) for 1 day at 30°C (A) and flushed with water to expose invasive growth (B).
(TIF)
Microarray analysis of descendants after prolonged nitrogen limitation. Six single clones were collected after chemostat cultivation with a single limited nitrogen source of either glutamine or ammonium. All other nutrients were in excess. After growth to mid-exponential phase in liquid YPD, microarray analysis was performed. Gene expression data are shown for the glutamine-limited descendants Sfl1Q320STOP, Ptr3647::C…N and Δgap1 (RB12) and for the ammonium-limited gap1S388P mep2G352S (RB16), Δgap1 gdhD206E (RB17) and no confirmed SNPs for RB18 (-). Asterisk denotes descendants that grow invasively. Transcripts are clustered using Pearson correlation and data are presented as log2-transformed ratios. The color code bar illustrates the fold-change in gene expression in descendants relative to the progenitor strain CEN.PK113-7D.
(TIF)
Acknowledgments
We would like to thank David Botstein for experimental support and valuable discussions; Owen Ryan and Charles Boone are acknowledged for ∑1276b strains and Peter Kötter for CEN.PK strains.
Funding Statement
Support for this work was received from the Danish Agency for Science Technology and Innovation FNU 09-062220 and 272-06-0287, http://en.fi.dk/; a National Institutes of Health Research Grant GM046406, http://www.nihr.ac.uk/Pages/default.aspx; the National Institute of General Medical Sciences Center for Quantitative Biology (GM071508), http://www.nigms.nih.gov/News/Results/quantitative_bio_center.htm. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mitchell AP (1994) Control of meiotic gene expression in Saccharomyces cerevisiae . Microbiol Rev 58: 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, et al. (2004) “Sleeping Beauty”: Quiescence in Saccharomyces cerevisiae . Microbiol Mol Biol Rev 68: 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR (1992) Unipolar Cell Divisions in the Yeast Saccharomyces-Cerevisiae Lead to Filamentous Growth - Regulation by Starvation and Ras. Cell 68: 1077–1090. [DOI] [PubMed] [Google Scholar]
- 4. Roberts RL, Fink GR (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 8: 2974–2985. [DOI] [PubMed] [Google Scholar]
- 5. Reynolds TB, Fink GR (2001) Bakers’ Yeast, a Model for Fungal Biofilm Formation. Science 291: 878–881. [DOI] [PubMed] [Google Scholar]
- 6. Lambrechts MG, Bauer FF, Marmur J, Pretorius IS (1996) Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation ináyeast. Proc Natl Acad Sci 93: 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Douglas LM, Li L, Yang Y, Dranginis AM (2007) Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot Cell 6: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorenz MC, Heitman J (1998) The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae . EMBO J 17: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Braus GH, Grundmann O, Bruckner S, Mosch HU (2003) Amino Acid Starvation and Gcn4p Regulate Adhesive Growth and FLO11 Gene Expression in Saccharomyces cerevisiae . Mol Biol Cell 14: 4272–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gagiano M, van Dyk D, Bauer FF, Lambrechts MG, Pretorius IS (1999) Msn1p/Mss10p, Mss11p and Muc1p/Flo11p are part of a signal transduction pathway downstream of Mep2p regulating invasive growth and pseudohyphal differentiation in Saccharomyces cerevisiae . Mol Microbiol 31: 103–116 doi: 10.1046/j.1365-2958.1999.01151.x. [DOI] [PubMed] [Google Scholar]
- 11. van Nuland A, Vandormael P, Donaton M, Alenquer M, Lourenco A, et al. (2006) Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol Microbiol 59: 1485–1505. [DOI] [PubMed] [Google Scholar]
- 12. Grenson M, Hou C, Crabeel M (1970) Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J Bacteriol 103: 770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iraqui I, Vissers S, Bernard F, De Craene JO, Boles E, et al. (1999) Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol 19: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu X, Garrett J, Schreve J, Michaeli T (1996) GNP1, the high-affinity glutamine permease of S. cerevisiae . Curr Genet 30: 107–114. [DOI] [PubMed] [Google Scholar]
- 15. Regenberg B, During-Olsen L, Kielland-Brandt MC, Holmberg S (1999) Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae . Curr Genet 36: 317–328. [DOI] [PubMed] [Google Scholar]
- 16. Lasko PF, Brandriss MC (1981) Proline transport in Saccharomyces cerevisiae . J Bacteriol 148: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klasson H, Fink GR, Ljungdahl PO (1999) Ssy1p and Ptr3p Are Plasma Membrane Components of a Yeast System That Senses Extracellular Amino Acids. Mol Cell Biol 19: 5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forsberg H, Ljungdahl PO (2001) Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol Cell Biol 21: 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brückner S, Mösch HU (2012) Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae . FEMS Microbiol Rev 36: 25–58. [DOI] [PubMed] [Google Scholar]
- 20. Pan X, Heitman J (2002) Protein Kinase A Operates a Molecular Switch That Governs Yeast Pseudohyphal Differentiation. Mol Cell Biol 22: 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR (2009) Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A 106: 18321–18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robertson LS, Fink GR (1998) The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proceedings of the National Academy of Sciences of the United States of America 95: 13783–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conlan RS, Tzamarias D (2001) Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J Mol Biol 309: 1007–1015. [DOI] [PubMed] [Google Scholar]
- 24. Tarassov K, Messier V, Landry CR, Radinovic S, Molina MMS, et al. (2008) An in Vivo Map of the Yeast Protein Interactome. Science 320: 1465–1470. [DOI] [PubMed] [Google Scholar]
- 25. Vanhalewyn M, Dumortier F, Debast G, Colombo S, Ma P, et al. (1999) A mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1K1876M, specifically affects glucose- and acidification-induced cAMP signalling and not the basal cAMP level. Mol Microbiol 33: 363–376. [DOI] [PubMed] [Google Scholar]
- 26. Mösch HU, Kübler E, Krappmann S, Fink GR, Braus GH (1999) Crosstalk between the Ras2p-controlled Mitogen-activated Protein Kinase and cAMP Pathways during Invasive Growth of Saccharomyces cerevisiae . Mol Biol Cell 10: 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coffman JA, Rai R, Cooper TG (1995) Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae . Journal of Bacteriology 177: 6910–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Donaton MC, Holsbeeks I, Lagatie O, Van ZG, Crauwels M, et al. (2003) The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae . Mol Microbiol 50: 911–929. [DOI] [PubMed] [Google Scholar]
- 29. Fichtner L, Schulze F, Braus GH (2007) Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288c. Mol Microbiol 66: 1276–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo B, Styles CA, Feng Q, Fink GR (2000) A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A 97: 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teunissen AW, van den Berg JA, Steensma HY (1995) Transcriptional regulation of flocculation genes in Saccharomyces cerevisiae . Yeast 11: 435–446. [DOI] [PubMed] [Google Scholar]
- 32. Halme A, Bumgarner S, Styles C, Fink GR (2004) Genetic and Epigenetic Regulation of the FLO Gene Family Generates Cell-Surface Variation in Yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- 33. Veelders M, Brückner S, Ott D, Unverzagt C, Mösch HU, et al. (2010) Structural basis of flocculin-mediated social behavior in yeast. Proceedings of the National Academy of Sciences 107: 22511–22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, et al. (2010) Genotype to phenotype: a complex problem. Science 328: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, et al. (2010) Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A 107: 18551–18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Regenberg B, Hansen J (2000) GAP1, a novel selection and counter-selection marker for multiple gene disruptions in Saccharomyces cerevisiae . Yeast 16: 1111–1119. [DOI] [PubMed] [Google Scholar]
- 37.Guthrie C, Fink GR(1991) Methods in Enzymology, vol. 194, Guide to Yeast Genetics and Molecular Biology. New York: Academic Press. [PubMed] [Google Scholar]
- 38. Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucl Acids Res 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Usaite R, Patil KR, Grotkjaer T, Nielsen J, Regenberg B (2006) Global Transcriptional and Physiological Responses of Saccharomyces cerevisiae to Ammonium, L-Alanine, or L-Glutamine Limitation. Appl Environ Microbiol 72: 6194–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, et al. (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- 41. Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, et al. (2006) Genome-Wide Detection of Polymorphisms at Nucleotide Resolution with a Single DNA Microarray. Science 311: 1932–1936. [DOI] [PubMed] [Google Scholar]
- 42. Rupp S (2007) Interactions of the fungal pathogen Candida albicans with the host. Future Microbiology 2: 141–151 doi:10.2217/17460913.2.2.141. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press.
- 44. Delforge J, Messenguy F, Wiame JM (1975) The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR- mutations and the general control of amino-acid biosynthesis. Eur J Biochem 57: 231–239. [DOI] [PubMed] [Google Scholar]
- 45. Regenberg B, Holmberg S, Olsen LD, Kielland-Brandt MC (1998) Dip5p mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate in Saccharomyces cerevisiae . Curr Genet 33: 171–177. [DOI] [PubMed] [Google Scholar]
- 46. Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, et al. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci 101: 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, et al. (2000) An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enz Microb Technol 26: 706–714. [DOI] [PubMed] [Google Scholar]
- 48. Rupp S, Summers E, Lo HJ, Madhani H, Fink G (1999) MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J 18: 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, et al. (2008) The Repertoire and Dynamics of Evolutionary Adaptations to Controlled Nutrient-Limited Environments in Yeast. PLoS Genet 4 (12) e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid permeases are essential for invasive growth in Sfl1Q320STOP, Ptr3647::C…N and ∑1276b. The role of amino acid permeases on invasive growth was tested in several strains: a ura3 auxotroph version of the CEN.PK wildtype (CEN.PK113-5D), the mutants Sfl1Q320STOP ura3 (RB20) and Ptr3647::C…N ura3 (RB428) and the Σ1276b strain (10560-2B). In the ura3 Sfl1Q320STOP background we deleted dip5 (RB317), gnp1 (RB197) and gap1 (RB531). In the ura3 Ptr3647::C…N background we deleted dip5 (RB468), gnp1 (RB469) and gap1 (RB547). In the ∑1278b background, we tested invasive growth ability of dip5 (RB598), gnp1 (RB599), gap1 (RB600) and gnp1 dip5 (RB157). All strains were grown as dot spots on rich complex medium (YPD) for 1 day at 30°C (A) and flushed with water to expose invasive growth (B).
(TIF)
Microarray analysis of descendants after prolonged nitrogen limitation. Six single clones were collected after chemostat cultivation with a single limited nitrogen source of either glutamine or ammonium. All other nutrients were in excess. After growth to mid-exponential phase in liquid YPD, microarray analysis was performed. Gene expression data are shown for the glutamine-limited descendants Sfl1Q320STOP, Ptr3647::C…N and Δgap1 (RB12) and for the ammonium-limited gap1S388P mep2G352S (RB16), Δgap1 gdhD206E (RB17) and no confirmed SNPs for RB18 (-). Asterisk denotes descendants that grow invasively. Transcripts are clustered using Pearson correlation and data are presented as log2-transformed ratios. The color code bar illustrates the fold-change in gene expression in descendants relative to the progenitor strain CEN.PK113-7D.
(TIF)