Abstract
The heterologous expression of a highly functional xylose isomerase pathway in Saccharomyces cerevisiae would have significant advantages for ethanol yield, since the pathway bypasses cofactor requirements found in the traditionally used oxidoreductase pathways. However, nearly all reported xylose isomerase-based pathways in S. cerevisiae suffer from poor ethanol productivity, low xylose consumption rates, and poor cell growth compared with an oxidoreductase pathway and, additionally, often require adaptive strain evolution. Here, we report on the directed evolution of the Piromyces sp. xylose isomerase (encoded by xylA) for use in yeast. After three rounds of mutagenesis and growth-based screening, we isolated a variant containing six mutations (E15D, E114G, E129D, T142S, A177T, and V433I) that exhibited a 77% increase in enzymatic activity. When expressed in a minimally engineered yeast host containing a gre3 knockout and tal1 and XKS1 overexpression, the strain expressing this mutant enzyme improved its aerobic growth rate by 61-fold and both ethanol production and xylose consumption rates by nearly 8-fold. Moreover, the mutant enzyme enabled ethanol production by these yeasts under oxygen-limited fermentation conditions, unlike the wild-type enzyme. Under microaerobic conditions, the ethanol production rates of the strain expressing the mutant xylose isomerase were considerably higher than previously reported values for yeast harboring a xylose isomerase pathway and were also comparable to those of the strains harboring an oxidoreductase pathway. Consequently, this study shows the potential to evolve a xylose isomerase pathway for more efficient xylose utilization.
INTRODUCTION
Efficient utilization of all available carbon in lignocellulosic biomass is one of the major challenges preventing economically viable biofuel production (1, 45). Organisms commonly used for biofuel production, such as the yeast Saccharomyces cerevisiae, are unable to natively utilize the pentose sugars that comprise a substantial portion of lignocellulosic biomass (29, 49). Among these pentose sugars, xylose is the most abundant in commonly studied biomass sources. Thus, the ability to improve xylose catabolism and conversion in a recombinant host, such as S. cerevisiae, would substantially improve the prospects for biofuel and biochemical production.
Recently, our group has improved xylose (d-xylose) catabolism through the introduction of heterologous molecular transporters, a key rate-determining step in xylose catabolism, especially at low concentrations (50). Once this xylose is transported into the cell, one of two main heterologous pathways can be utilized (49). The first pathway, with oxidoreductase-based chemistry, has been well established in yeast through the heterologous expression of (at a minimum) xylose reductase (XR) and xylitol dehydrogenase (XDH), leading to the ability of S. cerevisiae to utilize xylose (3, 11, 13, 14, 26, 33, 36). However, this pathway is inherently limited by a cofactor imbalance with the xylose reductase-utilizing NADPH and the xylitol dehydrogenase-utilizing NAD+, which leads to diversion of metabolic flux toward undesired products as a compensation reaction and decreases the ethanol yield (49). Recent work has focused on modifying the cofactor preferences of these enzymes to make them more compatible and to establish an oxidation-reduction cycle (37, 47). However, even with matching cofactor specificities, the oxidoreductase pathway requires cofactors that may limit overall pathway throughput. In all of these cases, the yield of ethanol from xylose still remains suboptimal compared with native xylose utilizers.
A second, alternative pathway for xylose catabolism mainly exists in bacteria and rarely in yeasts. This isomerase-based pathway has no cofactor requirements and thus could lead to higher theoretical yields (0.51 g ethanol/g xylose), since no by-product is necessarily produced to compensate for cofactor imbalance. In comparison, experimental ethanol yields using the oxidoreductase and xylose isomerase pathways under anaerobic conditions have been shown to be between 0.09 and 0.23 (6, 29) and near 0.43 g ethanol/g xylose, respectively (16). For this reason, there is considerable interest in improving a xylose isomerase-based pathway in S. cerevisiae with a particular focus on improving both the cell growth rate and the xylose consumption rate. Recent reports of successful expression of xylose isomerase genes from Piromyces sp. (19), Orpinomyces sp. (28), and Clostridium phytofermentans (8) in S. cerevisiae raise the prospect of efficient xylose fermentation. Furthermore, researchers have applied adaptive evolutionary engineering (21), optimized metabolic flux by introducing/overexpressing xylose transporter and/or overexpressing the downstream pathway (20, 27), and employed bioprospecting to identify other putative xylose isomerase enzymes (8, 34). In all of these cases, extensive downstream overexpression and/or evolutionary engineering is required to improve cell growth and xylose consumption. Even so, these levels are not yet comparable with those of strains expressing an oxidoreductase pathway (4, 16, 45).
Beyond the assembly of xylose catabolic pathways, xylose isomerase is an important enzyme for the food industry, especially in the production of high-fructose corn syrup. For these applications, xylose isomerase has been extensively studied (5) to improve the thermal stability (30, 42), pH optimum (23), and substrate preference (31). However, these studies were mainly focused on obtaining a xylose isomerase that (i) has an optimum temperature and a pH range (60 to 80°C and pH 7.0 to 9.0, respectively) (44) different from those of conventional ethanol fermentation, (ii) is expressed in Escherichia coli rather than S. cerevisiae (2), and (iii) is found to be unsuccessfully expressed (40) or to be inactive at mesophilic temperature (46) in S. cerevisiae, mainly due to protein misfolding (12). Moreover, later attempts to improve the xylose isomerases for ethanol fermentation, such as cold adaptation (25) and optimizing expression levels (35), were unsatisfactory in constructing a functional xylose catabolic pathway in yeast.
Here, we report the first directed-evolution study of a xylose isomerase gene (xylA from Piromyces sp.) for improved specific enzyme activity under the conditions tested, cell growth, xylose consumption rate, and ethanol production, in the yeast S. cerevisiae. Directed evolution is an efficient approach for tailoring proteins that require refined functions, such as higher stability, tolerance, substrate specificity, and product selectivity of the protein. The iterative application of this method allows proteins with significantly improved function to be easily obtained in a short time (7). To this end, we subjected the xylA gene to iterative rounds of random mutagenesis (aided by error-prone PCR), followed by selection for increased cell growth on xylose as a sole carbon source. After three rounds of mutagenesis and selection, we obtained an improved mutant of xylose isomerase that can offer a promising starting point for further strain engineering to improve xylose catabolism.
MATERIALS AND METHODS
Strains and culture conditions.
S. cerevisiae strain BY4741-S1 with gre3 deleted (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YHR104w::kanMX4 p415TEF-tal1) was used as a host strain in this study. The strain was obtained by transforming an S. cerevisiae BY4741 gre3 knockout strain (supplied by Zhihua Li, University of Texas at Austin) with tal1 from Saccharomyces stipitis cloned into a p415 vector under the control of the TEF promoter (32). Yeast strains were routinely propagated at 30°C in yeast synthetic complete (YSC) medium composed of 6.7 g/liter yeast nitrogen base; 20 g/liter glucose; and CSM-Leu-Trp-Ura (complete synthetic medium without Leu, Trp, or Ura), CSM-Leu-His-Ura, or CSM-His-Leu-Trp-Ura (MP Biomedicals, Solon, OH). Mutant selection, growth characterization, and ethanol fermentation were conducted in identical media, except that 20 g/liter or 40 g/liter xylose was added as a carbon source. E. coli strain DH10β was used for all cloning and plasmid propagation. DH10β was grown at 37°C in Luria-Bertani (LB) broth supplemented with 50 μg/ml ampicillin. All strains were cultivated with orbital shaking at 225 rpm. Yeast and bacterial strains were stored at −80°C in 15% glycerol.
Construction of a xylA mutant library.
A library of randomly mutated xylose isomerase sequences was generated by error-prone PCR. A yeast codon-optimized version of the xylose isomerase gene (xylA) from Piromyces sp. was synthesized by Blue Heron (Bothell, WA) and cloned into the Mumberg plasmid p416-GPD (32) to form p416-GPD-XI. Error-prone PCR of the xylose isomerase gene was conducted with the primer set S004 xylA-f (5′-CCGGGCTGCAGGAATTCATG-3′) and S004 xylA-r (5′-GCGTGACATAACTAATTACATGACTCGAGTTA-3′) using a GeneMorph II Random Mutagenesis Kit (Stratagene, La Jolla, CA). According to the directions, libraries using low (0 to 4.5 mutations/kb), medium (4.5 to 9 mutations/kb), and high (9 to 16 mutations/kb) mutagenesis rates were cloned to achieve a library size of 1 × 105. The randomly amplified xylose isomerase genes were digested with EcoRI and XhoI (New England BioLabs, Ipswich, MA) and ligated into the EcoRI and XhoI sites of p416-GPD. The mutant plasmid library was transformed into E. coli DH10β using standard electroporation protocols (38). The resulting E. coli library was harvested from the petri dishes, isolated using the Zippy Plasmid Miniprep Kit (Zymo Research, Irvine, CA), and retransformed into S. cerevisiae BY4741-S1. Yeast transformation was conducted using a Frozen EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA) according to the manufacturer's instructions to achieve a one to three times coverage of the original mutagenesis library. This process and procedure for mutagenesis was repeated for each of the three rounds of directed evolution. The yeast libraries were allowed to grow for 3 days on plates before being scraped off and harvested for selection. The cells were then pooled and subjected to growth-based enrichment.
Mutant selection.
To enrich fast-growing transformants, cells from the yeast library were cultured and serially transferred into 20 ml of fresh YSC medium with xylose as a sole carbon source in a loosely closed 50-ml Falcon tube. Serial transfer was repeated every 4 or 5 days using 10% inocula. After seven rounds of serial transfers, cells from the final culture were plated onto YSC medium with xylose, and the largest colonies were isolated. In total, 40 cells were selected from these plates for further characterization. The cell growth of isolated variants was compared in 5 ml of YSC medium with xylose in a 14-ml culture tube, and the fastest growing variants were initially selected. The vectors from these promising mutants were isolated, sequenced, and retransformed. A second growth rate measurement against a control confirmed that the growth rate increase was due to the mutant xylose isomerase and not background adaptation of the host strain. The mutant xylose isomerase conferring the highest growth rate on xylose was used as the template for the next round of mutagenesis and selection. In total, two additional rounds of iterative random mutagenesis and selection were conducted. The best-performing mutants were termed xylA*1 (1st-round mutnat), xylA*2 (2nd-round mutant), and xylA*3 (3rd-round mutant), respectively, and the retransformed strains expressing these mutant xylose isomerases were termed S1A1, S1A2, and S1A3, respectively. The number of serial transfers was reduced from seven to five in the second and third rounds of the selection process.
Site-directed mutagenesis of identified mutations.
Site-directed mutations were conducted to confirm the beneficial mutations in the xylose isomerase mutant. A series of six individual back mutations was made using the third-round mutant xylose isomerase (xylA*3) and the Quikchange II kit (Stratagene, La Jolla, CA) and transformed into S. cerevisiae to identify beneficial mutations on the basis of cell growth. Once identified, the beneficial mutations were introduced into wild-type xylose isomerase to confirm the improved cell growth phenotype on xylose.
In vitro xylose isomerase activity measurements.
Xylose isomerase activities from cell extracts were assayed by measuring the decrease of NADH in 1 ml of reaction mixture at 340 nm using a spectrophotometer (18). The reaction mixture contained 100 mM Tris-HCl buffer (pH 7.5), 0.15 mM NADH, 10 mM MgCl2, and 2 U sorbitol dehydrogenase (Roche, Mannheim, Germany). The cell extracts were prepared from yeast transformants cultivated until the early exponential growth phase in selective medium by using YPER Plus Dialyzable Yeast Protein Extraction Reagent (Thermo Scientific, Rockford, IL) The protein contents of the cell extracts were determined by a Bradford assay using a Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Enzyme assays were performed in biological triplicate, and 25 to 500 mM xylose was used to determine kinetic parameters.
Fermentation assays and growth analysis.
Ethanol fermentations were performed at high yeast optical density (OD) under oxygen-limited and microaerobic conditions. For oxygen-limited conditions, cells were grown at 30°C in 50 ml of YSC medium containing 40 g/liter xylose as a sole carbon source in a 125-ml flask. For microaerobic conditions, cells were grown at 30°C in 40 ml of YSC medium with 20 g/liter xylose in a sealed 50-ml Falcon tube. For both of these fermentations, a 1-day-old preculture (grown on glucose) was pelleted and resuspended in xylose medium and inoculated at an initial OD of 20. Aerobic growth rate analysis was performed at low yeast optical density while culturing cells at 30°C in 5 ml of YSC medium with 20 g/liter xylose in a 14-ml culture tube. Xylose concentrations were measured using a YSI 7100 Multiparameter Bioanalytical System (YSI Life Sciences, Yellow Springs, OH), and ethanol concentrations were measured using an Ethanol Assay, UV-Method Kit (R-Biopharm, Darmstadt, Germany). Cell density was measured spectrophotometrically at 600-nm absorbance, from which the cell dry weight was calculated for the ethanol production and xylose consumption rates. One OD at 600 nm (OD600) unit was considered to be 0.17 g cells/liter (14). Fermentation and growth assays were performed in biological triplicate.
Protein structure prediction for xylose isomerase.
The three-dimensional structure of the xylose isomerase protein was predicted by Swiss-Model (http://swissmodel.expasy.org), a Web-based server for automated comparative modeling of three-dimensional protein structures (41). The crystal structure of Thermotoga neapolitana xylose isomerase (Protein Data Bank [PDB] code 1A0E) (9) was used as a template. The active site of xylose isomerase was predicted using the computer program PHYRE (a protein fold recognition server; http://www.sbg.bio.ic.ac.uk/phyre/) (17). The predicted protein model and mutations were visualized using Pymol version 1.3.
RESULTS
Identification of a xylose isomerase mutant conferring increased cell growth on xylose.
In this study, we sought to utilize directed evolution to improve the poor growth and xylose consumption rates usually associated with xylose isomerase-based pathways in heterologous S. cerevisiae. First, the xylose isomerase gene (xylA) from Piromyces sp. was synthesized as a codon-optimized version for S. cerevisiae to maximize translational efficiency. This optimal version was then used as a template for random mutagenesis afforded by error-prone PCR using the GeneMorph II Random Mutagenesis kit. Specifically, a library size of 1 × 105 members (as measured by independent E. coli colonies posttransformation) was created using a range of mutation rates. This library was cloned into a yeast expression vector behind the strong GPD (glyceraldehyde-3-phosphate dehydrogenase) promoter for high-level expression.
To create a suitable host strain for selection and testing of the xylose isomerase library, the standard haploid yeast strain BY4741 was chosen as a background. Two additional genetic changes were selected for this host strain. First, a gre3 knockout strain of BY4741 was chosen, as Gre3p is an aldose reductase that has been previously shown to inhibit xylose isomerase by nonspecifically producing xylitol from xylose (43). Second, the overexpression of a heterologous transaldolase, encoded by tal1, from S. stipitis was chosen, since this protein has been previously reported to be beneficial for xylose utilization in S. cerevisiae (14). The overexpression of downstream pathway enzymes to ensure proper flux is advantageous for selection phenotypes (37). To achieve overexpression of tal1, the gene was cloned behind a TEF promoter in the p415 plasmid. The resulting strain containing both the gre3 knockout and the heterologous tal1 expression was named BY4741-S1. Finally, the native chromosomal copy of the S. cerevisiae xylulokinase gene (XKS1) was maintained as opposed to overexpressing a heterologous copy from S. stipitis. Previous work has shown that strong overexpression of heterologous xylulokinase can be toxic to cells, especially with nonoptimal xylose catabolic pathways; thus, we sought to initially avoid this improper balance of enzyme levels in this strain (15).
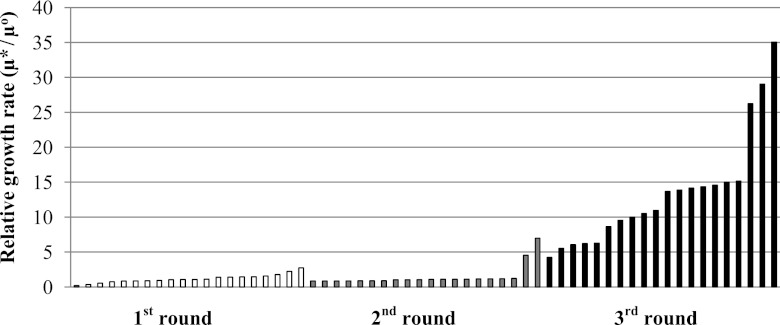
The mutant xylose isomerase library was transformed into S. cerevisiae BY4741-S1, and screening/selection were conducted on the basis of growth rate advantage. To this end, pools of cells were serially subcultured in xylose media for 5 to 7 serial transfers. Following this selection, many mutant strains were isolated and tested for growth (a sampling of 20 from each round is provided in Fig. 1). The mutant conferring the most improved growth phenotype was subjected to subsequent rounds of mutation and selection. This process was repeated for a total of three rounds of mutagenesis and selection. At each step, improved cell growth phenotypes of selected mutants were confirmed through retransformation of the xylose isomerase mutant gene into a fresh host strain to exclude any adaptive changes in the genomic DNA of the isolated strains. It should be noted that cell adaptation can provide an additional means of improving the xylose-utilizing cells created here. For example, the strain isolated after the third round of mutation showed almost double the growth rate of the retransformed strain. Thus, the ultimate production strain will utilize the synergy between pathway engineering and adaptive laboratory evolution.
Fig 1.
Comparison of the relative growth rates of isolated mutants (selected). The growth rates of strains isolated during the first (white bars), second (gray bars), and third (black bars) rounds of mutagenesis (μ*) were compared to those of the wild type (μ°). Twenty representative strains are shown out of the 40 tested for each round. The average growth rate of these isolated mutants increased progressively with the round of mutagenesis. The isolated strain with the highest growth rate in each round was selected.
The growth rates of the retransformed strains (S1A1, S1A2, and S1A3) expressing the best mutant xylose isomerase isolated from each round (xylA*1, xylA*2, and xylA*3 from the first, second, and third rounds, respectively) are shown in Table 1 compared to strains (S1A) harboring the wild-type xylose isomerase gene (xylA). Growth on xylose was not observed in a control strain (S1C) harboring a blank plasmid, as wild-type yeast cells lack a pathway to catabolize xylose. The growth rates on xylose progressively increased through the successive rounds of directed evolution (Table 1). S1A3 imparted a 9-fold increase in the growth rate compared to S1A.
Table 1.
Comparison of literature-based growth and fermentation rates with xylose as a sole carbon sourcea
| Strain | Strain description | Aerobic growth rate (h−1) | Xylose consumption rate (g · g−1 h−1) | Ethanol production rate (g · g−1 h−1) | Ethanol yield (g · g−1) | Reference |
|---|---|---|---|---|---|---|
| BY4741-S1A | Piromyces XI-expressing strain overexpressing tal1 and Δgre3 | 0.001 | 0.007 | 0.0032 | 0.46 | This study |
| BY4741-S1A1 | Piromyces XI mutant (xylA*1) strain overexpressing tal1 and Δgre3 | 0.001 | 0.0082 | 0.0041 | 0.50 | This study |
| BY4741-S1A2 | Piromyces XI mutant (xylA*2) strain overexpressing tal1 and Δgre3 | 0.003 | 0.0123 | 0.0060 | 0.49 | This study |
| BY4741-S1A3 | Piromyces XI mutant (xylA*3) strain overexpressing tal1 and Δgre3 | 0.009 | 0.0126 | 0.0056 | 0.45 | This study |
| BY4741-S2A3K | Piromyces XI mutant (xylA*3) strain overexpressing tal1, XKS1, and Δgre3 | 0.061 | 0.057 | 0.024 | 0.42 | This study |
| CEN.PK-RWB 202 | Piromyces XI-expressing strain | 0.005 | NAb | NA | NA | 19 |
| CEN.PK-RWB202-AFX | Evolved strain from CEN.PK-RWB 202 for 160 days in xylose medium | 0.18 | 0.340 | 0.140 | 0.42 | 22 |
| CEN.PK- RWB217 | Piromyces XI-expressing strain overexpressing XKS1, TKL1, RPE1, RKI1, and Δgre3 | 0.22 | 1.060 | 0.456 | 0.43 | 20 |
| CEN.PK- TMB3066 | Piromyces XI-expressing strain overexpressing XKS1, TKL1, RPE1, RKI1, HXT7, and Δgre3 | 0.02 | 0.005 | 0.002 | 0.43 | 16 |
| BWY10Xyl | Clostridium XI-expressing strain (evolved industrial strain in xylose medium) | 0.04 | 0.07 | 0.03 | 0.43 | 8 |
| CEN.PK2-1C-TMB 3424 | XR/XDH-expressing strain overexpressing XKS1, GAL2, TKL1, RPE1, RKI1, and Δgre3 | NA | 0.06 | 0.007 | 0.12 | 37 |
The fermentation performances of S. cerevisiae BY4741-S1A, -S1A1, -S1A2, -S1A3, and -S2A3 were compared with previously reported results. The strains developed here have 10- to 30-fold- and 3- to 10-fold-higher rates than previously reported engineered strains for xylose isomerase pathways and oxidoreductase pathways, respectively.
NA, not available.
Enzymatic assay of xylose isomerase mutants.
As demonstrated above, these mutant xylA genes were able to increase the growth rate of yeast cells on xylose; thus, we sought to understand the underlying mechanism of this improvement using in vitro kinetics assays. To do so, total protein was extracted from these strains, and enzyme assays were conducted using a spectrophotometry-based coupled-enzyme system (18). Similar to the growth rates, the enzyme activities of xylose isomerase mutants increased progressively with the rounds of mutagenesis and selection (Table 2). The xylA*3 gene had a 77% increase in Vmax compared to xylA (0.094 μmol min−1 mg protein−1 compared to 0.053 μmol min−1 mg protein−1 for xylA*3 and xylA, respectively). It should be noted that the Vmax value measured here for xylA is consistent with previously reported values (8). In contrast, the Km values did not show the expected trend of decrease, which refers to high substrate affinity leading to high enzyme activity, with increased cell growth (the Km for xylA*3 is significantly higher than the wild-type value). Nevertheless, these Km values (averaging around 100 mM) are quite high, highlighting the root cause of difficulties with the isomerase system. Therefore, the improved cell growth seems to be derived mainly from increased Vmax rather than decreased Km of the mutant xylose isomerase.
Table 2.
Enzyme kinetics and sequence analysis of xylA mutantsa
| Gene | Vmax (μmol mg protein−1 min−1) | Km (mM) | Mutations |
|---|---|---|---|
| xylA | 0.053 ± 0.007 | 86.97 ± 6.56 | |
| xylA*1 | 0.064 ± 0.001 | 89.98 ± 3.93 | E129D, V433I |
| xylA*2 | 0.083 ± 0.002 | 50.18 ± 7.08 | E129D, V433I, E15D, E114G |
| xylA*3 | 0.094 ± 0.006 | 168.39 ± 11.98 | E129D, V433I, E15D, E114G, T142S, A177T |
The wild-type (xylA) xylose isomerase gene and first-round (xylA*1), second-round (xylA*2), and third-round (xylA*3) mutants were expressed in S. cerevisiae, and in vitro xylose isomerase activity was measured and reported. Sequence analysis revealed the mutations identified in each round. The standard deviations of biological triplicates are indicated.
Identification of critical mutations in isolated xylose isomerase mutants.
In addition to enzyme analysis, the improved xylose isomerase genes were sequenced to identify the mutant residues responsible for improved activity. In each round of mutagenesis, two amino acid substitutions occurred, which resulted in six mutations in the xylA*3 third-round mutant: E15D, E114G, T142S, E129D, A177T, and V433I (Table 2).
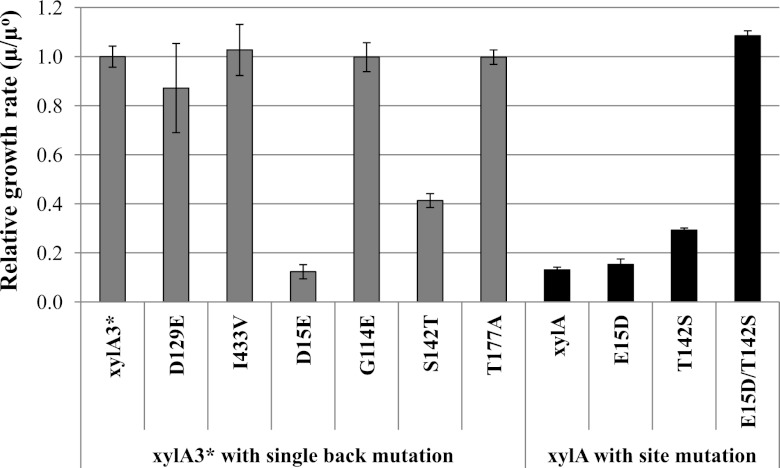
The cumulative amino acid substitutions obtained in this third-round mutant were next investigated to identify which were necessary and sufficient for the improved performance of xylA*3. To accomplish this, single back mutations of each amino acid substitution in xylA*3 were conducted to identify the essential nature of each of the six mutated sites. The strains expressing xylA*3 with single back mutations of amino acids at positions 15 and 142, which were obtained during the 2nd and 3rd rounds, respectively, showed 46% and 32% decreases in aerobic growth rates on xylose compared to the strain expressing xylA*3. This suggests that both E15D and T142S are necessary mutations that strongly contribute to the improved performance of mutant xylose isomerase (Fig. 2). The remaining four mutations appeared to be neutral mutations, as their back substitution did not have an impact on enzyme performance as measured by the resulting growth rate. The beneficial effects of the E15D and T142S mutations were confirmed by single and double point mutations using xylA as a template. This test was conducted to determine whether these mutations, either alone or in conjunction, were sufficient to obtain the same phenotype as the isolated mutant. As shown in Fig. 2, single substitution for either amino acid at position 15 or 142 was not enough to confer improved activity, but substitutions at both positions resulted in nearly the same performance as xylA3* in aerobic growth rates. As a result, the improved performance of xylA3* seems to be the result of synergistic mutations at positions 15 and 142.
Fig 2.
Identification of critical mutations by site-directed mutation. The aerobic cell growth rates (μ) of strains expressing the third-round mutant with a single back mutation and the wild type with a point (site) mutation were compared to that of the strain expressing the third-round mutant xylose isomerase (μ°). The back mutations at amino acid positions 15 and 142 resulted in 46% and 32% decreases in the growth rates. Combining these two positions improved the performance of wild-type xylose isomerase in aerobic growth on xylose. The error bars represent the standard deviations of biological triplicates.
Xylose isomerase mutants improve xylose consumption and ethanol production.
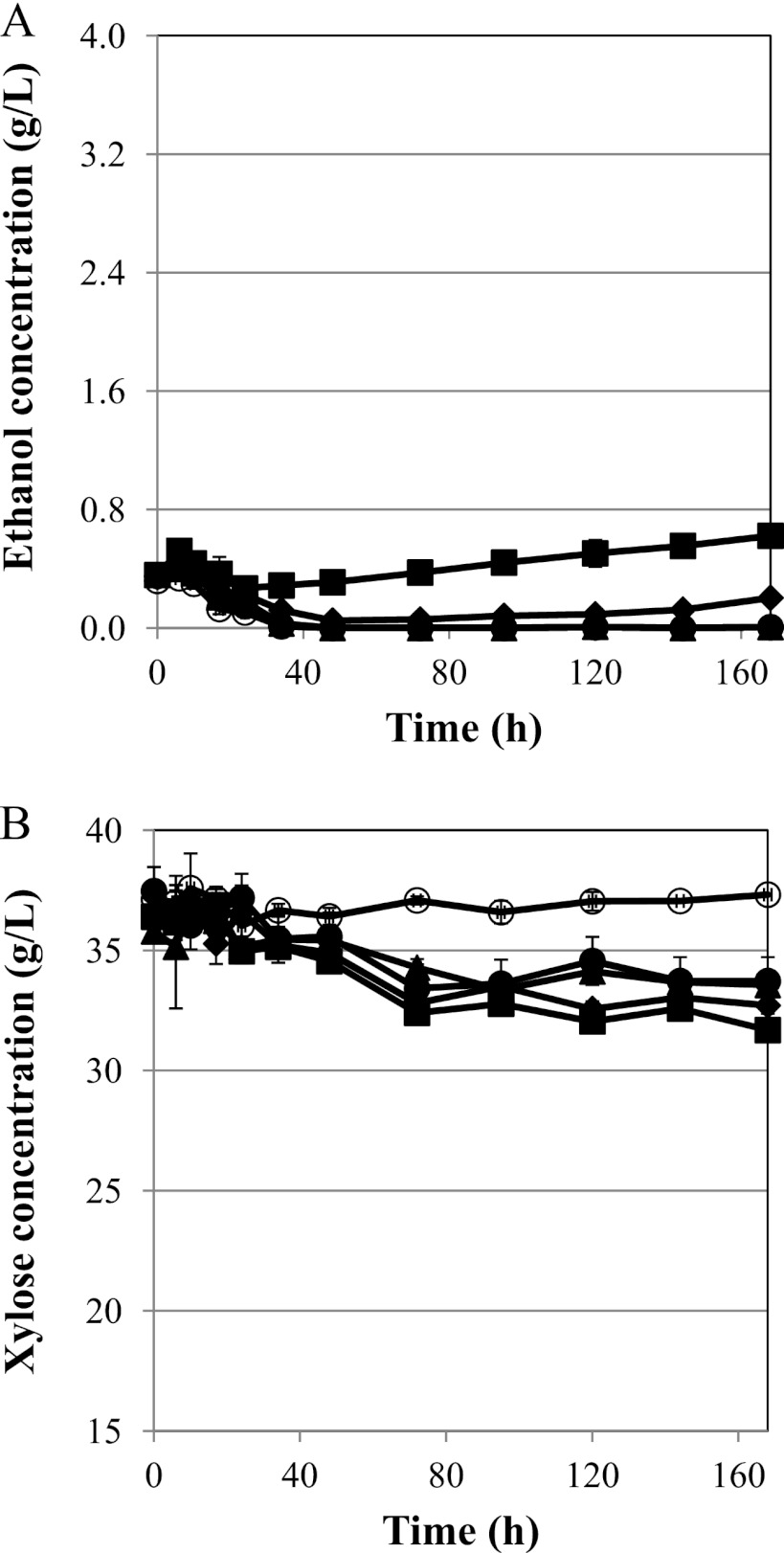
We next evaluated the xylose consumption and ethanol production rates enabled by these xylose isomerase mutants using a series of high-cell-density (OD = 20) batch fermentations under both oxygen-limited and microaerobic conditions. For the oxygen-limited conditions, 50 ml was cultured in a covered 125-ml flask. Under these conditions, xylose consumption rates increased progressively through the rounds of directed evolution, with S1A3 increasing xylose consumption rates by nearly 90% over S1A (Fig. 3B). As described above, the control strain (S1C) harboring a blank plasmid did not grow and thus consumed no xylose. Interestingly, only S1A2 and S1A3 produced measurable ethanol levels under these conditions (Fig. 3A). S1C, S1A, and S1A1 did not produce any ethanol (Fig. 3A). Furthermore, S1A3 had a relatively short lag time for ethanol production, whereas S1A2 started producing only after 95 h of fermentation (Fig. 3A). Thus, the identified mutant xylose isomerase gene (especially xylA*3) enables ethanol production capacity under these oxygen-limited conditions.
Fig 3.
Oxygen-limited fermentation tests with xylA mutants. Ethanol production (A) and xylose consumption (B) profiles were measured under oxygen-limited conditions for S. cerevisiae BY4741-S1C (○), S1A (●), S1A1 (▲), S1A2 (⧫), and S1A3 (■). Ethanol production was present only in the second- and third-round mutants. Xylose consumption was likewise increased in these strains. The error bars represent the standard deviations of biological triplicates.
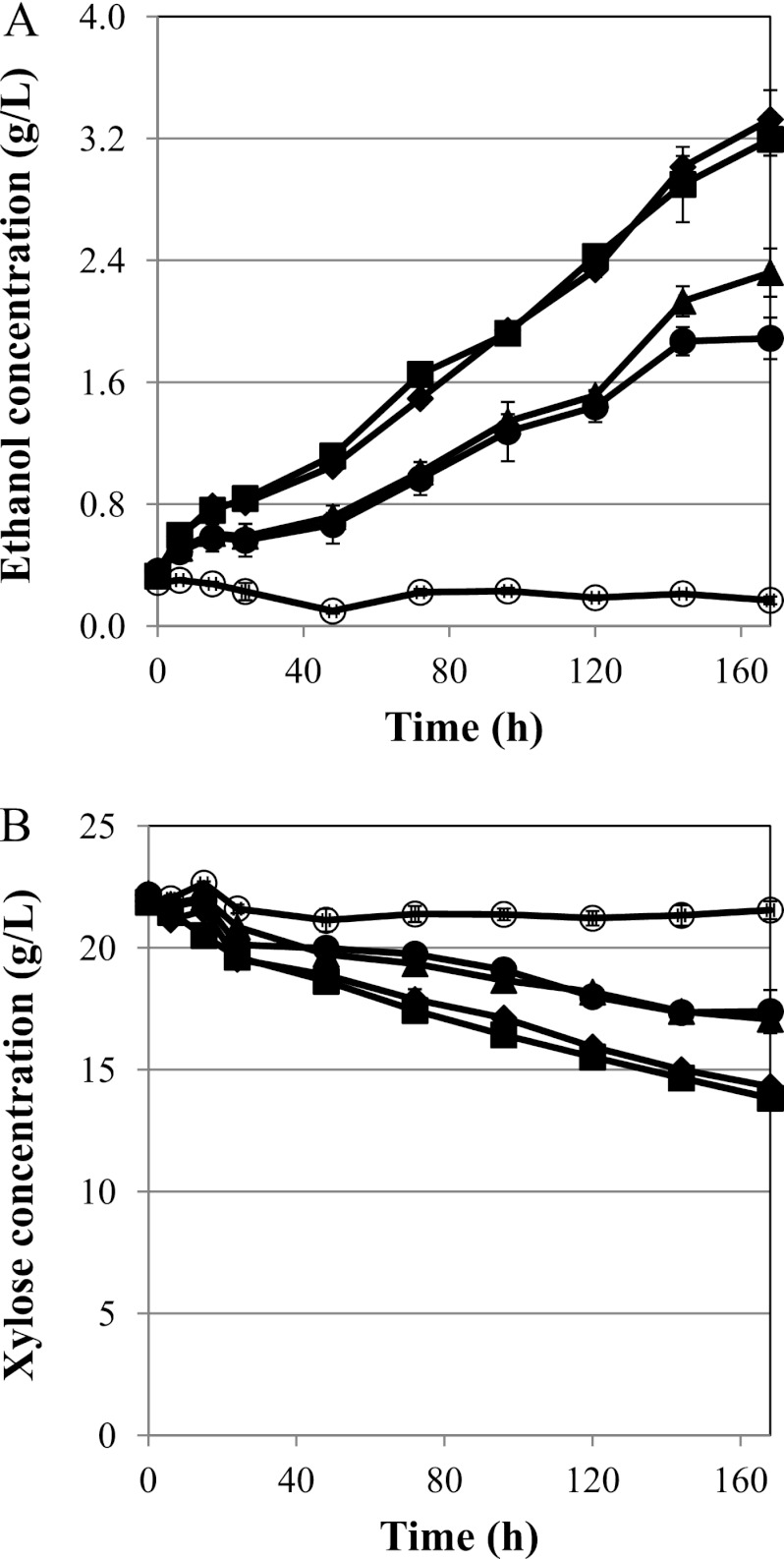
As a second test for ethanol production, we utilized less aerobic conditions (specifically, microaerobic conditions) afforded by culturing 40 ml in a 50-ml sealed vial. These conditions were expected to be more favorable for ethanol fermentation and resulted in all strains producing ethanol at significantly increased rates (Table 1). The ethanol production capacity was greatly increased in strains harboring these mutant versions of xylose isomerase (Fig. 4A), as were xylose utilization rates (Fig. 4B). S1A1 produced ethanol at the rate of 0.0041 g ethanol g cell−1 h−1 under these conditions, which represents an increase of 28% relative to that of S1A (0.0032 g ethanol g cell−1 h−1). In addition, the ethanol yield in S1A1 (0.50 g ethanol/g xylose) was close to the theoretical value of 0.51 g/g. Subsequent mutants of xylose isomerase resulted in higher rates of ethanol production with 88% and 75% increases over S1A for S1A2 and S1A3, respectively. It is interesting that under these more microaerobic conditions, the performances of S1A2 and S1A3 were quite similar. Although S1A3 produced ethanol at a slightly lower rate (0.0056 g ethanol g cell−1 h−1) than S1A2 (0.0060 g ethanol g cell−1 h−1), the xylose consumption rate was slightly higher in S1A3 (0.0126 versus 0.0123 g xylose g cell−1 h−1) (Fig. 4A and B). Despite the increased rate, ethanol yields in these strains were slightly lower than in S1A1 (0.49 and 0.45 g ethanol/g xylose for S1A2 and S1A3, respectively). Collectively, these results, along with the production profiles (Fig. 4), suggest that downstream metabolic flux is limited in the cells harboring these improved xylose isomerase genes under microaerobic conditions.
Fig 4.
Microaerobic fermentation tests with xylA mutants. Ethanol production (A) and xylose consumption (B) profiles were measured under microaerobic conditions for S. cerevisiae BY4741-S1C (○), -S1A (●), -S1A1 (▲), -S1A2 (⧫), and -S1A3 (■). Ethanol production was increased over oxygen-limited conditions, with both the second and third rounds having similar productivities. The total improvement in ethanol production was nearly 90% using these mutants. Xylose consumption was increased progressively with the round of directed evolution in these strains. The error bars represent the standard deviations of biological triplicates.
Improved performance of mutant xylose isomerase by additional engineering.
The expression of the improved xylose isomerase in the minimally engineered strain S. cerevisiae BY4741 still showed limited cell growth, xylose consumption, and ethanol production, presumably due to insufficient downstream metabolic flux. To further improve the xylose fermentation performance, the xylose isomerase enzymes (mutant and wild type) were cloned into a high-copy-number plasmid, p426-GPD (32), and cotransformed into S. cerevisiae BY4741-S1, along with an additional plasmid expressing the xylulokinase gene XKS1 driven by the TEF promoter (32). The resulting strains were named BY4741-S2AK, -S2A2K, and -S2A3K, expressing xylA, xylA*2, and xyl*3, respectively. It should be noted that although these strains have an expected higher downstream flux than S. cerevisiae BY4741-S1, they are still relatively minimally engineered compared with other strains commonly described in the literature that contain many pentose phosphate pathway enzyme overexpressions (Table 1). Therefore, this experiment was intended to demonstrate the potential of the improved xylose isomerase enzyme as a promising starting point for further strain engineering.
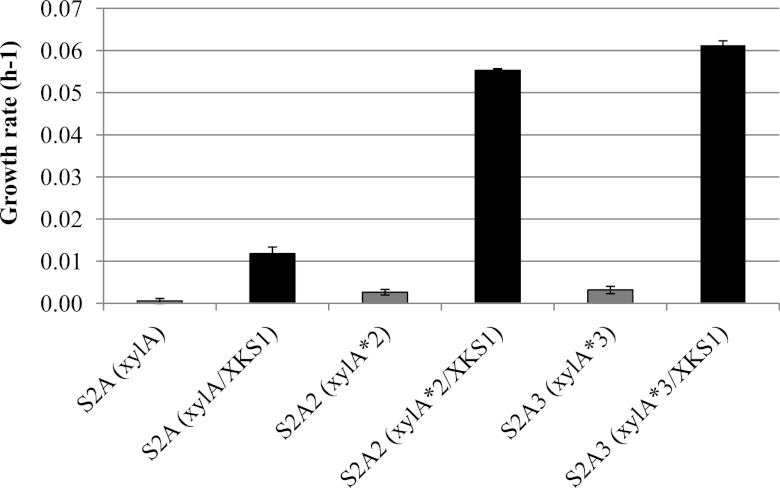
As shown in Fig. 5, the overexpression of xylulokinase significantly increased the aerobic growth rates of the strains expressing the various xylose isomerase enzymes (S2AK, S2A2K, and S2A3K expressing xylA, xylA*2, and xylA*3, respectively) compared to the strains without xylulokinase overexpression (S2A, S2A2, and S2A3 expressing xylA, xylA*2, and xylA*3, respectively). The improvement imparted by xylulokinase overexpression was most profound in strains harboring the mutant xylose isomerase variants. The aerobic growth rate of S2A3K (0.061 ± 0.00) was 20-fold higher than that of the strain without xylulokinase overexpression and nearly 61-fold higher than that of a strain simply overexpressing wild-type xylose isomerase.
Fig 5.
Aerobic growth rates of the strains expressing xylose isomerase with xylulokinase overexpression. Growth rates were measured for the strain expressing wild-type (xylA) and 2nd-round (xylA*2) and 3rd-round (xylA*3) mutant xylose isomerase with xylulokinase overexpression (black bars) and compared to those without xylulokinase overexpression (gray bars). The improved growth rate of the strain expressing xylA*3 (0.061 ± 0.001) was 5-fold higher than that of the strain expressing xylA (0.013 ± 0.001). The total improvement of the best strain compared with the strain simply expressing wild-type xylA was 61-fold. The error bars represent the standard deviations of biological triplicates.
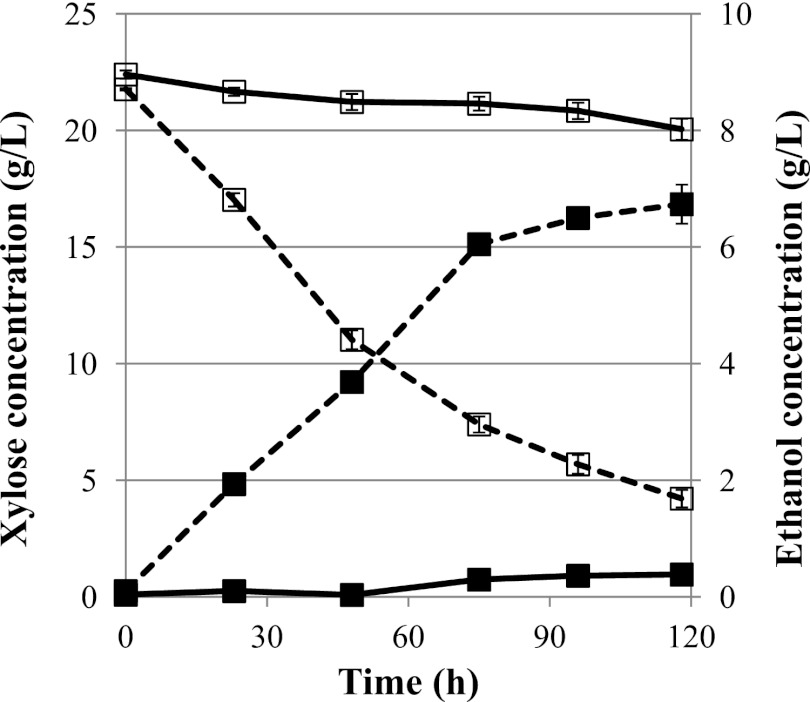
Finally, we tested the performance of the strain expressing the 3rd-round mutant xylose isomerase enzyme with xylulokinase overexpression (S2A3K) in a microaerobic xylose fermentation, as described above. In these high-cell-density batch fermentations, S2A3K produced ethanol at the rate of 0.024 g ethanol g cell−1 h−1 and consumed xylose at the rate of 0.057 g xylose g cell−1 h−1, which represents an increase of nearly 8-fold relative to those of S1A (0.0032 g ethanol g cell−1 h−1 and 0.007 g xylose g cell−1 h−1 S1A) (Fig. 6 and Table 1). These production rates are comparable to the rates obtained in strains harboring the traditional oxidoreductase pathway (Table 1). The ethanol yield in these fermentations was 0.42 g ethanol/g xylose, which is significantly higher than that of the oxidoreductase pathway.
Fig 6.
Microaerobic fermentation tests with xylulokinase overexpression. Ethanol production (solid squares) and xylose consumption (open squares) profiles were measured under microaerobic conditions for S. cerevisiae BY4741-S2A3 (solid lines) and S2A3K (dashed lines). Ethanol production was increased by xylulokinase overexpression. Total improvements in both ethanol production and xylose consumption rates were nearly 8-fold when the xylose isomerase mutant was coexpressed with heterologous xylulokinase. The error bars represent the standard deviations of biological triplicates.
DISCUSSION
This study demonstrates that a xylose isomerase pathway in S. cerevisiae can be improved by directed evolution. The minimally engineered strain expressing the improved xylose isomerase outperformed the strain expressing wild-type xylose isomerase in terms of ethanol production by nearly 90%, in terms of xylose consumption by 80%, and in terms of the aerobic growth rate by 9-fold. These improvements were achieved by a 77% increase in the in vitro catalytic activity of the enzyme. Combined with overexpression of one additional downstream enzyme (xylulokinase), the strain expressing the mutant xylose isomerase exhibited a 61-fold increase in aerobic growth and an 8-fold increase in both ethanol production and xylose consumption rates. These improvements clearly show the potential of the mutant as a promising starting point for further strain engineering for efficient xylose fermentation.
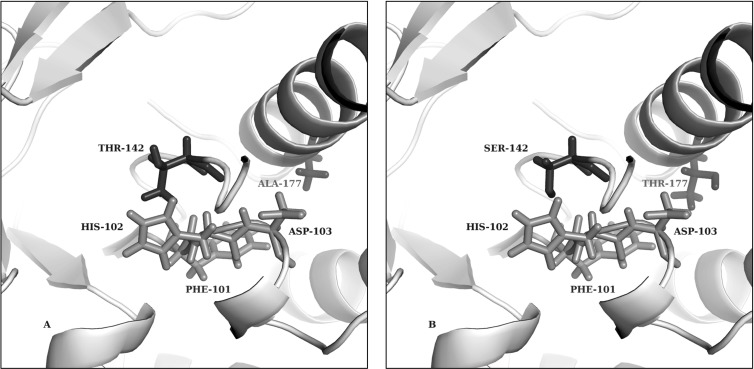
The increased enzyme activity seems to have resulted from a series of identified mutations in both the active-site and monomer-binding contacts. Figure 7 shows three-dimensional protein structure modeling predicted based on the T. neapolitana xylose isomerase (PDB code 1A0E [9]), which was available and shows high sequence similarity (65%) and identity (51%) to the xylA product, suggested that improved enzyme activity is likely due to two main factors: (i) increased substrate-enzyme interaction by mutations (Thr142 and Ala177) near the active site (His102) and (ii) increased enzyme stability caused by mutations near the monomer-binding contacts (Glu15 and Val433). This theory is supported by the fact that T142S and E15D were both necessary for the improved xylose isomerase performance of xylA*3 (Fig. 2). It is interesting that mutations near the monomer-binding contacts occurred in the early rounds of mutagenesis and selection, prior to active-site mutations. Previous reports suggest that xylose isomerase is active only when in the forms of a dimer and a tetramer (24). Based on the evolutionary trajectory identified in this work, it seems that increased stability of the xylose isomerase dimer/tetramer through mutations near the monomer-binding sites was a prerequisite for increased enzyme activity and active-site mutations. The instability of dimerized xylose isomerase is one of the possible explanations for the previous poor expression and activity of this enzyme in S. cerevisiae (44).
Fig 7.
Structure predictions for the xylose isomerase active site in the wild type and a mutant (xylA*3). The smaller size of SER-142 in the xylose isomerase mutant could loosen structural inhibition. In addition, THR-177 in the xylose isomerase mutant seems to open the active site, thereby allowing xylose to more easily interact with this binding pocket. Shown are the mutations near the active site of xylose isomerase in predicted three-dimensional structures of wild-type (A) and mutant (B) xylose isomerase. Shown are the mutation sites (SER-142 and THR-177) and their wild-type counterpart residues and the PHE-101, HIS-102, and ASP-103 active-site residues.
The performance of the identified xylose isomerase mutant was tested in two high-cell-density fermentation assays—oxygen-limited and microaerobic conditions. Under microaerobic conditions, the xylose consumption and ethanol production rates of S1A3 (0.0126 g xylose g cell−1 h−1 and 0.0056 g ethanol g cell−1 h−1, respectively) were considerably higher than previously reported values for yeast cells harboring a xylose isomerase pathway (Table 1). As an example, when wild-type Piromyces sp. xylA was expressed in an S. cerevisiae CEN.PK (TMB3066) background with XKS1, TAL1, TKL1, RPE1, RKI1, and HXT7 overexpression and gre3 deletion, the xylose consumption and ethanol production rates were reported as 0.005 g xylose g cell−1 h−1 and 0.002 g ethanol g cell−1 h−1, respectively (16). Thus, the results presented here—obtained strictly by overexpression of a mutant xylose isomerase, along with tal1 from S. stipitis and gre3 deletion—are over 2- to 3-fold higher in these metrics. Moreover, the ethanol production rate of S1A3 is also comparable to those of strains expressing an oxidoreductase pathway (Table 1). Most studies on ethanol fermentation with S. cerevisiae expressing oxidoreductase pathways have reported ethanol production rates in the range of 0.007 to 0.043 g ethanol g cell−1 h−1 under anaerobic conditions, which depend on the host strains, downstream gene overexpression, and reactor type (37, 45), though the highest reported ethanol production rate of 0.23 g ethanol g cell−1 h−1 was achieved by random mutagenesis of xylose reductase (37). Based on the efforts reported here, the ethanol production rates of a xylose isomerase-based pathway are now comparable to early reports on the oxidoreductase pathway.
The improvements in xylose consumption and ethanol production of strains expressing mutant xylose isomerase are substantial, especially since they were achieved without the need for an extensive pathway and metabolic engineering in the host strain. This implies that the xylose consumption rate and ethanol production rate are expected to be further increased by additional metabolic/evolutionary engineering. In previous research, as shown in Table 1, the xylose consumption and ethanol production rates of strains expressing wild-type xylose isomerase from Piromyces sp. were improved by overexpressing downstream genes (3-fold increase in the ethanol production rate) or evolutionary engineering (36-fold increase in the aerobic growth rate) in a bioreactor. By overexpressing one additional enzyme, xylulokinase, we successfully demonstrated that the additional metabolic engineering would significantly improve xylose fermentation performance (Table 1 and Fig. 5 and 6). S2A3K showed a higher aerobic growth rate than the evolved industrial strain expressing xylose isomerase from Clostridium sp. (7) and the strain expressing wild-type xylose isomerase from Piromyces sp. with extensive engineering of downstream genes and xylose transporter (15). Moreover, the ethanol production rate of S2A3K (0.024 g ethanol g cell−1 h−1) was comparable to those of strains with an oxidoreductase pathway, which are usually in the range of 0.007 to 0.043 g ethanol g cell−1 h−1 under anaerobic conditions (35, 44). Given that the better performance was obtained without extensive strain engineering, it is expected that rates and yields could be further improved by the overexpression of other enzymes involved in the pentose phosphate pathway or xylose transporters. Thus, the improved xylose isomerase reported here could serve as a critical starting point for further strain engineering to boost ethanol yields and productivity.
Under oxygen-limited conditions, the improvement of ethanol production is also important, as this mutant pathway enabled ethanol production. S. cerevisiae strains S1A2 and S1A3 produced ethanol, whereas S1A did not produce any ethanol. Moreover, xylose consumption rates under these conditions were increased by nearly 90% with S1A3. Ethanol fermentation is typically an anaerobic process that converts sugar to ethanol. Although S. cerevisiae can produce ethanol under aerobic conditions by respirofermentation, ethanol is usually not the by-product under aerobic conditions (48). As a result, efficient industrial ethanol fermentors are mainly operated under anaerobic conditions (39); however, intermittent aerations are required to maintain S. cerevisiae viability and ethanol productivity during long periods of fermentation. Under aerobic or oxygen-limited conditions created by aeration, ethanol productivity decreases (39). Hence, an ideal strain for ethanol fermentation will produce high concentrations of ethanol under anaerobic conditions and is less vulnerable to lower yields under aerobic or oxygen-limited conditions. The S. cerevisiae strain with an improved xylose isomerase pathway reported here exhibited both of these properties, with rapid initiation of ethanol production under oxygen-limited conditions and strong ethanol production under more anaerobic conditions. Thus, this pathway could form the foundation of a promising host for industrial ethanol fermentation.
This study produced a significantly improved xylose isomerase pathway in S. cerevisiae through the use of directed evolution without the need for extensive pathway engineering. The xylose isomerase-based pathway is an advantage for yeast fermentation, as it bypasses the cofactor imbalance issues that plague the oxidoreductase pathway. The third-round mutant created here obviated xylose isomerase as the rate-limiting step in this xylose catabolic pathway. Beyond this enzyme, it will be necessary to optimize the downstream genes involved in the xylose metabolic pathway (downstream approach) and to introduce an efficient xylose transporter (upstream approach) to further improve the xylose fermentation efficiency of engineered S. cerevisiae (29, 49). In further strain engineering, it will be important to balance the upstream and downstream enzymatic activities, since isomerization of xylose to xylulose is reversible and xylose formation is more favorable than xylulose formation at equilibrium by 80:20 (10). Regardless, the xylose isomerase pathway has long been considered an attractive alternative to the oxidoreductase pathway for expression in yeast. The results presented here demonstrate that this enzyme can be evolved for improved function in yeast, leading to more efficient cell growth, xylose uptake rates, and ethanol fermentation rates—three phenotypes that were limited with the wild-type enzyme.
ACKNOWLEDGMENTS
We acknowledge financial support for this project from the University of Texas at Austin Research Grant Program.
We thank Zhihua Li (Marcotte laboratory, University of Texas at Austin) for providing the gre3 host strain and Nathan Crook (Alper laboratory, University of Texas at Austin) for assistance with protein structure prediction.
Footnotes
Published ahead of print 8 June 2012
REFERENCES
- 1. Alper H, Stephanopoulos G. 2009. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential. Nat. Rev. Microbiol. 7:715–723 [DOI] [PubMed] [Google Scholar]
- 2. Amore R, Wilhelm M, Hollenberg C. 1989. The fermentation of xylose—an analysis of the expression of Bacillus and Actinoplanes xylose isomerase genes in yeast. Appl. Microbiol. Biotechnol. 30:351–357 [Google Scholar]
- 3. Bera A, Ho N, Khan A, Sedlak M. 2011. A genetic overhaul of Saccharomyces cerevisiae 424A(LNH-ST) to improve xylose fermentation. J. Ind. Microbiol. Biotechnol. 38:617–626 [DOI] [PubMed] [Google Scholar]
- 4. Bettiga M, Hahn-Hagerdal B, Gorwa-Grauslund M. 2008. Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnol. Biofuels 1:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhosale S, Rao M, Deshpande V. 1996. Molecular and industrial aspects of glucose isomerase. Microbiol. Rev. 60:280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bikard D, Julié-Galau S, Cambray G, Mazel D. 2010. The synthetic integron: an in vivo genetic shuffling device. Nucleic Acids Res. 38:e153 doi:10.1093/nar/gkq511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bloom JD, Arnold FH. 2009. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. U. S. A. 106:9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brat D, Boles E, Wiedemann B. 2009. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75:2304–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chayen NE, Conti E, Vielle C, Zeikus JG. 1997. Crystallization and initial X-ray analysis of xylose isomerase from Thermotoga neapolitana. Acta Crystallogr. D 53:229–230 [DOI] [PubMed] [Google Scholar]
- 10. Chiang L-C, Hsiao H-Y, Ueng PP, Tsao GT. 1981. Enzymatic and microbial preparation of D-xylulose from D-xylose. Appl. Environ. Microbiol. 42:66–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia Sanchez R, et al. 2010. Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol. Biofuels 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gardonyi M, Hahn-Hagerdal B. 2003. The Streptomyces rubiginosus xylose isomerase is misfolded when expressed in Saccharomyces cerevisiae. Enzyme Microbiol. Technol. 32:252–259 [Google Scholar]
- 13. Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund M. 2007. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 108:147–177 [DOI] [PubMed] [Google Scholar]
- 14. Jin Y-S, Alper H, Yang Y-T, Stephanopoulos G. 2005. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl. Environ. Microbiol. 71:8249–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin Y-S, Ni H, Laplaza JM, Jeffries TW. 2003. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl. Environ. Microbiol. 69:495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karhumaa K, Sanchez R, Hahn-Hagerdal B, Gorwa-Grauslund M-F. 2007. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 18. Kersters-Hilderson H, Callens M, Van Opstal O, Vangrysperre W, De Bruyne CK. 1987. Kinetic characterization of d-xylose isomerases by enzymatic assays using d-sorbitol dehydrogenase. Enzyme Microbiol. Technol. 9:145–148 [Google Scholar]
- 19. Kuyper M, et al. 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae. FEMS Yeast Res. 4:69–78 [DOI] [PubMed] [Google Scholar]
- 20. Kuyper M, et al. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5:399–409 [DOI] [PubMed] [Google Scholar]
- 21. Kuyper M, et al. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 5:925–934 [DOI] [PubMed] [Google Scholar]
- 22. Kuyper M, Winkler AA, van Dijken JP, Pronk JT. 2004. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res. 4:655–664 [DOI] [PubMed] [Google Scholar]
- 23. Lambeir AM, et al. 1992. Protein engineering of xylose (glucose) isomerase from Actinoplanes missouriensis. 2. Site-directed mutagenesis of the xylose binding site. Biochemistry 31:5459–5466 [DOI] [PubMed] [Google Scholar]
- 24. Lavie A, Allen KN, Petsko GA, Ringe D. 1994. X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis. Biochemistry 33:5469–5480 [DOI] [PubMed] [Google Scholar]
- 25. Lönn A, Gárdonyi M, van Zyl W, Hahn-Hägerdal B, Otero RC. 2002. Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Eur. J. Biochem. 269:157–163 [DOI] [PubMed] [Google Scholar]
- 26. Lu C, Jeffries T. 2007. Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 73:6072–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Madhavan A, et al. 2009. Alcoholic fermentation of xylose and mixed sugars using recombinant Saccharomyces cerevisiae engineered for xylose utilization. Appl. Microbiol. Biotechnol. 82:1037–1047 [DOI] [PubMed] [Google Scholar]
- 28. Madhavan A, et al. 2009. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol. Appl. Microbiol. Biotechnol. 82:1067–1078 [DOI] [PubMed] [Google Scholar]
- 29. Matsushika A, Inoue H, Kodaki T, Sawayama S. 2009. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl. Microbiol. Biotechnol. 84:37–53 [DOI] [PubMed] [Google Scholar]
- 30. Meng M, Bagdasarian M, Zeikus JG. 1993. Thermal stabilization of xylose isomerase from Thermoanaerobacterium thermosulfurigenes. Nat. Biotechnol. 11:1157–1161 [Google Scholar]
- 31. Meng M, Lee C, Bagdasarian M, Zeikus JG. 1991. Switching substrate preference of thermophilic xylose isomerase from D-xylose to D-glucose by redesigning the substrate binding pocket. Proc. Natl. Acad. Sci. U. S. A. 88:4015–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mumberg D, Müller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122 [DOI] [PubMed] [Google Scholar]
- 33. Nevoigt E. 2008. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 72:379–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parachin N, Gorwa-Grauslund M. 2011. Isolation of xylose isomerases by sequence- and function-based screening from a soil metagenomic library. Biotechnol. Biofuels 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parachin NS, Bergdahl B, van Niel EWJ, Gorwa-Grauslund MF. 2011. Kinetic modelling reveals current limitations in the production of ethanol from xylose by recombinant Saccharomyces cerevisiae. Metab. Eng. 13:508–517 [DOI] [PubMed] [Google Scholar]
- 36. Richard P, Toivari MH, Penttilä M. 2000. The role of xylulokinase in Saccharomyces cerevisiae xylulose catabolism. FEMS Microbiol. Lett. 190:39–43 [DOI] [PubMed] [Google Scholar]
- 37. Runquist D, Hahn-Hagerdal B, Bettiga M. 2010. Increased ethanol productivity in xylose-utilizing Saccharomyces cerevisiae via a randomly mutagenized xylose reductase. Appl. Environ. Microbiol. 76:7796–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sambrook J, Russell D. 2011. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Sánchez ÓJ, Cardona CA. 2008. Trends in biotechnological production of fuel ethanol from different feedstocks. Bioresour. Technol. 99:5270–5295 [DOI] [PubMed] [Google Scholar]
- 40. Sarthy A, et al. 1987. Expression of the Escherichia coli xylose isomerase gene in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 53:1996–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sriprapundh D, Vieille C, Zeikus JG. 2000. Molecular determinants of xylose isomerase thermal stability and activity: analysis of thermozymes by site-directed mutagenesis. Protein Eng. 13:259–265 [DOI] [PubMed] [Google Scholar]
- 43. Traff K, Otero Cordero R, van Zyl W, Hahn-Hagerdal B. 2001. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl. Environ. Microbiol. 67:5668–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Maris A, et al. 2007. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv. Biochem. Eng. Biotechnol. 108:179–204 [DOI] [PubMed] [Google Scholar]
- 45. Van Vleet JH, Jeffries TW. 2009. Yeast metabolic engineering for hemicellulosic ethanol production. Curr. Opin. Biotechnol. 20:300–306 [DOI] [PubMed] [Google Scholar]
- 46. Walfridsson M, Anderlund M, Bao X, Hahn-Hagerdal B. 1997. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl. Microbiol. Biotechnol. 48:218–224 [DOI] [PubMed] [Google Scholar]
- 47. Watanabe S, et al. 2007. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology 153:3044–3054 [DOI] [PubMed] [Google Scholar]
- 48. Wiebe MG, et al. 2008. Central carbon metabolism of Saccharomyces cerevisiae in anaerobic, oxygen-limited and fully aerobic steady-state conditions and following a shift to anaerobic conditions. FEMS Yeast Res. 8:140–154 [DOI] [PubMed] [Google Scholar]
- 49. Young E, Lee S-M, Alper H. 2010. Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol. Biofuels 3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Young E, Poucher A, Comer A, Bailey A, Alper H. 2011. Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Appl. Environ. Microbiol. 77:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]