Abstract
Leptospirosis is a potentially deadly zoonotic disease that afflicts humans and animals. Leptospira interrogans, the predominant agent of leptospirosis, encounters diverse conditions as it proceeds through its life cycle, which includes stages inside and outside the host. Unfortunately, the number of genetic tools available for examining the regulation of gene expression in L. interrogans is limited. Consequently, little is known about the genetic circuits that control gene expression in Leptospira. To better understand the regulation of leptospiral gene expression, the L. interrogans kdp locus, encoding homologs of the P-type ATPase KdpABC potassium transporter with their KdpD sensors and KdpE response regulators, was selected for analysis. We showed that a kdpE mutation in L. interrogans prevented the increase in kdpABC mRNA levels observed in the wild-type L. interrogans strain when external potassium levels were low. To confirm that KdpE was a positive regulator of kdpABC transcription, we developed a novel approach for constructing chromosomal genetic fusions to the endogenous bgaL (β-galactosidase) gene of the nonpathogen Leptospira biflexa. We demonstrated positive regulation of a kdpA′-bgaL fusion in L. biflexa by the L. interrogans KdpE response regulator. A control lipL32′-bgaL fusion was not regulated by KdpE. These results demonstrate the utility of genetic fusions to the bgaL gene of L. biflexa for examining leptospiral gene regulation.
INTRODUCTION
Pathogenic species of the spirochete Leptospira encounter a variety of environmental conditions throughout their life cycles (1, 18). The natural reservoirs for Leptospira are typically small rodents, which release the spirochetes from their renal tubules into the surrounding environment during urination. The leptospires contaminating the soil or water enter humans or animals through skin abrasions or mucous membranes and hematogenously disseminate to a number of organs, where they can cause severe systemic illness (1, 18). The genome sequences of four pathogenic and two nonpathogenic Leptospira strains revealed a large number of genes encoding potential transcriptional regulatory proteins, which are thought to regulate the expression of leptospiral genes as the spirochetes encounter these different environments (14). Whole-transcriptome studies and analyses of individual genes have identified a number of Leptospira interrogans genes that are regulated by culture conditions and the host environment (2). However, a true understanding of the genetic circuitry underlying the response of Leptospira to environmental stimuli cannot be obtained until the regulatory proteins or RNAs involved in the control of specific genes are identified.
Genetic approaches to identifying and examining trans-acting factors that regulate gene expression in pathogenic Leptospira are scant (18). Targeted gene disruption is extremely difficult and has been accomplished for only ligB and fliY (11, 21). Random insertional mutagenesis of pathogenic Leptospira can be performed by introduction of the mariner-based Himar1 transposon by transformation or conjugation (31, 33). Some of the Himar1 insertion mutations disrupted genes encoding potential transcriptional regulatory proteins and can aid the study of genetic regulatory systems in L. interrogans (31). For example, an L. interrogans mutation in a gene encoding a PerR homolog has revealed its role in the peroxide stress response and the genes controlled by the regulatory protein (23).
Many genetic studies have been performed with the nonpathogen Leptospira biflexa, since it is more amenable to genetic manipulation than L. interrogans. Targeted gene disruptions and high-frequency random Himar1 mutagenesis have been performed on L. biflexa (24, 34, 39). L. biflexa is easily transformed with a plasmid carrying the origin of replication of the LE1 bacteriophage (37). The plasmid allows investigations of the functional properties of L. interrogans gene products in L. biflexa. In a recent study, expression of the L. interrogans adhesins LigA and LigB in L. biflexa enabled L. biflexa to serve as a surrogate host for studies of leptospiral binding to fibronectin and fibrinogen (9). Recently, a plasmid-based green fluorescent protein (GFP) reporter was developed to measure promoter activities of leptospiral genes in L. biflexa (3, 8). When the L. biflexa hsp20 and groES heat shock promoters were fused to gfp, GFP levels increased with temperature (3). The gfp reporter was also used to assess the promoter activities of the L. interrogans genes lig, sph2, and lipL41 in L. biflexa grown under different culture conditions (8).
Here, we describe a novel β-galactosidase reporter system for examining the regulation of expression of leptospiral genes in L. biflexa by trans-acting factors. We selected the L. interrogans kdp locus to test the reporter because of the well-understood role of the E. coli KdpE response regulator in activating transcription of the kdp operon encoding a P-type ATPase transporter in which translocation of the essential mineral potassium across the transmembrane KdpA transporter subunit is coupled to the transient phosphorylation of an invariant aspartate residue in the KdpB ATPase subunit (5). When E. coli is starved for potassium, the KdpE response regulator is phosphorylated by the KdpD histidine kinase and activates transcription of the kdpFABC operon encoding the subunits of the high-affinity potassium uptake system, KdpFABC (15). We found that a Himar1 insertion mutation in the L. interrogans kdpE gene prevented the increase in kdpABC mRNA levels observed in the wild-type L. interrogans strain in response to low potassium levels, indicating that KdpE was a positive regulator of kdpABC transcription when potassium is limiting. We next fused the kdpE promoter and translation initiation region from L. interrogans kdpA to the gene encoding the endogenous β-galactosidase on the chromosome of L. biflexa. Expression of L. interrogans KdpE in trans significantly increased β-galactosidase expression from the fusion, confirming that KdpE is a positive regulator of kdp expression. This reporter system should have broad applicability to the investigation of leptospiral gene expression.
MATERIALS AND METHODS
Strains and culture conditions.
L. interrogans serovar Manilae strain L495, L. interrogans serovar Copenhageni strain Fiocruz L1-130, and L. biflexa servoar Patoc strain Patoc 1 (Paris) have been described previously (17, 35). The L. interrogans serovar Manilae mutant strain M45 has the transposon Himar1 inserted into the kdpE gene (Fig. 1) (31). The L. interrogans L495 and M45 strains were generous gifts from Ben Adler and Gerald Murray. The leptospiral culture medium EMJH was assembled as described previously and included lactalbumin hydrolysate (Becton Dickinson), superoxide dismutase, sodium pyruvate, and 100 μg/ml 5-fluorouracil (43). L. interrogans Fiocruz L1-130 was cultivated in EMJH supplemented with 1% heat-inactivated rabbit serum. The serum was omitted when cultivating L. biflexa and L. interrogans L495 and M45. For experiments comparing the properties of the L495 and M45 strains, “low-K+” EMJH was made by replacing KH2PO4 with NaH2PO4, and “standard” EMJH was made by adding potassium chloride to low-K+ EMJH to achieve a final concentration of 2 mM. Leptospira cultures were placed in Erlenmeyer flasks and incubated on a rotary shaker at 30°C. The growth of cultures was monitored by counting the leptospires under a dark-field microscope.
Fig 1.
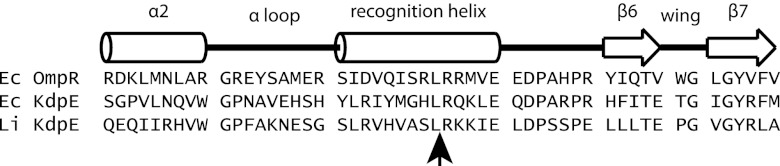
Location of the Himar1 insertion in the predicted structure of L. interrogans KdpE. The alignment of the amino acid sequence of the carboxy terminus of L. interrogans KdpE with that of the winged helix-turn-helix of E. coli OmpR was aided by an earlier published alignment of E. coli KdpE and OmpR (26). The secondary structure of OmpR is depicted with cylinders (α helix) and open arrows (β strand). The location of the Himar1 insertion in the L. interrogans M45 strain is marked by an arrow below the KdpE sequence.
Frozen competent Escherichia coli NEB5α was obtained from New England BioLabs. LB broth and LB agar powders were supplied by Genesee Scientific (San Diego, CA) and reconstituted with water as recommended by the manufacturer prior to autoclaving. Kanamycin and spectinomycin were added to LB medium to a final concentration of 40 μg/ml for selection of E. coli transformants.
Animals.
To obtain low-passage-number spirochetes for the in vitro experiments, L. interrogans was passaged through hamsters (13). Female Golden Syrian hamsters were purchased from Harlan Laboratories (Indianapolis, IN), and 1,000 motile L. interrogans strain L495 and strain M45 spirochetes in 0.5 ml of EMJH were injected by the intraperitoneal route into the hamsters. The animals were euthanized when they displayed signs of illness. Their kidneys were removed, ground in phosphate-buffered saline (PBS), and inoculated into semisolid EMJH (0.2% agarose). The cultures were incubated at 30°C. All animal procedures were approved by the West Los Angeles Veterans Affairs Institutional Animal Care and Use Committee.
Reagents.
5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was supplied by Genesee Scientific. ortho-Nitrophenyl-β-d-galactopyranoside (ONPG), spectinomycin dihydrochloride, chloroform, isopropanol, and kanamycin monosulfate were obtained from Sigma. Nuclease-free water was obtained from Ambion. All other chemicals were purchased from Fisher Scientific.
RNA analysis.
RNA was extracted from L. interrogans strains with TRIzol reagent (Invitrogen) as described previously (29). Absorbances were read with the NanoVue spectrophotometer (GE Healthcare). Two micrograms of each RNA was converted into cDNA with Superscript II reverse transcriptase (Invitrogen) and random nonamers (Sigma) following the manufacturer's instructions. The amounts of specific cDNA molecules were determined by quantitative PCR using the Bio-Rad iQ5 Real-time System. Each reaction mixture contained cDNA derived from 10 ng of RNA, 0.4 μM each primer, and 12.5 μl iQ SYBR green Supermix raised to a total volume of 25 μl with nuclease-free water. The assay was performed on three independent sets of samples, and each sample was assayed in triplicate. The ratio of kdp transcript to flaA2 transcript was determined by the 2−ΔΔCT method (22).
To determine whether adjacent genes in kdpABC are cotranscribed, the cDNA was amplified with the primer pairs kdpA-4F/kdpB-3R and kdpB4F/kdpC-6R to amplify the kdpA-kdpB and kdpB-kdpC intergenic regions, respectively (Table 1). PCR was performed with the HotStarTaq Master Mix from Qiagen.
Table 1.
PCR primers
| Oligonucleotide namea | Sequence (5′ to 3′)b | Coordinatesc | Purposed |
|---|---|---|---|
| bgaL(Bm)-1F | ATCggatccTCCTcatatgATCTTTGGAGCCTGTTATTACCCA | 1, 27 | Plasmid |
| bgaL(Sc)-2R | ACAgagctcGCGTGAGTATTGTTGCGTGGCAGTGAC | 879, 853 | Plasmid |
| kdpp(Xh)-1F | CTTATActcgagATCTTAACAATTTCTTAACTTCCTT | −214, −190 | Plasmid |
| kdpp(Nd)-2R | CCTATCcatatgAATTCCTTAAAACTCTTCCGGTCTA | −4, −28 | Plasmid |
| lipL32p(Kp)-1F | AGCGACggtaccTCAATTTGTGTCTGAGATTTGA | −231, −210 | Plasmid |
| lipL32p(Xh)-2R | AGActcgagTTCcatatgCTCTCCTTAGTTAGGAAAATCACG | −4, −27 | Plasmid |
| tufp(Kp)-1F | TCCAGGggtaccTAGTTGGTCCTATTTCACATTTG | −307, −285 | Plasmid |
| tufp(Xh)-2R | ATActcgagTTCcatatgGACTTTTTAATCTCCTTACTTTTGAAC | −4, −30 | Plasmid |
| flaB1p(Kp)-1F | CTCGCTggtaccGATCGAACCTAAGATTAGCTCA | −312, −291 | Plasmid |
| flaB1p(Xh)-2R | GTCctcgagTTCcatatGTTTCCTCCTTGAAACTGATC | −4, −24 | Plasmid |
| kdpE(Nd)-1F | CCTCTAcatatgAATCCTAAAATTCTGGTGGC | 1, 23 | Plasmid |
| kdpE(Xh)-4R | AATGctcgagTTATAAATGAATTGCAAGCCTATAACCTAC | 684, 655 | Plasmid |
| pGKlep4-2F | ATCGGTGCGGGCCTCT | 5422, 5437 | Verification |
| bgaL-4R | TCTACAATCTCTGGACCAAACATC | 1430, 1407 | Verification |
| kdpA-1F | TGCTGACCGTATTTTTAAGCGGGATA | 1139, 1164 | qRT-PCR |
| kdpA-2R | AAACCGTGAGGGCCTCGATTTGT | 1334, 1302 | qRT-PCR |
| kdpB-1F | TTGCCGCAGAAGCGGGAGTG | 1457, 1476 | qRT-PCR |
| kdpB-2R | TGCGCTAATGCAGGCGCATC | 1598, 1579 | qRT-PCR |
| kdpC-3F | TTTCCATTCGGCTTCTTTTG | 26, 45 | qRT-PCR |
| kdpC-4R | TACTTCCGCTGGATCGAAAC | 133, 114 | qRT-PCR |
| flaA2-5F | CATCTTACTTGCTGGACTGCTCTGC | 15, 39 | qRT-PCR |
| flaA2-6R | ATCTGGGTTTGCCCCTGTTTG | 123, 103 | qRT-PCR |
| kdpp-1F | ATCTTAACAATTTCTTAACTTCCTT | −214, −189 | Cotranscription |
| kdpA-3R | TCGGAAAAAGTTTGTACAACTCCA | 590, 567 | Cotranscription |
| kdpA-4F | GGAAATAATGGAAGTGCTTTTGCT | 1369, 1392 | Cotranscription |
| kdpB-3R | CCCGGATAATAACATCGTAAGAGC | 663, 640 | Cotranscription |
| kdpB-4F | GCTTTGTTCGGAAGTTTTTATGCT | 1792, 1815 | Cotranscription |
| kdpC-6R | GCCTTCCGTTAGTTTACGTGACTT | 447, 424 | Cotranscription |
F, forward primer; R, reverse primer.
Restriction sites are in lowercase; start and stop codons are underlined.
Position of the 5′-most and 3′ nucleotide complementary to the target, relative to the first nucleotide in the start codon.
Plasmid, for plasmid construction; Verification, for verification of proper integration of bgaL fusions into the L. biflexa chromosome; qRT-PCR, quantitative RT-PCR.
Plasmids.
All plasmid purifications, DNA fragment purification from gel slices, and DNA purification from liquids were performed with the Zyppy Plasmid Miniprep Kit, Zyppy Gel Extraction Kit, and Zyppy DNA Clean & Concentrator-5 Kit, respectively, from Zymo Research (Irvine, CA). Genomic DNA was purified from L. interrogans Fiocruz L1-130 and L. biflexa with the Wizard genomic DNA purification kit from Promega and served as the PCR template for the plasmid constructions. Synthetic oligonucleotides were purchased from Invitrogen. Restriction enzymes, linkers, Klenow fragment, and Quick T4 DNA ligase were purchased from New England BioLabs. Phusion DNA polymerase was used for all PCRs done for plasmid constructions and was purchased from New England BioLabs. Ladder I DNA molecular weight standards were obtained from Genesee. All plasmid insertions were sequenced by Laragen (Culver City, CA).
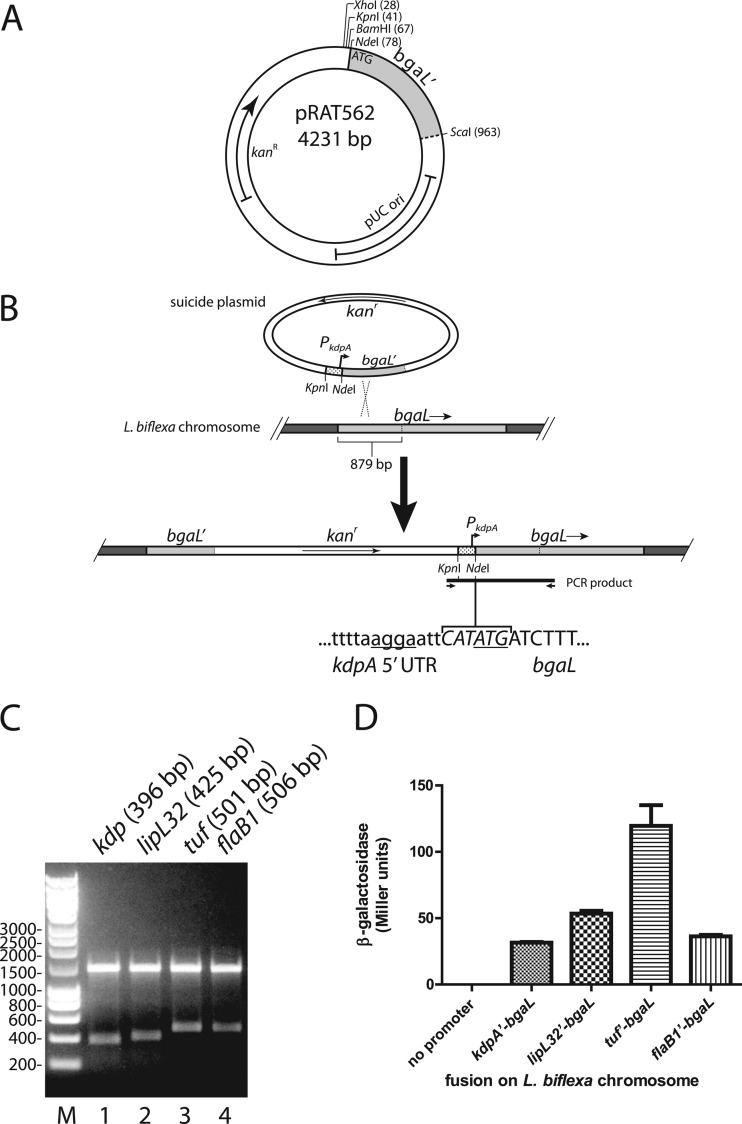
pRAT562 harbors the 5′ fragment of bgaL (for beta-galactosidase of Leptospira) that targets the plasmid for integration into the bgaL gene of L. biflexa. To construct pRAT562, the first 293 codons of the L. biflexa bgaL gene were amplified from the genomic DNA of L. biflexa by PCR with the primers bgaL(Bm)-1F and bgaL(Sc)-2R (Table 1). The bgaL amplicon was then digested with BamHI and SacI. pGKlep4 was digested with BamHI and SacI to remove all leptophage sequences and attached to the digested bgaL DNA to create pRAT562, which encodes resistance to kanamycin (see Fig. 4A).
Fig 4.
Integration of bgaL fusions into the chromosome of L. biflexa. (A) Sequence features of pRAT562. Restriction sites used for cloning are shown. The first 293 of the 661 codons of L. biflexa bgaL are present in pRAT562. (B) Scheme for integration of the kdpA promoter and translation initiation signals upstream of the bgaL coding region on the chromosome of L. biflexa. The kdpA promoter and 5′ untranslated region cloned into pRAT562 are depicted by the stippled segment. The bgaL sequences are shaded in light gray. Homologous recombination between the plasmid and chromosome occurred within the first 879 bp of bgaL. The integrated sequence was detected by PCR with a forward primer that annealed within the plasmid vector sequence and a reverse primer that annealed within the bgaL sequence downstream of the cloned segment. The nucleotide sequences of the NdeI recognition site and flanking regions are shown. The Shine-Dalgarno sequence and start codon are underlined. Lowercase, kdpA sequence; italics, NdeI recognition sequence; uppercase, second and third bgaL codons. (C) PCR products were digested with NdeI and subjected to electrophoresis in a 1.0% agarose gel. The expected sizes of the smaller fragments are indicated in parentheses. (D) β-Galactosidase activities measured from various chromosomal bgaL fusions. Each bar represents the mean β-galactosidase expression from three independent cultures. The error bars denote the standard deviations.
To fuse the promoters and translation initiation regions of various L. interrogans genes to the 5′ end of the bgaL sequence in pRAT562, the sequences upstream of the start codons of kdpA (LIC10990), lipL32 (LIC11352), tuf (EF-Tu, LIC12875), and flaB1 (LIC11890) were amplified from Fiocruz L1-130 genomic DNA by PCR with the oligonucleotide pairs kdpp(Xh)-1F/kdpp(Nd)-2R, lipL32p(Kp)-1F/lipL32p(Xh)-2R, tufp(Kp)-1F/tufp(Xh)-2R, and flaB1p(Kp)-1F/flaB1p(Xh)-2R, respectively (Table 1). The kdp PCR product was digested with XhoI and NdeI and inserted into pRAT562 that was digested with the same enzymes (see Fig. 4B). The lipL32, tuf, and flaB1 amplicons were digested with KpnI and NdeI and similarly inserted into pRAT562. These plasmids were transformed into L. biflexa for integration into the bgaL gene.
pRAT575 was the vector used to express the L. interrogans KdpE protein from an autonomous plasmid in L. biflexa. pRAT575 was constructed by inserting a KpnI linker (5′-GGGTACCC-3′) and an XhoI linker (5′-CCTCGAGG-3′) into the PvuII site and the NgoMIV site of the shuttle plasmid pSLe94 (4) (a generous gift from Mathieu Picardeau), respectively. The NgoMIV 5′ overhangs were filled in with Klenow fragment prior to insertion of the XhoI linker. pRAT575 encodes resistance to spectinomycin.
The shuttle plasmids expressing L. interrogans KdpE from heterologous promoters were constructed in two steps. The promoters and translation initiation regions of lipL32, tuf, and flaB1 were fused to kdpE (LIC10994) on pGKlep4, and the kdpE fusions were subsequently transferred to pRAT575 (4). For the first step, the lipL32, tuf, and flaB1 promoter/translation initiation regions were amplified by PCR with the primer pairs listed above (Table 1). The PCR products and pGKlep4 were digested with KpnI and XhoI, and the promoter fragments were inserted into the pGKlep4 backbone. The reverse primers contained an NdeI recognition site near the XhoI site (Table 1). The kdpE protein-coding region was amplified by PCR with the primers kdpE(Nd)-1F and kdpE(Xh)-4R, digested with NdeI and XhoI, and inserted downstream of the promoters that were cloned into pGKlep4. Finally, the promoter-kdpE fusions were excised by digestion with KpnI and XhoI and transferred to pRAT575 to generate the plasmids used (see Fig. 5).
Fig 5.
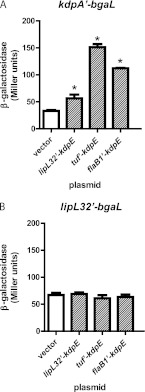
Positive regulation of the kdpA′-bgaL fusion by KdpE. (A) The L. biflexa kdpA′-bgaL strain was transformed with pSLe94 (vector) or a KdpE expression plasmid. (B) Same as panel A, except the L. biflexa strain harbored the lipL32′-bgaL fusion. Each bar represents the mean β-galactosidase expression from three independent cultures. The error bars denote the standard deviations. *, P < 0.05 (1-way ANOVA and Dunnett's multiple-comparison posttest).
Integration of fusions into the L. biflexa chromosome.
The plasmids harboring the bgaL fusions were denatured to promote homologous recombination with the bgaL gene in the chromosome (34), and 20 μl of each plasmid (4 to 5 μg) was denatured by adding 2 μl of 2 M NaOH and 2 mM EDTA and incubating the mixture for 30 min at 37°C. The mixtures were neutralized by addition of 2 μl 3 M sodium acetate (NaOAc), pH 5.2. To deplete the salt, the DNA was drop dialyzed against 40 ml water for 60 min (28). The denatured plasmid was then removed from the filter and transformed into L. biflexa by electroporation (37). Leptospires that had the plasmid integrated into the chromosome were selected by plating onto EMJH plates containing 30 μg/ml kanamycin. The plates also contained 20 μg/ml X-Gal to qualitatively assess the β-galactosidase expression of the transformants.
To verify integration of the fusion at the correct location, genomic DNA was extracted with the DNeasy Blood and Tissue Kit (Qiagen) and amplified with HotStarTaq (Qiagen) with the pGKlep4-2F and bgaL-4R primers (Table 1). The amplicons were digested with NdeI, and the digestion products were analyzed by electrophoresis in a 1.0% agarose gel (Sea-Kem LE agarose from Lonza).
β-Galactosidase assay.
Three colonies from each L. biflexa transformation were incubated in EMJH with 30 μg/ml kanamycin and 40 μg/ml spectinomycin. L. biflexa carrying the bgaL fusions was grown to an optical density at 420 nm (OD420) of 0.1 to 0.4. β-Galactosidase expression levels were determined essentially as described by Miller (30). In brief, cultures were chilled on ice for 20 min. The assay buffer comprised 9.6 ml of Z buffer (100 mM sodium phosphate, 10 mM KCl, 1 mM MgSO4, pH 7.0), 0.4 ml of 0.1% SDS, and 27 μl of β-mercaptoethanol. Five hundred microliters of the chilled cultures was mixed by vortexing with 500 μl assay buffer and 15 μl chloroform in 13- by 100-mm borosilicate glass tubes (VWR) for 10 s, and the mixtures were placed in a 28°C water bath for 5 min. Control reaction mixtures containing EMJH instead of the culture were included for background subtraction. The reaction was initiated by adding 200 μl of 4 mg/ml o-nitrophenol-β-d-galactoside and allowed to proceed for at least 25 min. When the reaction mixtures turned yellow, the reactions were terminated by the addition of 500 μl of 1 M Na2CO3, and the reaction time was recorded. Slow reactions were terminated at 90 min. All reaction mixtures were clarified by centrifugation at 16,000 × g for 3 min in an Eppendorf 5424 microcentrifuge, and the absorbance of the supernatant fluid was read at 420 nm (A420) with the Ultrospec 2000 (GE Healthcare). Miller units were calculated with the following equation (30): Miller units = (1,000 × A420)/(t × v × OD420), where t is the reaction time in minutes, v is the volume of culture mixed with the assay buffer (0.5 ml), and OD420 is the optical density of the culture.
Statistical analysis.
Statistical analysis was performed with GraphPad (La Jolla, CA) Prism 5. The statistical tests used are described in the figure legends. Growth rates during the logarithmic phase of growth were calculated with the Trendline function in Microsoft Excel 2000.
RESULTS
We first verified that KdpE was a regulator of kdpABC expression in L. interrogans before testing the L. biflexa reporter system with the kdp genes. An L. interrogans kdpE mutant designated M45 was one of the Himar1 insertion mutants isolated by Murray et al. (31). The insertion point of the transposon in M45 follows codon 199 in kdpE (Fig. 1) (31). To predict which structural element of the KdpE response regulator was disrupted by the transposon, the amino acid sequence of LIC10994 was aligned with the DNA-binding domain of E. coli OmpR, a response regulator whose crystal structure had been solved (26). Alignment was aided by earlier published alignments of the DNA-binding domains of OmpR and E. coli KdpE (27). The alignment indicates that the transposon was inserted into the recognition helix of the putative winged helix-turn-helix of KdpE (Fig. 1).
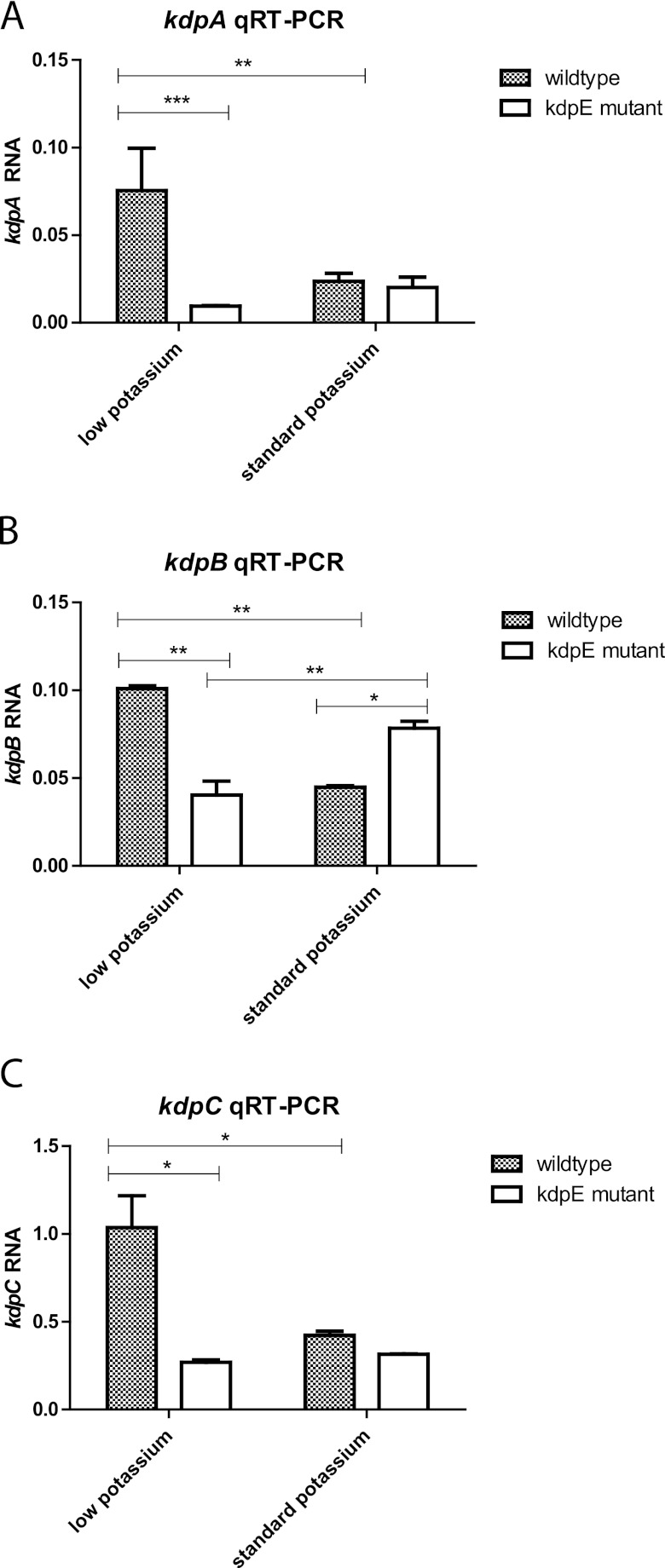
To determine whether KdpE stimulated transcription of kdpABC during growth at a low concentration of potassium, the wild-type and M45 strains were cultivated in “low-K+” EMJH, which was assembled by replacing potassium phosphate with sodium phosphate in the EMJH recipe, and “standard” EMJH, which was obtained by adding potassium to a final concentration of 2 mM (see Materials and Methods). Over three independent experiments, there were no significant differences in the growth rates among the four cultures (one-way analysis of variance [ANOVA]; P = 0.7964). RNA from the wild-type and M45 strains grown in standard and low-K+ EMJH was extracted and analyzed by quantitative reverse transcriptase PCR (RT-PCR) with primers specific for kdpA (Table 1). A basal level of kdpA transcript was observed in the M45 strain under both growth conditions. When the wild-type strain was grown in low-K+ EMJH, kdpA transcript levels were 3-fold higher than in standard EMJH, indicating that kdp transcription was increased in response to low potassium (Fig. 2A). In contrast, kdpA transcript levels failed to increase when strain M45 was grown in low-K+ medium instead of standard EMJH. In low-K+ medium, kdpA transcript levels were an average of 8-fold higher in the wild-type strain than in strain M45, while in standard EMJH medium there was no significant difference in kdpA transcript levels between the wild-type and M45 strains. These results are consistent with the notion that L. interrogans responds to limiting potassium levels by activating the KdpE response regulator, which then enhances transcription from the kdp promoter so that higher levels of the subunits for the high-affinity Kdp-ATPase potassium transporter can be expressed.
Fig 2.
Effect of the kdpE mutation on kdp transcript levels in low-potassium medium. Transcript levels were determined by quantitative reverse transcriptase (qRT) PCR as described in Materials and Methods. Signals from the kdpABC mRNA were normalized to flaA2 transcript levels. Representative results from three independent experiments are shown. The error bars denote the standard deviation. *, P < 0.01; **, P < 0.001; ***, P < 0.0001 (2-way ANOVA).
We also examined transcript segments encoding the other two subunits of the Kdp transporter. The transcript levels for the kdpB and kdpC sequences were significantly higher (2.3- and 3.0-fold, respectively) in the wild-type strain than in M45 during growth in low-K+ EMJH (Fig. 2B and C). The kdpE mutation had no significant effect on kdpC transcript levels in standard EMJH. However, kdpB transcript levels were 1.7-fold higher in the kdpE mutant than in the wild-type strain (Fig. 2B) (P < 0.01; 2-way ANOVA). The biological significance of this modest effect, if any, is unclear. It is possible that KdpE functions as a negative regulator of kdpB when potassium is abundant. Nevertheless, the results suggest that KdpE is a positive regulator of kdpABC transcription in L. interrogans when the external potassium concentration is low.
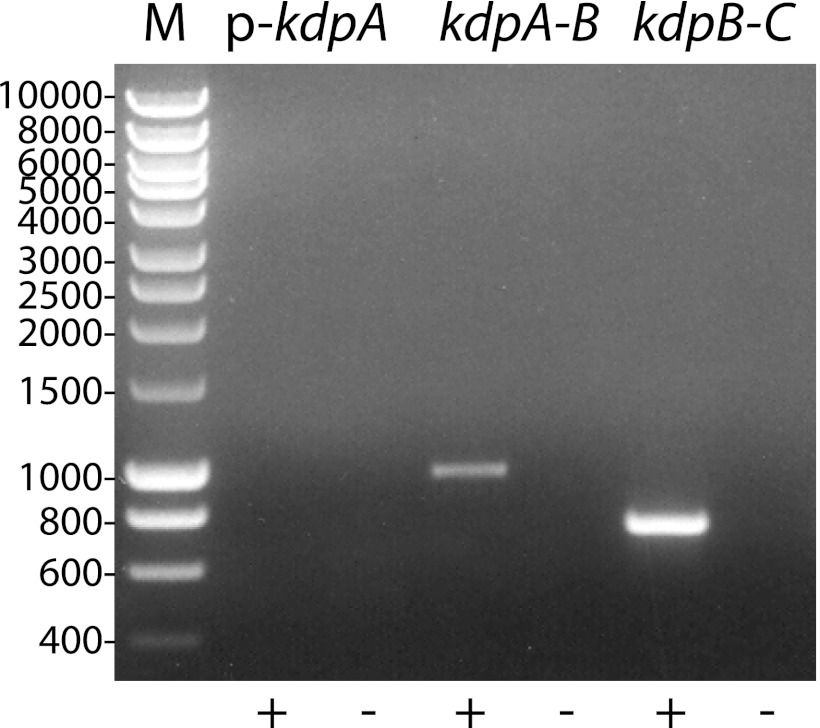
The intergenic regions of the kdpABC sequence were analyzed by RT-PCR to determine whether the genes were cotranscribed. Figure 3 shows that primers flanking the kdpAB and kdpBC intergenic regions successfully generated amplicons by RT-PCR. The negative-control primer pair with forward and reverse primers annealing to the kdp promoter region and kdpA, repectively, failed to generate an amplicon by RT-PCR (Fig. 3, lane 2). All primer pairs tested generated amplicons from L. interrogans genomic DNA (data not shown). These results indicate that adjacent genes in kdpABC are cotranscribed.
Fig 3.
Cotranscription of adjacent genes in kdpABC. Total RNA was extracted from L. interrogans grown under low-potassium conditions. Primer pairs flanking the intergenic regions of kdpABC were used for reverse transcriptase PCR. The primer pairs that included the forward oligonucleotide targeting the kdp promoter region served as a negative control.
We decided to monitor expression of kdp by fusing kdpA to a β-galactosidase reporter to examine the effects of expressing KdpE in trans on kdp expression. We selected the nonpathogen L. biflexa as the host strain for our experiments because of its ease of genetic manipulation compared to pathogenic strains of Leptospira and because it does not encode Kdp orthologs that may interfere with regulation of kdpA expression.
L. biflexa formed blue colonies on EMJH plates containing X-Gal, indicating that functional β-galactosidase was already being produced by the spirochete. A single gene in the L. biflexa genome (LEPBIa0024) is annotated as encoding a β-galactosidase. The L. biflexa and well-characterized E. coli β-galactosidases share little sequence identity and belong to different families of glycosyl hydrolases. E. coli β-galactosidase is a member of glycosyl hydrolase family 2, whereas L. biflexa β-galactosidase is a member of family 42. Despite their unrelated protein sequences, the catalytic domains of the two β-galactosidase variants share the TIM barrel superfold that includes the two glutamate residues that participate in catalysis (16).
To exploit the endogenous β-galactosidase gene of L. biflexa, we replaced the sequence upstream of the start codon of LEPBIa024 with the corresponding region from various genes of L. interrogans. Since only a single plasmid capable of replicating in L. biflexa was available, the fusion was placed on the chromosome so that the plasmid remained available for expression of KdpE and other potential regulatory gene products. The first 293 of the 661 codons of LEPBIa0024 were cloned into a suicide plasmid unable to replicate in L. biflexa, creating the vector pRAT562 (Fig. 4A). The sequences upstream of the kdpA, lipL32, tuf (EF-Tu), and flaB1 protein-coding regions were then inserted in front of the partial LEPBIa0024 coding sequence in pRAT562. The cloned segments comprised the sequence between the protein-coding region of the upstream gene and the start codon of the four genes. The five plasmids, including pRAT562, were next denatured with sodium hydroxide and transformed into L. biflexa by electroporation (Fig. 4B). The plasmid was treated with sodium hydroxide rather than UV light to minimize mutagenesis of the DNA. After a 24-hour period of outgrowth in liquid EMJH, transformants were selected on EMJH plates containing kanamycin and X-Gal. Because the plasmid DNA lacked an origin of replication that could function in L. biflexa, colonies could be recovered only if the plasmid integrated into the chromosome by homologous recombination at the 5′ end of LEPBIa0024. The colonies that grew following integration of pRAT562 were white on X-Gal plates, confirming that LEPBIa0024 was the sole source of β-galactosidase in L. biflexa during growth on EMJH. On the other hand, chromosomal integration of the plasmids harboring the kdpA, lipL32, tuf, and flaB1 fusions resulted in blue colonies on X-Gal plates. Because LEPBIa0024 encoded β-galactosidase activity, the gene was named bgaL (for beta-galactosidase of Leptospira), following the convention adopted for naming β-galactosidase reporters belonging to glycosyl hydrolase family 42, including bgaB from Bacillus stearothermophilus and bgaH from Haloferax lucentense (12, 38).
Colonies were inoculated into EMJH medium containing kanamycin and grown to mid- to late log phase. PCR was employed to confirm that the plasmid properly integrated into the bgaL gene on the chromosome. Genomic DNA was purified from the spirochetes and analyzed by PCR with a forward primer annealing upstream of the L. interrogans sequence within the vector sequence and a reverse primer annealing downstream of the bgaL segment that was cloned into the plasmid (Fig. 4B). A PCR product would be observed only if the plasmid integrated into the chromosome at the expected location. Amplicons of the expected sizes were obtained (data not shown). Digestion of the amplicons with NdeI released fragments containing the upstream regions of kdpA, lipL32, tuf, and flaB1 of the sizes anticipated (396 bp, 425 bp, 501 bp, and 506 bp, respectively) (Fig. 4B and C).
We measured the β-galactosidase activity expressed by the L. biflexa bgaL fusion strains essentially as described by Miller's standard protocol (see Materials and Methods). β-Galactosidase activity was not detected from a strain with the promoterless bgaL construct (Fig. 4D). A range of β-galactosidase activities was expressed by the strains that carried fusions of the 5′ flanking regions of L. interrogans genes to bgaL, with tuf producing the highest levels (Fig. 4D).
Following the successful demonstration of β-galactosidase expression driven by different L. interrogans promoters, we determined whether the kdpA′-bgaL fusion could be regulated by L. interrogans KdpE in trans. We cloned the protein-coding region of kdpE from L. interrogans immediately downstream of the lipL32, tuf, and flaB1 promoters in the pSLe94 shuttle vector, which carries the LE1 bacteriophage replication origin and a gene encoding resistance to spectinomycin. The plasmids were then transformed into the L. biflexa strain carrying the kdpA′-bgaL fusion, and colonies were selected on EMJH plates containing kanamycin and spectinomycin. As a control, the pSLe94 vector was also transformed into L. biflexa. The colonies were transferred to standard liquid EMJH containing kanamycin and spectinomycin for growth.
Bar graphs of the results are shown in Fig. 5A. As expected, the expression levels of the kdpA′-bgaL strain transformed with pSLe94 were similar to those expressed without the plasmid (Fig. 4D and 5A). The plasmid expressing KdpE from the lipL32 promoter and translation initiation region increased β-galactosidase expression 1.7-fold, from 33 to 56 Miller units. When KdpE expression was driven by tuf, β-galactosidase activity from the kdpA′-bgaL fusion increased 4.5-fold to 151 units. Similarly, when KdpE was expressed in trans from flaB1 upstream signals, β-galactosidase expression increased 3.4-fold to 112 units. These results confirmed that KdpE of L. interrogans is a positive regulator of kdp expression. When the same kdpE expression plasmids were introduced into an L. biflexa strain harboring the control lipL32′-bgaL fusion, which was not expected to be regulated by KdpE, no effect of KdpE on fusion expression was discerned (Fig. 5B).
DISCUSSION
We developed a novel reporter gene system that enables translational fusion of leptospiral genes directly to the chromosomal copy of bgaL in L. biflexa (Fig. 4B). This system can be deployed to examine the regulation of leptospiral genes by trans-acting factors. Since complementation of L. interrogans mutations is difficult, this approach also provides an independent genetic approach to support conclusions derived from the study of L. interrogans mutations that disrupt trans-acting regulatory factors that control gene expression. To demonstrate an application of the bgaL reporter, we constructed a kdpA′-bgaL fusion to show that leptospiral KdpE positively regulates the expression of the P-type ATPase transporter Kdp (Fig. 4B and 5).
The chromosomal bgaL reporter has several advantages over the plasmid-based green fluorescent protein reporter described in earlier studies (3, 8). First, since only one plasmid capable of replicating stably in L. biflexa is available, placing the fusion on the chromosome allows genes encoding candidate regulatory factors to be cloned easily into the plasmid to test whether they can control fusion expression in trans. Second, background levels of β-galactosidase in L. biflexa with its bgaL gene disrupted is extremely low (Fig. 4D), permitting the study of weakly expressed genes. In contrast, the relatively high background fluorescence of L. biflexa may obscure changes in the expression of weak promoters fused to gfp (8). Third, the copy number of the bgaL fusion remains stable, since the fusion is located on the chromosome while the copy number of plasmid-borne fusions may be affected by culture conditions. Finally, the 5′ untranslated region of the fusion transcripts originated from the L. interrogans gene being tested. This should permit examination of small RNAs and other regulatory proteins that target the 5′ untranslated regions of mRNAs to control gene expression posttranscriptionally. Leptospira is likely to express small RNAs, since all the leptospiral genomes sequenced to date harbor a gene encoding a CsrA homolog, a protein that binds to specific regulatory small RNAs (19).
A minor limitation of our fusion constructs as currently designed is that translational initiation at the artificial fusion junction may be blocked by secondary structures, as would be the case with translational fusions to any reporter. This appears be the case with flaB1, which expressed low levels of β-galactosidase when fused to bgaL (Fig. 4D) yet was able to exert 4-fold positive control on the kdpA′-bgaL fusion when driving kdpE expression (Fig. 5A). When the 60 nucleotides encompassing the translation initiation region of flaB1′-bgaL was folded using Mfold (44), most of the Shine-Dalgarno sequence was located in double-stranded RNA, whereas the Shine-Dalgarno sequence remained single stranded when the corresponding regions flanking the start codons of flaB1 and the flaB1′-kdpE transcripts were folded (data not shown). Moving the fusion junction downstream to include part of the coding region of the test gene might minimize problems with reporter sequences causing formation of double-stranded RNA within the translation initiation region.
LipL32 is the most abundant protein in L. interrogans (25), yet the β-galactosidase level expressed from the lipL32′-bgaL fusion was barely higher than that of the kdpA′-bgaL fusion in the absence of KdpE (Fig. 4D). Moreover, when its expression was driven by lipL32, KdpE barely enhanced β-galactosidase expression from the kdpA′-bgaL fusion (Fig. 5A). A recent study has shown that lipL32 expression is altered during the interaction of L. interrogans with macrophage-derived cell lines in vitro (41). Strong expression of lipL32′-bgaL may require a transcriptional regulator that is not present in L. biflexa. Alternatively, some of the lipL32 promoter elements may be missing from the fusion. Further studies with additional lipL32′-bgaL fusion constructs may allow us to distinguish between these possibilities.
Expression of the L. interrogans kdpE gene in L. biflexa growing in EMJH activated expression of the kdpA′-bgaL fusion despite the plentiful potassium in the culture medium and the absence of KdpD. Although KdpE must be phosphorylated to activate transcription from the kdp promoter in E. coli (15), phosphorylation of KdpE may not be necessary for the response regulator to enhance transcription of the kdp genes in Leptospira. Alternatively, L. interrogans KdpE may be phosphorylated somehow when expressed in L. biflexa. In other bacteria, a response regulator may be phosphorylated by a noncognate histidine kinase when the gene encoding the cognate sensor is deleted (20). In many cases, a phosphatase activity of the cognate sensor prevents cross talk under noninducing conditions by removing the phosphate from the response regulator. In E. coli, KdpD possesses a phosphatase activity that dominates over its kinase when potassium is plentiful in the culture medium (6). Therefore, the absence of KdpD in L. biflexa may permit stable phosphorylation of KdpE by aberrant cross talk from a noncognate histidine kinase. Although acetylphosphate can phosphorylate KdpE in vitro, it is unlikely to be the phosphate donor in vivo, since L. biflexa lacks the genes encoding phosphate acetyltransferase and acetate kinase, which are necessary to convert acetyl-coenzyme A (CoA) or acetate to acetylphosphate (40).
The role of the kdp gene products in potassium metabolism of L. interrogans, if any, remains unknown. The growth rate of the L. interrogans kdpE mutant was not affected even when the potassium concentration in the culture medium was minimized. Residual potassium in one or more of the EMJH components may have allowed growth of the kdpE mutant. It is also possible that the kdp genes are not required for growth when the potassium concentration is limited, as is the case for Staphylococcus aureus (42). Free-living bacteria with the Trk potassium transporter can lack Kdp orthologs (10). Neither of the two sequenced L. biflexa strains possesses the kdp genes, but they, along with the four sequenced pathogenic Leptospira strains, encode orthologs of the TrkA and TrkH subunits of Trk (7, 32, 35, 36), which may be capable of transporting enough potassium into the leptospires to support their growth even when potassium is limited. These observations suggest that the kdp gene products provide a function for L. interrogans beyond satisfying the spirochete's need for potassium. The absence of the kdp locus from the genomes of the two sequenced Leptospira borgpetersenii Hardjo strains, which lack a significant host-free environmental phase, suggest that their gene products are involved in the adaptation of L. interrogans following entry into or exit from the host (7, 29).
We are currently exploiting the L. interrogans kdpE mutant and the kdpA′-bgaL reporter strain to search for other L. interrogans genes that are regulated by KdpE. The bgaL reporter system described here can also be readily adapted to study the functions of other transcriptional or posttranscriptional regulators.
ACKNOWLEDGMENTS
We are grateful to Ben Adler and Gerald Murray for providing us with the L. interrogans L495 and M45 strains and the kdp sequences and to Mathieu Picardeau for providing us with the plasmid pSLe94. We thank David Haake, Henry Choy, and Jane Babbitt for helpful discussions. We also thank Alice Shapiro for technical support.
This study was supported by VA Medical Research Funds (to J.M.).
J.M conceived and designed the experiments, performed the experiments, analyzed the data, and wrote the paper. J.M. and M.L.C. contributed reagents, materials, and analysis tools.
Footnotes
Published ahead of print 8 June 2012
REFERENCES
- 1. Adler B, de la Peña Moctezuma A. 2010. Leptospira and leptospirosis. Vet. Microbiol. 140:287–296 [DOI] [PubMed] [Google Scholar]
- 2. Adler B, Lo M, Seemann T, Murray GL. 2011. Pathogenesis of leptospirosis: the influence of genomics. Vet. Microbiol. 153:73–81 [DOI] [PubMed] [Google Scholar]
- 3. Aviat F, Slamti L, Cerqueira GM, Lourdault K, Picardeau M. 2010. Expanding the genetic toolbox for Leptospira species by generation of fluorescent bacteria. Appl. Environ. Microbiol. 76:8135–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bauby H, Saint Girons I, Picardeau M. 2003. Construction and complementation of the first auxotrophic mutant in the spirochaete Leptospira meyeri. Microbiology 149:689–693 [DOI] [PubMed] [Google Scholar]
- 5. Bramkamp M, Altendorf K, Greie JC. 2007. Common patterns and unique features of P-type ATPases: a comparative view on the KdpFABC complex from Escherichia coli (review). Mol. Membr. Biol. 24:375–386 [DOI] [PubMed] [Google Scholar]
- 6. Brandon L, Dorus S, Epstein W, Altendorf K, Jung K. 2000. Modulation of KdpD phosphatase implicated in the physiological expression of the Kdp ATPase of Escherichia coli. Mol. Microbiol. 38:1086–1092 [DOI] [PubMed] [Google Scholar]
- 7. Bulach DM, et al. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cerqueira GM, et al. 2011. Development of transcriptional fusions to assess Leptospira interrogans promoter activity. PLoS One 6:e17409 doi:10.1371/journal.pone.0017409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choy HA, et al. 2011. The multifunctional LigB adhesin binds homeostatic proteins with potential roles in cutaneous infection by pathogenic Leptospira interrogans. PLoS One 6:e16879 doi:10.1371/journal.pone.0016879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corratgé-Faillie C, et al. 2010. Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell. Mol. Life Sci. 67:2511–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Croda J, et al. 2008. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect. Immun. 76:5826–5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gregor D, Pfeifer F. 2001. Use of a halobacterial bgaH reporter gene to analyse the regulation of gene expression in halophilic archaea. Microbiology 147:1745–1754 [DOI] [PubMed] [Google Scholar]
- 13. Haake DA. 2006. Hamster model of leptospirosis. Curr. Protoc. Microbiol. 12:12E.2 doi:10.1002/9780471729259.mc12e02s02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haake DA, Matsunaga J. 2010. Leptospira: a spirochaete with a hybrid outer membrane. Mol. Microbiol. 77:805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heermann R, Jung K. 2010. The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. FEMS Microbiol. Lett. 304:97–106 [DOI] [PubMed] [Google Scholar]
- 16. Hidaka M, et al. 2002. Trimeric crystal structure of the glycoside hydrolase family 42 β-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose. J. Mol. Biol. 322:79–91 [DOI] [PubMed] [Google Scholar]
- 17. Ko AI, et al. 1999. Urban epidemic of severe leptospirosis in Brazil. Lancet 354:820–825 [DOI] [PubMed] [Google Scholar]
- 18. Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lapouge K, Schubert M, Allain FH, Haas D. 2008. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67:241–253 [DOI] [PubMed] [Google Scholar]
- 20. Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145 [DOI] [PubMed] [Google Scholar]
- 21. Liao S, et al. 2009. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 9:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 23. Lo M, et al. 2010. Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect. Immun. 78:4850–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Louvel H, Saint Girons I, Picardeau M. 2005. Isolation and characterization of FecA- and FeoB-mediated iron acquisition systems of the spirochete Leptospira biflexa by random insertional mutagenesis. J. Bacteriol. 187:3249–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malmstrom J, et al. 2009. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460:762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martínez-Hackert E, Stock AM. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109–124 [DOI] [PubMed] [Google Scholar]
- 27. Martínez-Hackert E, Stock AM. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301–312 [DOI] [PubMed] [Google Scholar]
- 28. Marusyk R, Sergeant A. 1980. A simple method for dialysis of small-volume samples. Anal. Biochem. 105:403–404 [DOI] [PubMed] [Google Scholar]
- 29. Matsunaga J, et al. 2007. Response of Leptospira interrogans to physiologic osmolarity: relevance in signaling the environment-to-host transition. Infect. Immun. 75:2864–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 31. Murray GL, et al. 2009. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect. Immun. 77:810–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nascimento AL, et al. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Picardeau M. 2008. Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl. Environ. Microbiol. 74:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Picardeau M, Brenot A, Saint Girons I. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189–199 [DOI] [PubMed] [Google Scholar]
- 35. Picardeau M, et al. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607 doi:10.1371/journal.pone.0001607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ren SX, et al. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893 [DOI] [PubMed] [Google Scholar]
- 37. Saint Girons I, et al. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 182:5700–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schrogel O, Allmansberger R. 1997. Optimisation of the BgaB reporter system: determination of transcriptional regulation of stress responsive genes in Bacillus subtilis. FEMS Microbiol. Lett. 153:237–243 [DOI] [PubMed] [Google Scholar]
- 39. Tchamedeu Kameni AP, Couture-Tosi E, Saint Girons I, Picardeau M. 2002. Inactivation of the spirochete recA gene results in a mutant with low viability and irregular nucleoid morphology. J. Bacteriol. 184:452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wolfe AJ. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xue F, et al. 2010. Transcriptional responses of Leptospira interrogans to host innate immunity: significant changes in metabolism, oxygen tolerance, and outer membrane. PLoS Negl. Trop. Dis. 4:e857 doi:10.1371/journal.pntd.0000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xue T, You Y, Hong D, Sun H, Sun B. 2011. The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect. Immun. 79:2154–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zuerner RL. 2005. Laboratory maintenance of pathogenic Leptospira. Curr. Protoc. Microbiol. 12:12E.11 doi:10.1002/9780471729259.mc12e01s00 [DOI] [PubMed] [Google Scholar]
- 44. Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]