Abstract
The genus Arcobacter has been associated with human illness and fecal contamination by humans and animals. To better characterize the health risk posed by this emerging waterborne pathogen, we investigated the occurrence of Arcobacter spp. in Lake Erie beach waters. During the summer of 2010, water samples were collected 35 times from the Euclid, Villa Angela, and Headlands (East and West) beaches, located along Ohio's Lake Erie coast. After sample concentration, Arcobacter was quantified by real-time PCR targeting the Arcobacter 23S rRNA gene. Other fecal genetic markers (Bacteroides 16S rRNA gene [HuBac], Escherichia coli uidA gene, Enterococcus 23S rRNA gene, and tetracycline resistance genes) were also assessed. Arcobacter was detected frequently at all beaches, and both the occurrence and densities of Arcobacter spp. were higher at the Euclid and Villa Angela beaches (with higher levels of fecal contamination) than at the East and West Headlands beaches. The Arcobacter density in Lake Erie beach water was significantly correlated with the human-specific fecal marker HuBac according to Spearman's correlation analysis (r = 0.592; P < 0.001). Phylogenetic analysis demonstrated that most of the identified Arcobacter sequences were closely related to Arcobacter cryaerophilus, which is known to cause gastrointestinal diseases in humans. Since human-pathogenic Arcobacter spp. are linked to human-associated fecal sources, it is important to identify and manage the human-associated contamination sources for the prevention of Arcobacter-associated public health risks at Lake Erie beaches.
INTRODUCTION
Arcobacter is a fastidious Gram-negative, non-spore-forming, motile, spiral-shaped organism that belongs to the family Campylobacteraceae (55, 56). Since the new genus Arcobacter was first described in 1991 (55), Arcobacter has been detected in sewage, drinking water, surface water, and groundwater sources (9, 10, 14, 19, 21, 37, 38). This indicates that water can be a vehicle for human exposure (9, 25). Besides environmental sources, researchers have observed Arcobacter spp. such as Arcobacter butzleri, A. cryaerophilus, and A. skirrowii in patients with diarrhea and/or food-borne illness (13, 30, 33, 49, 60), which has led to these microbes being characterized as potential food- and waterborne pathogens (1, 26). Moreover, the presence of Arcobacter in livestock and processed meat (28, 29, 48) suggests that animals may also serve as possible reservoirs for Arcobacter species. Therefore, the relative abundance, sources, and routes of human exposure particular to Arcobacter need to be considered a serious public health concern.
Water recreational activities are among the most prominent outdoor pursuits enjoyed by Americans. According to the Nationwide Survey on Recreation and the Environment (38a), 59.6% of individuals visit beaches annually. Lake Erie has 62 public access beaches along its Ohio shoreline (42). Despite serving as a valuable recreational destination, the beach water of Lake Erie continues to be contaminated with point and nonpoint sources originating from humans, livestock, and wildlife, which can elevate the infectious disease risk for Ohio's beachgoers (41). To maintain the safety of Ohio's beaches, beach advisories are issued when a single beach water sample exceeds the daily maximum bacterial standard (235 Escherichia coli CFU/100 ml) (41). The beach advisory is a recommendation to the public to avoid swimming in water that has exceeded the maximum bacterial standards for water quality for the purpose of reducing swimming-related illnesses, although an advisory does not officially close a beach to the public (53). In 2010, routine testing by the Ohio Department of Health (ODH) showed that 21% of beach water samples resulted in beach advisories (41). We monitored the Euclid, Villa Angela, and East and West Headlands beaches in Ohio, which were among the most visited beaches in 2010 (43). The Euclid and Villa Angela beaches are highly contaminated urban beaches exhibiting water quality results exceeding the daily maximum bacterial standard for 44% and 40% of beach water samples, respectively, whereas the East and West Headlands beaches are in a less densely populated area, with percentages of excess of 15% and 16%, respectively (41). Given the apparent fecal contamination in Lake Erie, concerns pertaining to Arcobacter spp. have merit. With respect to Lake Erie, Arcobacter was isolated in a gastrointestinal illness outbreak at South Bass Island (44, 46). The outbreak was attributed to the transport of microbial contaminants from wastewater treatment facilities and septic tanks to the lake and subsurface water on the island, which resulted in contamination of the drinking water wells (19). Moreover, Arcobacter is significantly more prevalent in water exhibiting high levels of fecal contamination (10). Thus, the evidence suggests that the Euclid and Villa Angela beaches are locations where Arcobacter is likely to be present and to pose a significant human health risk via human-water contact.
In order to understand possible associations between the presence of Arcobacter, fecal contamination levels, and Arcobacter sources, we determined the frequency of occurrence and density of Arcobacter spp. in conjunction with densities of E. coli and other genetic markers for fecal bacteria by using culture-based methods, quantitative PCR (qPCR) assays, and sequence analysis. This is the first study to investigate the presence of Arcobacter at Lake Erie beaches and provides evidence that recreational water is a potential transmission route enabling human exposure.
MATERIALS AND METHODS
Site description and water sample collection.
This study was conducted at four Ohio freshwater public beaches located on Lake Erie. The Euclid (41°35′9″N, 81°34′1″W) and Villa Angela (41°35′2″N, 81°34′9″W) beaches are urban beaches located at Cleveland Lakefront State Park, which was the most visited state park in Ohio in 2010 (9,285,452 total visitor occasions in 2010) (43). Euclid Beach is a 200-m-long beach, and Villa Angela offers a 300-m-long beach extending from Euclid Beach to the mouth of Euclid Creek (8). Both beaches are protected by breakwaters, and possible sources of fecal contamination include a large population of birds using the east section of the bathing area, storm sewer outfalls feeding into Euclid Creek, and combined sewer overflows (CSOs) and sanitary sewer overflows (SSOs) occurring throughout the Euclid Creek drainage (5, 20). Villa Angela is suspected to be impacted more greatly by water from Euclid Creek, which is immediately adjacent to the east of the bathing area. The East Headlands (41°45′33″N, 81°17′21″W) and West Headlands (41°45′21″N, 81°17′37″W) beaches are located in a less densely populated area, but due to the 1.6-km-long, heavily used natural sand beaches at Headlands Beach State Park, this park was the second most visited state park in Ohio in 2010 (4,367,619 total visitor occasions in 2010) (43). Sources of fecal contamination include septic systems, wastewater treatment plant effluent, and storm water runoff (20).
Water samples were collected 35 times, once daily, from each of the four study beaches during the 2010 swimming season (from 13 July to 16 September 2010), with support of the Ohio Division of Parks and Recreation. Water samples were collected from beach waters in accordance with the Ohio Department of Health sampling guidelines for beach waters (40). Water collection at all beaches occurred at locations where the water was approximately 1 m deep, in the center of the beach, and in the same vicinity each collection day. Water samples were collected in 750-ml Whirl-Pak bags by sweeping the container 30 cm below the water surface, and the samples were transported on ice to a temporary field lab at the Ohio State University Extension office in Painesville (Lake County, OH) and filtered within an hour. Physicochemical parameters (water temperature, turbidity, pH, dissolved oxygen [DO], and water conductivity) were measured in situ using a YSI water quality data sonde (Yellow Springs Instruments, Yellow Springs, OH) and a Hach turbidimeter (Hach Company, Loveland, CO). The data for wave height and precipitation were obtained from the National Oceanic and Atmospheric Administration's National Weather Service (http://www.weather.gov).
Enumeration of indicator bacteria.
From the beach water samples, 100, 50, and 20 ml of water were filtered through a mixed cellulose ester filter (0.45-μm pore size and 47-mm diameter; Millipore, Bedford, MA) to enumerate fecal indicator bacteria (E. coli and enterococci). E. coli was cultured on modified m-TEC agar (Aquacheck Laboratory, Inc., Weathersfield, VT). The plates were preincubated at 35°C for 2 h and then incubated at 44.5°C for 18 to 20 h. Red/magenta-colored colonies were counted as E. coli (52). Enterococci were cultured on m-EI agar (Aquacheck Laboratory, Inc., Weathersfield, VT) at 41 ± 0.5°C for 24 ± 2 h. Colonies with a blue halo were counted as enterococci (54).
Concentration and DNA extraction.
Beach water samples (200 ml) were prefiltered through 20-μm-pore-size nylon filter membranes (Osmonics, Minnetonka, MN) to remove algae and debris and then filtered through a 0.45-μm-pore-size mixed cellulose ester filter membrane (Pall Corporation, Ann Arbor, MI). The membrane was transferred into a 50-ml sterile tube that contained 1.4 ml of ASL buffer from a QIAamp DNA stool kit (Qiagen, Valencia, CA) and 0.1-mm- and 0.5-mm-diameter autoclaved glass beads (0.3 mg each; Biospec Products, Bartlesville, OK). After bead beating with a Mini-Beadbeater 96 apparatus (BioSpec Products, Bartlesville, OK) at 2,100 oscillations/min for 1 min, the supernatant was transferred to a new 2-ml microcentrifuge tube, followed by DNA extraction using a QIAamp DNA stool kit according to the manufacturer's instructions, and then suspended in 200 μl of elution buffer. The eluates were used immediately or stored at −80°C until further processing.
Quantification of Arcobacter, fecal indicators, and tetracycline resistance genes by real-time qPCR.
Real-time PCR assays were carried out with an ABI 48-well StepOne Real Time system (Applied Biosystems, Foster City, CA), using the extracted DNA samples. SYBR green-based real-time qPCR was performed in duplicate, targeting the Arcobacter 23S rRNA gene (Arco) and the tetracycline resistance gene tetQ (TetQ) as described previously by Bastyns et al. (3), González et al. (23), and Nikolich et al. (39), with minor modifications (Table 1). The real-time qPCR mixture consisted of a total volume of 20 μl containing the DNA templates (2 μl [2-ml portion of water sample] for Arco and 5 μl [5-ml portion of water sample] for TetQ), 10 μl of SYBR green PCR master mix (Applied Biosystems, Foster City, CA), and 500 nM (each) primers (Table 1). Thermal cycling consisted of an initial cycle of 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 3 min. After amplification, melting curve analysis was performed by heating samples to 95°C for 15 s, cooling them to 60°C for 1 min for TetQ and for 3 min for Arco, and then heating them to 95°C at a rate of 1.0°C/s. TaqMan-based real-time qPCR analysis was performed in duplicate, targeting the 16S rRNA gene of Bacteroides-Prevotella (HuBac), the uidA gene of E. coli (UidA), and the 23S rRNA gene of Enterococcus spp. (Ent) as previously described by Bernhard and Field (4), Okabe et al. (45), Chern et al. (6), and Ludwig and Schleifer (36), with minor modifications. The real-time qPCR mixture consisted of a total volume of 20 μl containing 5 μl (5-ml portion of water sample) of the DNA template, 10 μl of TaqMan Universal PCR master mix (Applied Biosystems, Foster City, CA), 500 nM (each) primers, and 250 nM probe (Table 1). The PCR protocol included an initial cycle of 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min for UidA and 2 min for Ent and at 62°C for 1 min for HuBac. Cycle threshold (CT) values for real-time qPCR results were determined with thresholds of 0.2 for Arco and “auto” for the other genetic markers. The negative control for each qPCR assay consisted of a mixture of all PCR reagents with nuclease-free water. The following were used as positive controls: A. butzleri DSM 8739 (ATCC 49616) for Arco, Bacteroides fragilis ATCC 25285 for HuBac, Enterococcus faecium ATCC 19434 for Ent, Prevotella melaninogenica ATCC 25845 for TetQ (American Type Culture Collection, Manassas, VA), and E. coli DH5α for UidA.
Table 1.
Primers and probes used in this study
| Genetic marker | Primer or probe | Sequence (5′–3′)a | Amplicon size (bp) | Amplified gene | Target(s) | Reference(s) |
|---|---|---|---|---|---|---|
| HuBac | qHS601F | GTTGTGAAAGTTTGCGGCTCA | 150 | 16S rRNA | Bacteroides-Prevotella | 4, 45 |
| qHS624MGB | FAM-CGTAAAATTGCAGTTGA-MGB | |||||
| qBac725R | CAATCGGAGTTCTTCGTGATATCTA | |||||
| UidA | UidAF | CAACGAACTGAACTGGCAGA | 130 | uidA | E. coli | 6 |
| UidAP | FAM-CCCGCCGGGAATGGTGATTAC-MGB | |||||
| UidAR | CATTACGCTGCGATGGAT | |||||
| TetQ | tetQF | CATGGATCAGCAATGTTCAATATCGG | 460 | tetQ | tetQ gene | 39 |
| tetQR | CCTGGATCCACAATGTATTCAGAGCGG | |||||
| Ent | ECST784F | AGAAATTCCAAACGAACTTG | 91 | 23S rRNA | Enterococcus spp. | 36 |
| GPL813TQ | FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-MGB | |||||
| ENC854R | CAGTGCTCTACCTCCATCATT | |||||
| Arco | ArcoI | GTCGTGCCAAGAAAAGCCA | 331 | 23S rRNA | Arcobacter spp. | 3, 23 |
| ArcoII | TTCGCTTGCGCTGACAT |
R, A or G; FAM, 6-carboxyfluorescein; MGB, minor groove binder.
Quantification of genetic markers.
To create a standard curve for Arco quantification, amplicons generated from partial sequences of the 23S rRNA gene of A. butzleri DSM 8739 were amplified with forward and reverse primers (Table 1), purified by use of a QIAquick PCR purification kit (Qiagen, Valencia, CA), ligated into the pGEM-T vector (Promega Co., Madison, WI), and transformed into E. coli DH5α competent cells by use of a MicroPulser electroporator (25-μF capacitance, 1.25 kV, 200-Ω resistance; Bio-Rad, Hercules, CA). Plasmid DNA was prepared using a QIAprep Spin miniprep kit (Qiagen, Valencia, CA). The concentration of purified plasmid was measured with a NanoDrop system (NanoDrop Technologies, Wilmington, DE), which used a standard curve derived from 10-fold serial dilutions of plasmid DNA (1.19 × 107 to 1.19 × 10−1). Real-time PCR assays were performed three times independently in triplicate to make standard curves by plotting CT values from the real-time PCR assays versus the log of the bacterial gene copy numbers and drawing a trend line through these points. For UidA quantification, 10-fold serial dilutions of E. coli DH5α were prepared, and then a plot was made using the CT values from qPCR and log gene copy numbers. The concentrations of the other genetic markers were determined using standard curves generated in our previous study (32) and in the present study (data not shown). The detection limits of Arco, HuBac, UidA, TetQ, and Ent real-time qPCRs were 11.9, 240, 20.5, 10.0, and 24.0 gene copies/reaction, respectively. Samples measuring at or below the detection limit were replaced with a value equal to the detection limit for statistical analyses.
Seminested PCR and sequence analysis.
In order to evaluate the genetic diversity of Arcobacter sequences detected, a seminested PCR assay targeting the 16S rRNA gene was performed. The PCR procedure used was that described by Harmon and Wesley (24), with some modifications. The reaction mix was prepared with 2 μl of template (DNA from each sampling site), 2.5 μl of 10× PCR buffer, 0.5 U of Taq DNA polymerase, 0.75 mM MgCl2, a mix containing a 100 μM concentration of deoxynucleoside triphosphate (dNTP), and 200 nM (each) forward (Arco1, 5′-AGAGATTRGCCTGTATTGTAT-3′; R = A or G) (modified from the work of Harmon and Wesley [24]) and reverse (Arco2, 5′-AGAGATTGGCCTGTATTGTAT-3′) (24) primers that amplify a 1,201-bp region of the Arcobacter 16S rRNA gene, based on the sequence of Arcobacter butzleri CCUG 10373 (GenBank accession no. L14626), to make up a final volume of 25 μl. The following cycling protocol was used: initial denaturation at 94°C for 4 min; 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 7 min. For seminested PCR amplification, 0.5 μl of each first PCR product was added to a 20-μl (final volume) PCR mixture containing 2 μl of 10× PCR buffer, 0.5 U of Taq DNA polymerase, 1.5 mM MgCl2, a mix containing a 100 μM concentration of dNTP, and 500 nM (each) forward (338f, 5′-ACTCCTACGGGAGGCAGCAG-3′) (31) and reverse (Arco2, 5′-AGAGATTGGCCTGTATTGTAT-3′) (24) primers that amplify a 1,087-bp region of the Arcobacter 16S rRNA gene, based on the sequence of Arcobacter butzleri CCUG 10373 (GenBank accession no. L14626). The seminested PCR protocol was the same as the first PCR protocol. The seminested PCR results were analyzed by electrophoresis on a 1.5% agarose gel and staining with ethidium bromide.
Randomly selected Arcobacter-positive qPCR products (n = 13) and all of the Arcobacter-positive seminested PCR products (n = 20) were purified using a QIAquick PCR purification kit. They were sequenced at the Plant-Microbe Genomics Facility of the Ohio State University (http://pmgf.biosci.ohio-state.edu/), using a forward primer (Arco1 for the qPCR products and 338f for the seminested PCR products) and an ABI Prism 3730 DNA analyzer (Applied Biosystems). The sequences were compared with those available in the GenBank databases by using the PubMed NCBI BLAST program to identify sequenced products. To obtain accurate molecular identification of sequenced products, the partial 16S rRNA sequences of 20 seminested PCR products were aligned with those of Arcobacter reference strains from GenBank as proposed by Levican et al. (34), with some modification, and the sequences were aligned with Arcobacter reference strains. Preliminary alignment was done with Clustal X (version 1.8) (51), with multiple alignments set to default. Regions of ambiguous alignment were excluded. Campylobacter jejuni subsp. jejuni ATCC 33560 (GenBank accession no. M59298) was used as an outgroup. Phylogenetic relationships were inferred using the DNAPARS and DNADIST programs, followed by NEIGHBOUR, of the PHYLIP package (version 3.57) (15). A phylogenetic tree was determined using the Kimura two-parameter distance and neighbor-joining methods. The phylogenetic tree was plotted with TREEVIEW (version 1.6.1) (47). The statistical significance of phylogenies was estimated using the SEQBOOT program for bootstrap analysis with 1,000 pseudoreplicate data sets; only those having grouped with bootstrap values of >50% were considered significant.
Statistical analysis.
All statistical analyses were performed with the IBM SPSS (ver. 19.0.0; SPSS Inc.) statistics package. The data from the real-time qPCR assays and counting methods were log transformed for all analyses. The data obtained in this study were not normally distributed, except for the number of E. coli CFU per 100 ml and the pH data (Kolmogorov-Smirnov test); therefore, statistical analyses were conducted using nonparametric methods. The Kruskal-Wallis test and the Mann-Whitney U test were used to compare Arco levels between sites or dates. Spearman's correlation analysis was used to test the possible correlations between Arco, other genetic markers, and physicochemical parameters. To assess the correlation between Arco densities and the occurrence of a beach advisory, E. coli data were treated as binary, that is, a code of 0 was assigned when the level of E. coli was below 235 CFU/100 ml, and a code of 1 was assigned when the level of E. coli was above 235 CFU/100 ml.
Nucleotide sequence accession numbers.
The original sequences obtained in this study were deposited in the GenBank database under sequence accession numbers JQ302194 to JQ302206 and JQ754652 to JQ754671.
RESULTS
Water quality parameters and enumeration of fecal indicator bacteria.
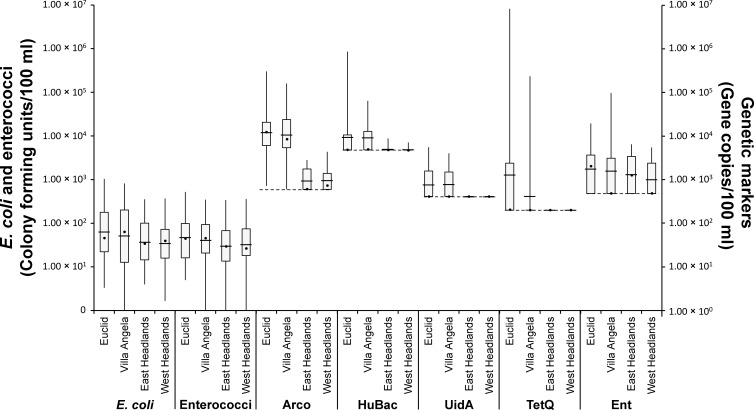
At each sampling site, physicochemical parameters were measured in situ or retrieved from the National Oceanic and Atmospheric Administration's National Weather Service (Table 2). The median values for most physicochemical parameters were similar among sampling sites. Only the median values for turbidity were significantly different among sampling sites (P = 0.007 by the Kruskal-Wallis test). This difference was particularly noticeable between the Euclid and Headlands beaches (P = 0.022; Mann-Whitney U test) and between the Villa Angela and Headlands beaches (P = 0.006; Mann-Whitney U test). In September, water temperature dropped sharply for all four beaches (P < 0.005 by the Kruskal-Wallis test and the Mann-Whitney U test). The mean levels and median levels of E. coli and enterococci at each sampling site are shown in Fig. 1; the levels of these indicator bacteria at the Euclid and Villa Angela beaches were higher than those at the Headlands beaches. However, no statistical significance of differences among sampling sites was observed (P > 0.05 by the Kruskal-Wallis test). In addition, 20.6% of the samples from the Euclid and Villa Angela beaches exceeded Ohio's daily maximum bacterial standard, whereas only one sample and two samples resulted in advisory conditions at the East and West Headlands beaches, respectively (Table 3). There was a positive correlation between E. coli and enterococci according to Spearman's correlation analysis (r = 0.580; P < 0.001). Of the physicochemical parameters, the only observed correlation with these fecal indicators, albeit weak, was with water temperature (r = 0.252 and P = 0.004 for E. coli; r = 0.217 and P = 0.014 for enterococci).
Table 2.
Summary of water quality parametersa
| Parameter | Statisticb | Value |
|||
|---|---|---|---|---|---|
| Euclid | Villa Angela | East Headlands | West Headlands | ||
| Water temp (°C) | Mean | 24.1 | 24.1 | 24.1 | 24.3 |
| Median | 25.1 | 25.1 | 25.2 | 25.3 | |
| Range | 19.7–27.0 | 19.5–27.0 | 19.0–28.0 | 19.3–27.2 | |
| Turbidity (NTUc) | Mean | 9.45 | 7.59 | 15.8 | 15.8 |
| Median | 2.70 | 2.70 | 7.40 | 7.00 | |
| Range | 1.10–37.6 | 1.30–41.7 | 1.80–99.9 | 1.20–99.9 | |
| pH | Mean | 8.49 | 8.46 | 8.50 | 8.52 |
| Median | 8.50 | 8.45 | 8.52 | 8.51 | |
| Range | 8.14–8.98 | 7.81–8.97 | 8.02–8.83 | 8.18–8.84 | |
| Conductivity (μS/cm) | Mean | 289 | 295 | 310 | 313 |
| Median | 310 | 308 | 311 | 318 | |
| Range | 149–333 | 182–347 | 255–413 | 227–416 | |
| Dissolved oxygen (mg/liter) | Mean | 10.9 | 11.9 | 11.7 | 12.4 |
| Median | 10.8 | 11.3 | 10.3 | 10.3 | |
| Range | 4.55–21.3 | 6.37–21.8 | 5.65–17.1 | 3.85–25.9 | |
| Wave ht (m) | Mean | 0.253 | 0.275 | 0.326 | 0.343 |
| Median | 0.100 | 0.160 | 0.190 | 0.230 | |
| Range | 0.00500–1.15 | 0.0100–1.20 | 0.0200–1.30 | 0.0200–1.29 | |
For all four beaches, the mean (median, range) precipitation was 0.233 (0, 0–2.24) cm.
Means are arithmetic means.
NTU, nephelometric turbidity units.
Fig 1.
Box plots showing geometric means and medians of E. coli, enterococcus, and genetic marker levels at four Lake Erie beaches. The length of each box shows the interquartile range and 50% of cases of the variable. The line and the dot in the box indicate the geometric mean and the median, respectively, while extended lines from the box show maximum and minimum values.
Table 3.
Occurrence of genetic markers in beach water samples and of advisory days during the study period
| Beach | No. of positive samples/total no. of samples (%) |
Occurrence of advisory daysa | ||||
|---|---|---|---|---|---|---|
| Arco | HuBac | UidA | TetQ | Ent | ||
| Euclid | 32/32 (100) | 12/33 (36.4) | 12/33 (36.4) | 14/33 (42.4) | 23/33 (69.7) | 7/34 (20.6) |
| Villa Angela | 31/33 (93.9) | 17/33 (51.5) | 12/33 (36.4) | 5/33 (15.2) | 16/33(48.5) | 7/34 (20.6) |
| East Headlands | 15/33 (45.5) | 2/33 (6.06) | 0/33 (0.00) | 0/33 (0.00) | 19/33 (57.6) | 1/35 (2.86) |
| West Headlands | 19/31 (61.3) | 2/33 (6.06) | 0/33 (0.00) | 0/33 (0.00) | 15/33 (45.5) | 2/34 (5.88) |
| Total | 97/129 (75.2) | 33/132 (25.0) | 24/132 (18.2) | 19/132 (14.4) | 73/132 (55.3) | 17/137 (12.4) |
Number of water samples exceeding Ohio's daily maximum bacterial standard (235 E. coli CFU/100 ml)/total number of samples (%).
qPCR analysis of Arcobacter spp. and fecal bacteria.
A total of 129 beach water samples were analyzed for Arcobacter spp., using a real-time qPCR assay. Arcobacter was detected frequently in the water samples from the four beaches. Ninety-seven samples (75.2%) were positive for Arcobacter, which was observed at a higher frequency than the other measured genetic markers, which occurred in 14.4% to 55.3% of the samples (Table 3). Nearly all samples collected at the Euclid (32 of 32 samples [100%]) and Villa Angela (31 of 33 samples [93.9%]) beaches were positive for Arcobacter spp., while these microbes were detected in nearly half of the samples collected at the Headlands beaches (15 of 33 samples [45.5%] for East Headlands and 19 of 31 samples [61.3%] for West Headlands). With the exception of Ent, the other genetic markers were detected frequently in the Euclid and Villa Angela samples, whereas UidA and TetQ were not detected and HuBac was detected infrequently in the Headlands samples (Table 3).
The levels of the Arco marker ranged from <5.95 × 102 (below the detection limit) to 3.00 × 105 gene copies/100 ml. The geometric means (GMs) of the Arco levels in the Euclid and Villa Angela samples were 1.18 × 104 (geometric standard deviation [GSD] = 3.94) and 1.05 × 104 (GSD = 4.10) gene copies/100 ml, respectively, which were >10-fold higher than those in samples from East Headlands (9.32 × 102 gene copies/100 ml; GSD = 1.81) and West Headlands (9.63 × 102 gene copies/100 ml; GSD = 1.84) (Fig. 1). The median values of the Arco levels in the Euclid and Villa Angela samples were also >10-fold higher than those in East Headlands samples (Fig. 1). The observed densities for Arco were significantly different among the sampling sites (P < 0.001 by the Kruskal-Wallis test), especially between the Euclid and Headlands beaches and the Villa Angela and Headlands beaches (P < 0.001 by the Mann-Whitney U test). Like the case for the Arco marker, the median values for HuBac, UidA, and TetQ were higher in the Euclid and Villa Angela samples, whereas the GMs of Ent were statistically different only between the Euclid and West Headlands samples (P < 0.005 by the Mann-Whitney U test).
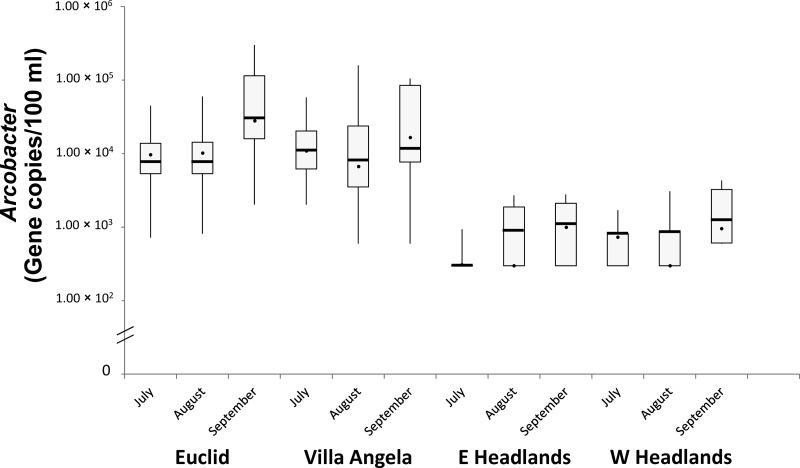
For the four beaches, the monthly GMs and medians for the Arco marker were higher in September than in the other months (Fig. 2), although the only significant month-to-month difference was shown for the Euclid samples (P = 0.039 by the Kruskal-Wallis test and P < 0.05 by the Mann-Whitney U test). It is noteworthy that the Arco levels in beach water samples were observed to be correlated weakly with water temperature (r = −0.245; P = 0.006) according to Spearman's correlation analysis (Table 4). With respect to temperature and Arco levels, a higher correlation coefficient was observed for Headlands beach waters (r = −0.346; P = 0.036) than for Euclid and Villa Angela beach waters (r = −0.284; P = 0.024) (Table 4). The occurrence of a beach advisory also had a weak correlation with the Arco level (r = 0.250; P = 0.005) (Table 4). Moreover, the Arco levels on beach advisory days (median value = 9.41 × 103 gene copies/100 ml) were higher than those on nonadvisory days (median value = 1.89 × 102 gene copies/100 ml) (P = 0.0052 by the Mann-Whitney U test). HuBac, UidA, and TetQ were negatively correlated with water temperature (r = −0.401 and P < 0.001 for HuBac, r = −0.206 and P = 0.020 for UidA, and r = −0.228 and P = 0.010 for TetQ). Meanwhile, according to Spearman's correlation analysis, Arco levels in the samples from the four beaches were correlated more strongly with the three genetic markers (r > 0.400; P < 0.001) than with the physicochemical parameters. This was most apparent in our observation of the stronger correlation between Arco and HuBac levels for Euclid and Villa Angela samples (r = 0.604; P < 0.001) (Table 4).
Fig 2.
Box plots showing monthly geometric means for Arcobacter levels at four Lake Erie beaches. The length of each box shows the interquartile range and 50% of cases of the variable. The line and the dot in the box indicate the geometric mean and the median, respectively, while extended lines from the box show maximum and minimum values.
Table 4.
Summary of Spearman's correlation analysis showing significant relationships between Arcobacter and other parameters
| Parameter | Spearman's correlation coefficient (P value) for relationship with Arco |
||
|---|---|---|---|
| Euclid and Villa Angela | East and West Headlands | Total | |
| HuBac | 0.604 (<0.001) | 0.592 (<0.001) | |
| UidA | 0.421 (<0.001) | ||
| TetQ | 0.306 (0.003) | 0.456 (<0.001) | |
| Water temp | −0.284 (0.024) | −0.346 (0.036) | −0.245 (0.006) |
| Turbidity | 0.278 (0.025) | 0.270 (0.031) | |
| Avg wind speed | 0.341 (0.005) | 0.287 (0.021) | 0.203 (0.021) |
| Wave ht | 0.303 (0.017) | ||
| Beach advisory | 0.250 (0.005) | ||
Genetic diversity of Arcobacter spp.
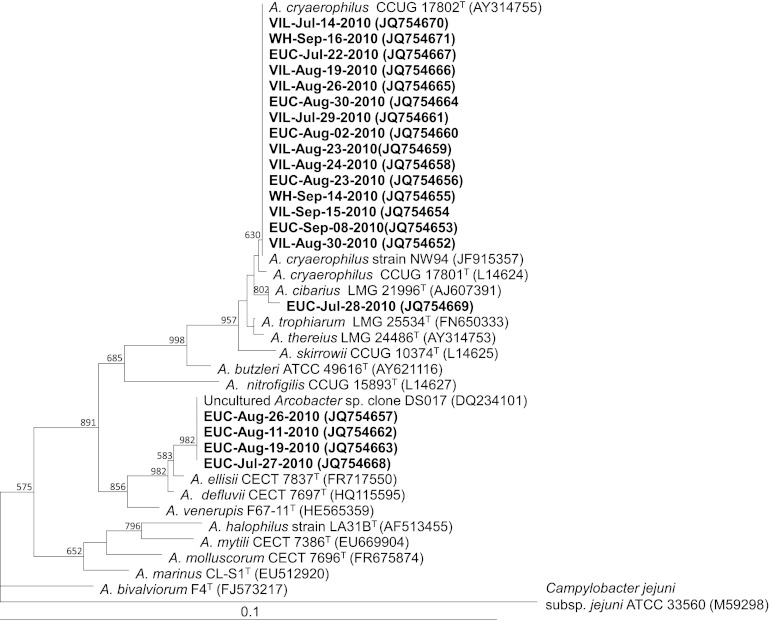
Of the 97 qPCR-positive samples (Table 3), only 20 samples (10 from Euclid, 8 from Villa Angela, and 2 from West Headlands) were positively detected with the seminested PCR targeting the partial 16S rRNA gene. Thirteen randomly selected qPCR products among the 97 qPCR-positive samples and the 20 seminested PCR-positive products were sequenced, and their sequences were identified as Arcobacter by comparison with those available in the GenBank databases, using the PubMed NCBI BLAST program (data not shown). To obtain more accurate identification of the sequenced products, phylogenetic analysis with the partial 16S rRNA sequences of seminested PCR products was performed. Three different Arcobacter phylotypes were found for the 20 Arcobacter sequences (451 bp) detected in this study (Fig. 3). Fifteen sequences (75.0%) were identical to or had one base pair difference from A. cryaerophilus strains, with significant bootstrap values (Fig. 3). Four sequences were identical to the uncultured environmental clone DS017 (GenBank accession no. DQ234101), detected in the Danshui River estuary in Northern Taiwan (35), with a 98% bootstrap value. These sequences were also closely related to A. ellisii (99.3% identity), which was recently added as an Arcobacter species (18). Lastly, one sequence showed high similarity to A. cibarius reference strains (99.8% identity), with an 80% bootstrap value.
Fig 3.
Phylogenetic tree constructed with the partial 16S rRNA gene sequences of 20 sequenced Arcobacter products. Campylobacter jejuni subsp. jejuni (GenBank accession no. M59298) was used as an outgroup. Bootstrap values of >50% are shown. The scale bar corresponds to 0.1 substitution per nucleotide position. The strain names are marked in bold letters: EUC, VIL, and WH denote Euclid, Villa Angela, and West Headlands, respectively, followed by the sampling dates. The numbers in parentheses are GenBank sequence accession numbers.
DISCUSSION
In this study, we investigated the occurrence of Arcobacter spp. in water from four Lake Erie beaches, which are popular destinations for local beachgoers and tourists in Ohio (43). Based on a report from the ODH (41), the four study sites are often contaminated with fecal pollution, and the Euclid and Villa Angela beaches are highly contaminated beaches, whereas the Headlands beaches are less contaminated. The previously reported ODH observations were supported by our results showing that the median levels of traditional fecal indicators (E. coli and enterococci) and both the occurrence and median levels of genetic markers (HuBac, UidA, and TetQ) were statistically higher in Euclid and Villa Angela waters than in Headlands waters (P < 0.005) (Fig. 1 and Table 3).
The Arcobacter results showed similar trends at these sites. Arcobacter was observed frequently at all of the beaches during the swimming season of 2010, but both the occurrence and median level of the Arco marker were higher for the Euclid and Villa Angela beaches than for the Headlands beaches (Fig. 1 and Table 3). The Arcobacter abundance in the beach waters with higher levels of fecal contamination was further supported by the statistical results. Arco levels in the samples from the four beaches were correlated with beach advisories (r = 0.250; P = 0.005) and with three genetic markers (HuBac, UidA, and TetQ) (r > 0.400; P < 0.001) (Table 4). Moreover, Arco levels were correlated significantly with HuBac levels at the highly contaminated beaches (Euclid and Villa Angela) (r = 0.604; P < 0.001). However, no significant relationship between Arco and HuBac levels was found at the less contaminated beaches (Headlands) (Table 4). The association between the Arco marker and fecal indicators in this study is consistent with previous findings demonstrating a positive association between Arcobacter spp. and bacterial indicators in freshwater and seawater (10, 11). Fong et al. (19) also reported that the Arcobacter-associated waterborne illness outbreak at Lake Erie's South Bass Island in 2004 was related to fecal contamination, whereby the highest density of Arcobacter spp. was observed in the water samples with the highest level of fecal contamination as determined by fecal indicator bacteria.
Our results showed that according to Spearman's correlation analysis, HuBac (a Bacteroides-Prevotella marker) was the genetic marker most strongly correlated with the Arco marker (r = 0.592; P < 0.001) among the fecal contamination markers (Table 4). It can be suggested that human fecal sources are likely to be a key contributor to Arcobacter contamination at these four Lake Erie beaches, since our Arcobacter results demonstrate an association with the HuBac marker, which was designed to be human specific (45). These results are interesting, particularly because E. coli and enterococcus levels measured by traditional culture-based methods, as well as the levels of the other two genetic markers (UidA and TetQ), which were designed to detect general fecal contamination (6, 39) without host specificity (2), did not show a significant correlation with Arco. In addition, beach advisories showed a weak relationship with Arco levels (r = 0.250; P = 0.005).
The abundance of Arcobacter was higher in September than in the other months (July and August) (Fig. 2) and was negatively correlated with water temperature (Table 4), which can be explained by previous research reporting that Arcobacter spp. may survive better at lower temperatures in water (17, 57). Therefore, it also can be suggested that water temperature is another key contributor to Arcobacter levels in Lake Erie beach water during the swimming season.
A general concern in the use of PCR detection for environmental monitoring is the potential presence of PCR inhibitors in water samples, as this may generate false-negative and/or biased results. In this study, the influence of any false-negative and/or biased results on our result interpretation could be negligible, because 122 of 129 beach water samples (94.6%) were positive for at least one of the genetic markers tested (data not shown). In future studies, the use of appropriate process controls should occur to reduce potential problems associated with PCR inhibition, as suggested by Wolf et al. (59).
Our results showed that 13 sequences from the qPCR products (partial 23S rRNA genes) were identified as Arcobacter sequences by comparison with those available in the GenBank databases, using the PubMed NCBI BLAST program. However, Arcobacter identification based on the 23S rRNA gene was less accurate than using other genetic markers, such as the 16S rRNA gene. The main reason that the identification of Arcobacter sequences by phylogenetic inference was limited was that there is very little information on 23S rRNA sequences of recently identified Arcobacter species, such as A. ellisii, A. defluvii, A. bivalviorum, and A. venerupis (12, 18, 34). Since the taxonomy of Arcobacter species has been based on analyses of the 16S rRNA gene (9, 58), a seminested PCR assay targeting the 16S rRNA gene was performed on the Arcobacter real-time qPCR-positive samples. Of 97 Arcobacter-positive samples, only 20 samples (10 for Euclid, 8 for Villa Angela, and 2 for West Headlands) showed visible bands on agarose gels (Fig. 3). Most of the 77 seminested PCR-negative samples did not seem to be falsely positive by qPCR, because 9 of 13 sequences from qPCR products could be obtained from seminested PCR-negative samples (data not shown). The lower recovery of Arcobacter sequences from the Headlands samples (2 seminested PCR-positive samples among 34 real-time qPCR-positive samples [5.9%]) than from the Euclid and Villa Angela samples (18/63 samples [28.6%]) may have been due to the low Arcobacter densities in Headlands samples (geometric mean, ∼20 gene copies/reaction [1,000 gene copies/100 ml]) (Fig. 3). The phylogenetic analysis showed that most of the Arcobacter sequences detected in the Lake Erie beach waters (15 of 20 sequences) were closely related to A. cryaerophilus (Fig. 3), which is prevalent in fecally contaminated water (9, 10) and frequently associated with human gastrointestinal diseases and animal diseases (8). This result was different from those of other studies showing that both A. butzleri and A. cryaerophilus were the most prevalent Arcobacter species in natural freshwater environments (10, 11, 16). The higher prevalence of A. cryaerophilus in this study can be explained by the use of qPCR; A. cryaerophilus in other water studies may have been underestimated due to the difficulty in isolating this organism (because of its susceptibility to antimicrobials used in the isolation medium) or because of a lower growth rate than that of A. butzleri (22) with the culture methods used in those studies. Among the other five sequences, one sequence was only one base pair different from reference strains of A. cibarius, which was first isolated from chicken broiler carcasses (27) and is associated with pigs and pig environments (7), suggesting that Arcobacter contamination in Lake Erie may originate partly from animal-associated fecal sources. The other four sequences were identical to the uncultured environmental clone DS017 (GenBank accession no. DQ234101), detected in the Danshui River estuary in Northern Taiwan (35), and were also closely related to A. ellisii (99.3% identity), which was isolated from mussels (18). These data indicate that Arcobacter species which have not yet been isolated or are not well studied may be considerably present in Lake Erie. Consequently, phylogenetic analysis suggests that Lake Erie beach water may serve as a possible reservoir for Arcobacter species that may originate from human and animal sources, some of which could be human pathogens.
In summary, our results demonstrate that human-pathogenic Arcobacter was prevalent in the water at four Lake Erie beaches during the 2010 swimming season. Since the major contamination sources of Arcobacter may originate from human-associated fecal contamination, it is important to identify and manage its sources to minimize public health risks linked to Arcobacter exposure in this region.
ACKNOWLEDGMENTS
This study was funded by research grants from the Lake Erie Protection Fund and the U.S. EPA Great Lakes Restoration Initiative (grant GL-00E00582).
References to commercial products or trade names are made with the understanding that no endorsement or discrimination by The Ohio State University and the funding sources is implied.
We thank Frank Lichtkoppler Scott and Randall Zondag at the Ohio State University Extension, Scott Fletcher at the Ohio Department of Natural Resources, and Nancy Niehus at the Lake County General Health District for their support.
Footnotes
Published ahead of print 1 June 2012
REFERENCES
- 1. Assanta MA, Roy D, Lemay M-J, Montpetit D. 2002. Attachment of Arcobacter butzleri, a new waterborne pathogen, to water distribution pipe surfaces. J. Food Prot. 65:1240–1247 [DOI] [PubMed] [Google Scholar]
- 2. Barnes B, Gordon DM. 2004. Coliform dynamics and the implications for source tracking. Environ. Microbiol. 6:501–509 [DOI] [PubMed] [Google Scholar]
- 3. Bastyns K, et al. 1995. A variable 23S rDNA region is a useful discriminating target for genus-specific and species-specific PCR amplification in Arcobacter species. Syst. Appl. Microbiol. 218:353–356 [Google Scholar]
- 4. Bernhard AE, Field KG. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bertke EE. 2007. Composite analysis for Escherichia coli at coastal beaches. J. Great Lakes Res. 33:335–341 [Google Scholar]
- 6. Chern EC, Brenner K, Wymer L, Haugland RA. 2009. Comparison of fecal indicator bacteria densities in marine recreational waters by QPCR. Water Qual. Expo. Health 1:203–214 [Google Scholar]
- 7. Chinivasagam HN, Corney BG, Wright LL, Diallo IS, Blackall PJ. 2007. Detection of Arcobacter spp. in piggery effluent and effluent-irrigated soils in southeast Queensland. J. Appl. Microbiol. 103:418–426 [DOI] [PubMed] [Google Scholar]
- 8. Cleveland Lakefront State Park 2011. Cleveland lakefront/headlands beach state parks. http://www.clevelandlakefront.org/
- 9. Collado L, Figueras MJ. 2011. Taxonomy, epidemiology and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24:174–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collado L, Inza I, Guarro J, Figueras MJ. 2008. Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ. Microbiol. 10:1635–1640 [DOI] [PubMed] [Google Scholar]
- 11. Collado L, et al. 2010. Occurrence and diversity of Arcobacter spp. along the Llobregat River catchment, at sewage effluents and in a drinking water treatment plant. Water Res. 44:3696–3702 [DOI] [PubMed] [Google Scholar]
- 12. Collado L, Levican A, Perez J, Figueras MJ. 2011. Arcobacter defluvii sp. nov., isolated from sewage. Int. J. Syst. Evol. Microbiol. 61:1895–1901 [DOI] [PubMed] [Google Scholar]
- 13. Engberg J, On SLW, Harrington CS, Gerner-Smidt P. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ertas N, Dogruer Y, Gonulalan Z, Guner A, Ulger I. 2010. Prevalence of Arcobacter species in drinking water, spring water, and raw milk as determined by multiplex PCR. J. Food Prot. 73:2099–2102 [DOI] [PubMed] [Google Scholar]
- 15. Felsenstein J. 1995. PHYLIP: phylogeny inference package, version 3.57c. University of Washington, Seattle, WA [Google Scholar]
- 16. Fera MT, et al. 2004. Detection of Arcobacter spp. in the coastal environment of the Mediterranean Sea. Appl. Environ. Microbiol. 70:1271–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fera MT, et al. 2010. Specific detection of Arcobacter spp. in estuarine waters of Southern Italy by PCR and fluorescent in situ hybridization. Lett. Appl. Microbiol. 50:65–70 [DOI] [PubMed] [Google Scholar]
- 18. Figueras MJ, Levican A, Collado L, Inza MI, Yustes C. 2011. Arcobacter ellisii sp. nov., isolated from mussels. Syst. Appl. Microbiol. 34:414–418 [DOI] [PubMed] [Google Scholar]
- 19. Fong TT, et al. 2007. Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie, South Bass Island, Ohio. Environ. Health Perspect. 115:856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Francy DS, Gifford AM, Darner RA. 2003. Escherichia coli at Ohio bathing beaches—distribution, sources, wastewater indicators, and predictive modeling. Water Resources Investigations report 02-4285. US Geological Survey, Columbus, OH [Google Scholar]
- 21. González A, Botella S, Montes RM, Moreno Y, Ferrús MA. 2007. Direct detection and identification of Arcobacter species by multiplex PCR in chicken and wastewater samples from Spain. J. Food Prot. 70:341–347 [DOI] [PubMed] [Google Scholar]
- 22. González A, Ferrús MA. 2011. Study of Arcobacter spp. contamination in fresh lettuces detected by different cultural and molecular methods. Int. J. Food Microbiol. 145:311–314 [DOI] [PubMed] [Google Scholar]
- 23. González A, Suski J, Ferrús MA. 2010. Rapid and accurate detection of Arcobacter contamination in commercial chicken products and wastewater samples by real-time polymerase chain reaction. Foodborne Pathog. Dis. 7:327–338 [DOI] [PubMed] [Google Scholar]
- 24. Harmon KM, Wesley IV. 1996. Identification of Arcobacter isolates by PCR. Lett. Appl. Microbiol. 23:241–244 [DOI] [PubMed] [Google Scholar]
- 25. Ho HTK, Lipman LJA, Gaastra W. 2006. Arcobacter, what is known about a potential foodborne zoonotic agent! Vet. Microbiol. 115:1–13 [DOI] [PubMed] [Google Scholar]
- 26. Houf K, et al. 2004. Antimicrobial susceptibility patterns of Arcobacter butzleri and Arcobacter cryaerophilus strains isolated from humans and broilers. Microb. Drug Resist. 10:243–247 [DOI] [PubMed] [Google Scholar]
- 27. Houf K, et al. 2005. Arcobacter cibarius sp. nov., isolated from broiler carcasses. Int. J. Syst. Evol. Microbiol. 55:713–717 [DOI] [PubMed] [Google Scholar]
- 28. Kabeya H, et al. 2003. Distribution of Arcobacter species among livestock in Japan. Vet. Microbiol. 93:153–158 [DOI] [PubMed] [Google Scholar]
- 29. Kabeya H, et al. 2004. Prevalence of Arcobacter species in retail meats and antimicrobial susceptibility of the isolates in Japan. Int. J. Food Microbiol. 90:303–308 [DOI] [PubMed] [Google Scholar]
- 30. Kiehlbauch JA, et al. 1991. Campylobacter butzleri sp. nov. isolated from humans and animals with diarrheal illness. J. Clin. Microbiol. 29:376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow MD. (ed), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom [Google Scholar]
- 32. Lee CS, Lee J. 2010. Evaluation of new gyrB-based real-time PCR system for the detection of B. fragilis as an indicator of human-specific fecal contamination. J. Microbiol. Methods 82:311–318 [DOI] [PubMed] [Google Scholar]
- 33. Lerner J, Brumberger V, Preac-Mursic V. 1994. Severe diarrhea associated with Arcobacter butzleri. Eur. J. Clin. Microbiol. Infect. Dis. 13:660–662 [DOI] [PubMed] [Google Scholar]
- 34. Levican A, et al. 2012. Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst. Appl. Microbiol. 35:133–138 [DOI] [PubMed] [Google Scholar]
- 35. Liao P-C, Huang BH, Huang S. 2007. Microbial community composition of the Danshui river estuary of Northern Taiwan and the practicality of the phylogenetic method in microbial barcoding. Microb. Ecol. 54:497–507 [DOI] [PubMed] [Google Scholar]
- 36. Ludwig W, Schleifer KH. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556–562 [DOI] [PubMed] [Google Scholar]
- 37. Moreno Y, et al. 2003. Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and fluorescent in situ hybridization. Appl. Environ. Microbiol. 69:1181–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morita Y, et al. 2004. Isolation and phylogenetic analysis of Arcobacter spp. in ground chicken meat and environmental water in Japan and Thailand. Microbiol. Immunol. 48:527–533 [DOI] [PubMed] [Google Scholar]
- 38a. Nationwide Survey on Recreation and the Environment 2004. Participation rates for outdoor activities in 2004. http://www.srs.fs.usda.gov/trends/RECUPDATES/recupdate0804.pdf
- 39. Nikolich MP, Hong G, Shoemaker NB, Salyers AA. 1994. Evidence for natural horizontal transfer of tetQ between bacteria that normally colonize humans and bacteria that normally colonize livestock. Appl. Environ. Microbiol. 60:3255–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ohio Department of Health 2009. Sampling procedures—beach monitoring. Ohio Department of Health, Columbus, OH: http://www.odh.ohio.gov/odhPrograms/eh/bbeach/beach_procedures.aspx [Google Scholar]
- 41. Ohio Department of Health 2011. Beach monitoring. Ohio Department of Health, Columbus, OH: http://www.odh.ohio.gov/odhPrograms/eh/bbeach/beachmon.aspx [Google Scholar]
- 42. Ohio Department of Natural Resources 2010. Ohio's Lake Erie public access guidebook. Ohio Department of Natural Resources, Columbus, OH: http://www.ohiodnr.com/LinkClick.aspx?fileticket=4NHRJGcaEOM%3d&tabid=21033 [Google Scholar]
- 43. Ohio Department of Natural Resources 2011. Ohio State Parks 2010 annual report. Ohio Department of Natural Resources, Columbus, OH: http://www.dnr.state.oh.us/Portals/2/annualreports/2010annualreport.pdf [Google Scholar]
- 44. Ohio Environmental Protection Agency 2005. South Bass Island, Ottawa County, gastrointestinal illness summer 2004. Ohio Environmental Protection Agency, Columbus, OH [Google Scholar]
- 45. Okabe S, Okayama N, Savichtcheva O, Ito T. 2007. Identification and quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwaters. Appl. Microbiol. Biotechnol. 74:890–901 [DOI] [PubMed] [Google Scholar]
- 46. O'Reilly CE, et al. 2007. A waterborne outbreak of gastroenteritis with multiple etiologies among resort island visitors and residents: Ohio, 2004. Clin. Infect. Dis. 44:506–512 [DOI] [PubMed] [Google Scholar]
- 47. Page RD. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 48. Rivas L, Fegan N, Vanderlinde P. 2004. Isolation and characterization of Arcobacter butzleri from meat. Int. J. Food Microbiol. 91:31–41 [DOI] [PubMed] [Google Scholar]
- 49. Samie A, Obi CL, Barrett LJ, Powell SM, Guerrant RL. 2007. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa: studies using molecular diagnostic methods. J. Infect. 54:558–566 [DOI] [PubMed] [Google Scholar]
- 50. Reference deleted.
- 51. Thompson JD, Gibson TJ, Jeanmougin F, Higgins DG. 1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. US Environmental Protection Agency 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). EPA-821-R-02-023 Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 53. US Environmental Protection Agency 2002. National beach guidance and required performance criteria for grants. EPA-823-B-02-004 Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 54. US Environmental Protection Agency 2006. Method 1600: enterococci in water by membrane filtration using membrane-enterococcus indoxyl-β-d-glucoside agar (mEI). EPA-821-R-06-009 Office of Water, US Environmental Protection Agency, Washington, DC [Google Scholar]
- 55. Vandamme P, De Ley J. 1991. Proposal for a new family, Campylobacteraceae. Int. J. Syst. Bacteriol. 41:451–455 [Google Scholar]
- 56. Vandamme P, et al. 1991. Revision of Campylobacter, Helicobacter and Wolinella taxonomy: emendation of generic description and proposal for Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41:88–103 [DOI] [PubMed] [Google Scholar]
- 57. Van Driessche E, Houf K. 2008. Survival capacity in water of Arcobacter species under different temperature conditions. J. Appl. Microbiol. 105:443–451 [DOI] [PubMed] [Google Scholar]
- 58. Wesley IV, Schroeder-Tucker L, Baetz AL, Dewhirst FE, Paster BJ. 1995. Arcobacter-specific and Arcobacter butzleri-specific 16S rRNA-based DNA probes. J. Clin. Microbiol. 33:1691–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolf S, Hewitt J, Greening GE. 2010. Viral multiple quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 76:1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wybo I, Breynaert J, Lauwers S, Lindenburg F, Houf K. 2004. Isolation of Arcobacter skirrowii from a patient with chronic diarrhea. J. Clin. Microbiol. 42:1851–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]