Abstract
The two metabolically versatile actinobacteria Rhodococcus opacus PD630 and R. jostii RHA1 can efficiently convert diverse organic substrates into neutral lipids mainly consisting of triacylglycerol (TAG), the precursor of energy-rich hydrocarbon. Neither, however, is able to utilize xylose, the important component present in lignocellulosic biomass, as the carbon source for growth and lipid accumulation. In order to broaden their substrate utilization range, the metabolic pathway of d-xylose utilization was introduced into these two strains. This was accomplished by heterogenous expression of two well-selected genes, xylA, encoding xylose isomerase, and xylB, encoding xylulokinase from Streptomyces lividans TK23, under the control of the tac promoter with an Escherichia coli-Rhodococcus shuttle vector. The recombinant R. jostii RHA1 bearing xylA could grow on xylose as the sole carbon source, and additional expression of xylB further improved the biomass yield. The recombinant could consume both glucose and xylose in the sugar mixture, although xylose metabolism was still affected by the presence of glucose. The xylose metabolic pathway was also introduced into the high-lipid-producing strain R. opacus PD630 by expression of xylA and xylB. Under nitrogen-limited conditions, the fatty acid composition was determined, and lipid produced from xylose by recombinants of R. jostii RHA1 and R. opacus PD630 carrying xylA and xylB represented up to 52.5% and 68.3% of the cell dry weight (CDW), respectively. This work demonstrates that it is feasible to produce lipid from the sugars, including xylose, derived from renewable feedstock by genetic modification of rhodococcus strains.
INTRODUCTION
Lipid consisting of fatty acids can be transformed through hydrotreatment into a range of hydrocarbons, including green diesel, or by transesterification into biodiesel as a renewable fuel replacement (11, 33, 43). As an alternative feedstock for the hydrocarbon, lipid produced by microorganisms has attracted much attention (34). It was reported that a variety of eukaryotic microorganisms, such as microalgae, yeasts, and fungi, accumulate a high content of mainly lipids, including triacylglycerol (TAG), under imbalanced nutrition conditions (11, 46, 55). In contrast, most bacteria are able to accumulate lipophilic storage products, such as polyhydroxyalkanoic acid (PHA) or poly(3-hydroxybutyric acid) (PHB) (12, 42). However, a few bacteria, mainly bacteria belonging to the genera Acinetobacter, Mycobacterium, Streptomyces, Nocardia, and Rhodococcus, can accumulate TAG during cultivation under nitrogen-limited conditions (4, 54). Strains of the Gram-positive bacterium Rhodococcus are especially of biotechnological importance due to the broad catabolic diversity of their enzymatic capabilities (37, 51). It was reported that the metabolically versatile bacterium Rhodococcus jostii RHA1, which was isolated from lindane-contaminated soil and which was initially known for its outstanding ability to degrade polychlorinated biphenyls (PCBs) (37), could accumulate up to 48.4% and 56.9% of the lipid of the cell dry weight (CDW) when it was growing on glucose and gluconate under nitrogen-starving conditions, respectively (22). Another oleaginous rhodococcus strain, R. opacus PD630, a super single-cell oil (SCO) producer with a lipid content of 76% of CDW, has been thoroughly studied, including by lipid body characterization, lipid content determination, fatty acid composition analysis, high cell density culture, pilot-scale fermentation, and whole-genome sequencing (3, 24, 32, 53, 55). Clearly, strains R. opacus PD630 and R. jostii RHA1 have shown the ability to produce a high content of lipids, the main components of which are TAGs, from carbon sources, including glucose, gluconate, and sucrose (22, 32, 53).
The feedstocks of lignocellulosic biomass are the most abundant renewable resources in the world (48). Fuels derived from lignocellulosic biomass will improve energy security, decrease greenhouse gas emissions, and move our industrial society toward sustainability (13, 50). The availability of genetic tools and complete genome sequences offers the opportunity to investigate the metabolic traits of the two Rhodococcus strains by genetic perturbation and functional genomics study (2, 22, 24, 35). Furthermore, the metabolic engineering approach can be used to improve microorganisms for utilization of carbohydrates from biomass (57). As discussed above, strains R. opacus PD630 and R. jostii RHA1 can efficiently convert carbon sources such as glucose into lipids, but there has been no report on improvement of their capabilities for utilization of the diverse sugars, including xylose, present in lignocellulosic biomass. The purpose of this work is to fill this gap by expanding the substrate range of Rhodococcus through implementation of the d-xylose metabolic pathway. In this study, the two well-characterized genes xylA and xylB, encoding d-xylose isomerase and xylulokinase from Streptomyces lividans TK23, respectively, were expressed in the Rhodococcus strains to provide them with a xylose utilization capability (20). The Rhodococcus recombinants were characterized for their enzyme activities, growth performance, and sugar consumption. The role of the genes encoding endogenous xylulokinase in R. jostii RHA1 was also assessed. Finally, the lipid contents of recombinants of R. opacus PD630 and R. jostii RHA1 and the fatty acid profiles of the cells grown on xylose were studied.
MATERIALS AND METHODS
Strains, plasmids, and cultivation conditions.
The Rhodococcus strains R. opacus PD630 and R. jostii RHA1 were grown at 30°C in W minimal salt medium containing sugar as the carbon source (41). The composition of W medium (per liter) included 0.85 g KH2PO4, 4.90 g Na2HPO4 · 12H2O, 0.50 g (NH4)2SO4, 0.10 g MgSO4 · 7H2O, 9.50 mg, FeSO4 · 7H2O, 10.75 mg MgO, 2.00 mg CaCO3, 1.44 mg ZnSO4 · 7H2O, 1.12 mg MnSO4 · 4H2O, 0.25 mg CuSO4 · 5H2O, 0.28 mg CoSO4 · 7H2O, 0.06 mg H3BO4 and 5.13 × 10−2 ml concentrated HCl. The stock solutions of the sugars, including glucose and d-xylose, were sterilized with a 0.22-μm-pore-size filter and added to the autoclaved medium. The growth of the strains on other carbon sources, including 30 g/liter of d-arabinose, glycerol, glucose, galactose, lactose, l-arabinose, fructose, mannose, sucrose, and gluconate, was also tested. Rhodococcus cells grown in lysogeny broth (LB) medium were harvested for electroporation (47). Escherichia coli TOP10 grown at 37°C was used as the host strain for gene cloning. LB medium supplemented with 50 μg/ml kanamycin or 100 μg/ml ampicillin was used for culture of E. coli carrying plasmids. For the cultivation of Rhodococcus recombinants, 50 μg/ml of neomycin was used.
In order for the cells to accumulate lipid, the wild-type strains and the recombinants were cultured under nitrogen-limited conditions (22). The Rhodococcus cells were grown in LB medium and harvested by centrifugation at 8,000 × g and 4°C for 10 min. Cell pellets were washed twice with W medium without sugar and nitrogen. After washing, the cells were inoculated into 100 ml of W medium containing 20 g/liter of glucose or xylose and 0.5 g/liter (NH4)2SO4. The culture was incubated at 30°C and 200 rpm in a 500-ml flask. The cells, grown for 96 h, were harvested and washed prior to lipid analysis.
DNA techniques.
Genomic DNA of the bacterium was isolated by using an UltraClean microbial DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA) according to the manual provided by the manufacturer. Plasmid DNA was extracted from E. coli by using a QIAprep miniprep kit from Qiagen (Valencia, CA). Restriction endonucleases were purchased from NEB (MA). PCR was performed with Phusion high-fidelity DNA polymerase from NEB. For amplification of the genes xylA and xylB by PCR, GC buffer was added into the reaction mixture per the manufacturer's instruction. Oligonucleotide primers were synthesized by Invitrogen (Grand Island, NY). The sequence and generated restriction sites of the primers are listed in Table S1 in the supplemental material. After the agarose gel was run, the PCR products and plasmids digested by restriction endonucleases were recovered and purified with a QIAquick gel extraction kit (Qiagen). The competent cells of E. coli were prepared with CaCl2 and transformed by the heat shock procedure (44). Rhodococcus cells were washed with water, treated with 10% glycerol, and then electroporated with the plasmids as previously described (47, 59). After electroporation, the cells were incubated on LB agar plates containing 50 μg/ml of neomycin.
Construction of plasmids carrying genes for xylose utilization.
The E. coli-Rhodococcus shuttle vector pNV18 was used to develop the expression vector in this study (10). A DNA fragment containing a polyhistidine (His) tag was generated with the primers HisF and HisR by PCR using plasmid pNV18 as the template (see Table S1 in the supplemental material). The PCR product was digested with XhoI and EcoRI and inserted into these two restriction sites of plasmid pNV18 to obtain plasmid pNVHis18. The tac promoter (Ptac) containing a ribosome binding site (RBS) was amplified with the primers TacF and TacR by PCR using plasmid pTAC-MAT-Tag-1 (Sigma, St. Louis, MO) as the template. Then, plasmid pNVHis18 was treated with PstI and HindIII and ligated with the digested PCR product to produce the expression vector pTACHis18. The 1.2-kb PCR product of xylA was obtained using genomic DNA of S. lividans TK23 as the template. The PCR product was digested with the restriction endonucleases PstI and KpnI. The digestion product was inserted into plasmid pTACHis18 to generate the xylA expression vector pXYLA. Similarly, the recombinant expression vector bearing the 1.5-kb xylB fragment from S. lividans TK23 was constructed and designated pXYLB. The expression cassette of xylB, including the coding sequence of xylB and tac promoter, was amplified by PCR with primers xyl-tac1 and xyl-tac2 using plasmid pXYLB as the template. The PCR product was digested with HindIII and inserted into plasmid pXYLA. The resulting 7.6-kb plasmid, pXYLAB, contained both xylA and xylB, and their expression was driven by Ptac individually (see Fig. S1 in the supplemental material).
Disruption of native RHA1 xylB genes.
Analysis of the genome sequence of R. jostii RHA1 revealed that the deduced sequence of the amino acids of two putative open reading frames (ORFs), RHA1_ro02812 (xylB1) and RHA1_ro02901 (xylB2), exhibited significant homologies to the xylulokinase amino acid sequence (37). To study the roles of these two candidates in xylose catabolism in the wild-type strain, these two genes were disrupted, and the double knockout was also generated by homologous integration between the fragments of the chromosome and the disrupted ORFs in the introduced plasmids (Table 1). The suicide vector pK18mobsacB was used to develop plasmids for the deletion of the potential xylB genes from R. jostii RHA1 as described below (45, 52). Two DNA fragments containing two possible xylB genes of R. jostii RHA1, xylB1 and xylB2, were amplified by PCR. The PCR products were digested with EcoRI and HindIII and cloned into plasmid pK18mobsacB to form the plasmids pKXYLB1and pKXYLB2, respectively. To disrupt the coding region of xylB1, plasmid pKXYLB1 was treated with SalI and then self-ligated. The resulting plasmid was pKB1-SalI (Table 1). Plasmid pKB2-PstI, containing a PstI-digested DNA fragment, was constructed to disrupt xylB2. Strain R. jostii RHA1 was transformed with the derivative plasmids of pK18mobsacB, pKB1-SalI, and pKB2-PstI by electroporation. The integration of plasmids introduced into the chromosome in the first crossover was selected for on LB agar plates containing kanamycin. The antibiotic-resistant cells were picked up and cultured overnight in LB medium and spread on LB agar plates containing 10% (wt/vol) sucrose without antibiotics. The second crossover of chromosomal DNA led to kanamycin-sensitive cells, but some of these cells were the recovered wild-type strains. Mutants with the disrupted gene were detected by colony PCR using the same primers used for the amplification of the target genes. The single knockouts of xylB1 and xylB2 were designated R. jostii ΔxylB1 and R. jostii ΔxylB2, respectively. xylB2 of the strain with a disrupted xylB1 was further deleted to construct the double-knockout strain R. jostii ΔxylB1B2 (Table 1).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S. lividans TK23 | Wild type, source of xylA and xylB | 20 |
| R. jostii RHA1 | Wild type | 37 |
| R. opacus PD630 | Wild type | 24 |
| R. jostii XYLA | R. jostii RHA1 bearing plasmid pXYLA | This study |
| R. jostii XYLAB | R. jostii RHA1 bearing plasmid pXYLAB | This study |
| R. jostii pNV18 | R. jostii RHA1 bearing plasmid pNV18 | This study |
| R. opacus XYLA | R. opacus PD630 bearing plasmid pXYLA | This study |
| R. opacus XYLAB | R. opacus PD630 bearing plasmid pXYLAB | This study |
| R. opacus pNV18 | R. opacus PD630 bearing plasmid pNV18 | This study |
| R. jostii ΔxylB1 | R. jostii RHA1 xylB1 knockout | This study |
| R. jostii ΔxylB2 | R. jostii RHA1 xylB2 knockout | This study |
| R. jostii ΔxylB1B2 | R. jostii RHA1 double knockout of xylB1 and xylB2 | This study |
| R. jostii ΔxylB1_xylA | R. jostii RHA1 xylB1 knockout bearing plasmid pXYLA | This study |
| R. jostii ΔxylB2_xylA | R. jostii RHA1 xylB2 knockout bearing plasmid pXYLA | This study |
| R. jostii ΔxylB1B2_xylA | R. jostii RHA1 double knockout of xylB1 and xylB2 bearing plasmid pXYLA | This study |
| Plasmids | ||
| pMAT-TAG | Ampr; E. coli expression vector, source of tac promoter | Sigma |
| pNV18 | Kmr; E. coli-Rhodococcus shuttle vector | 10 |
| pTACHis18 | Kmr; expression vector containing the tac promoter, derived from pNV18 | This study |
| pXYLA | Kmr; pTACHis18 containing xylA from S. lividans TK23 | This study |
| pXYLB | Kmr; pTACHis18 containing xylB from S. lividans TK23 | This study |
| pXYLAB | Kmr; pTACHis18 containing xylA and xylB genes | This study |
| pK18mobsacB | Kmr; sacB; cloning vector for mobilization into bacteria | 45 |
| pKXYLB1 | Kmr; pK18mobsacB containing the xylB1 fragment | This study |
| pKXYLB2 | Kmr; pK18mobsacB containing the xylB2 fragment | This study |
| pKB1-SalI | Kmr; pKXYLB1 containing disrupted xylB1 | This study |
| pKB2-PstI | Kmr; pKXYLB2 containing disrupted xylB2 | This study |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; sacB, gene encoding levansucrase.
Enzyme assays.
To investigate the function of the potential enzymes in Rhodococcus strains for xylose utilization and verify the expression of xylA and xylB from S. lividans TK23 in the recombinants, the enzyme activities of xylose isomerase and xylulokinase were determined by using the cell extract. The chemicals used in the enzymatic activity determination were obtained from Sigma (St. Louis, MO). The cell extracts of Rhodococcus were prepared by using an ultrasonic homogenizer (sonicator 3000; Misonix, CT), and all of the operations were performed on ice (29). The assay for the activity of xylose isomerase of the whole-cell lysate was carried out in a 1-ml reaction volume containing 70 mM d-xylose, 20 mM MgCl2, 5 mM MnSO4, and 2 mM dithiothreitol in 100 mM Tris buffer (pH 7.5) (29). The reaction mixture was incubated for 30 min at 30°C, and then 0.5 M HClO4 was added to stop the reaction. The d-xylulose formed was quantified by using the cysteine-carbazole-sulfuric acid assay with measurement of the absorbance at 540 nm (A540) (17, 18). The enzyme activity of xylulose kinase was determined by the reduction of d-xylulose in the reaction mixture as described before (29). One unit of enzyme activity was defined as the amount of enzyme which generated 1 μmol of the product or consumed the substrate, d-xylulose, per minute.
Analytical procedure.
The samples were collected and centrifuged, and the supernatant was used for the determination of the content of the residual sugar in the medium. Glucose and d-xylose were analyzed using a Dionex ICS-3000 ion chromatography system equipped with a CarboPac TM PA 20 analytical column (4 by 50 mm), and CarboPac TM PA 20 guard column (3 by 30 mm) (Dionex Corporation, CA) (61, 64). Cell growth was quantified by measuring the A600 of the culture using a Shimadzu UV-visible spectrophotometer (UV-2550). A standard curve of the A540 versus the concentration of xylulose was also generated by using this spectrophotometer. For quantification of the lipid and fatty acid profiles of the strains, the cells were harvested and freeze-dried overnight. The methods used for fatty acid methyl ester preparation and gas chromatography (GC) analysis were the same as those described in our previous publications (56, 61, 64).
DNA sequencing and analysis.
All of clones were sequenced by GENEWIZ, Inc. (NJ). The sequence of the Streptomyces xylA and xylB genes was obtained from GenBank (accession no. AF184899) (39). To elucidate the pathway for d-xylose metabolism, the database Kyoto Encyclopedia of Genes and Genomes (KEGG) was used (39). The genes from R. jostii RHA1 involved in d-xylose metabolism were retrieved from KEGG or the Rhodococcus Genome Project (http://www.rhodococcus.ca/) (37). The genome sequence of R. opacus PD630 was available at the following website: http://www.broadinstitute.org (24). The sequence was analyzed by using the BLAST server of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), when needed (36).
RESULTS
Growth of Rhodococcus strains with various carbon sources.
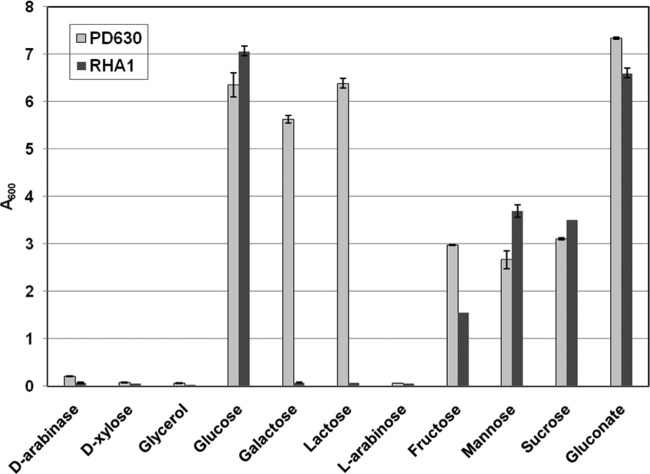
The growth performance of R. jostii RHA1 and R. opacus PD630 on diverse carbon sources in terms of the A600 of the culture at 120 h is shown in Fig. 1. The results showed that neither of the strains could utilize arabinose, xylose, or glycerol as the sole carbon source for growth. They also indicated that both strains could use glucose or gluconate as the sole carbon source for growth, and the A600s of R. jostii RHA1 and R. opacus PD630 reached 6.35 and 7.06, respectively, when 30 g/liter glucose was present in the medium. As shown in Fig. 1, the strain R. opacus PD630 could grow on galactose. However, R. jostii RHA1 could not utilize this sugar as the carbon source. The results showed that although strain R. jostii RHA1 could degrade a broad range of natural and xenobiotic compounds, including aromatic compounds, carboxylate compounds, and lignin, the capability of the strain for carbohydrate catabolism needs to be further improved (9, 37). Furthermore, our results also confirmed that there was a significant difference in catabolic phenotypes, including galactose utilization, in the closely related species R. jostii RHA1 and R. opacus PD630 (24).
Fig 1.
Growth of R. jostii RHA1 and R. opacus PD630 in W medium containing 30 g/liter of different sugars at 120 h. The cultures were first incubated in LB medium at 30°C and 200 rpm until late log phase. The cells were harvested by centrifugation, washed to remove the metabolites, and inoculated into 100 ml of W medium in a 500-ml flask. The initial A600 was adjusted to about 0.1. Incubation was carried out at 30°C and 200 rpm.
Construction and growth of xylA and xylB recombinants of R. jostii RHA1 on xylose.
The resulting vectors pXYLA and pXYLAB were introduced into R. jostii RHA1. The results of enzyme assay showed that d-xylose isomerase and xylulokinase were actively expressed in the recombinants R. jostii XYLA and R. jostii XYLAB (Table 2). Especially, the expression of these two enzymes by the recombinant was not repressed when glucose was present in the medium. As shown in Fig. 2A, the result showed that the recombinants R. jostii XYLA and R. jostii XYLAB could grow on 30 g/liter of xylose, whereas the control, wild-type strain R. jostii RHA1 bearing the empty vector pNV18 could not grow on this C5 sugar. Recombinant R. jostii XYLAB, expressing both xylA and xylB from S. lividans TK23, had a higher biomass yield than R. jostii XYLAB, carrying only xylA. However, according to the time course of growth, these two strains demonstrated similar growth rates on xylose. The specific growth rates (μ's) of R. jostii XYLA and R. jostii XYLAB on 30 g/liter of xylose were 0.052 h−1 and 0.054 h−1, respectively. Additionally, the biomass yield on different contents of d-xylose was assessed. As indicated in Fig. 2B, the optimal content of xylose for both strains was 40 g/liter, and the maximum A600 of the strain harboring the xlyA and xylB genes reached 5.5 at 120 h. The result showed that a higher content of sugars inhibited the growth of the strains. It also manifested that R. jostii XYLAB showed a higher biomass yield than R. jostii XYLA on d-xylose under the same culture conditions. To compare the growth of cells on different carbon sources, the A600 of the culture grown on d-xylose and glucose was determined. As indicated in Fig. 2C, although the final biomass yields of these two strains were similar, R. jostii XYLAB grew much slower on d-xylose than strain R. jostii/pNV18 grew on glucose. The specific growth rates of the strain grown on d-xylose and glucose were 0.07 h−1 and 0.19 h−1, respectively. Furthermore, the lag phase of the cell incubated on d-xylose was much longer. A similar phenomenon was also observed with other recombinants grown on xylose as the carbon source, especially when the culture seeds for the inoculum were cultured on glucose (29).
Table 2.
Specific activities of xylose isomerase and xylulokinase in R. jostii RHA1, R. opacus PD630, and their recombinantsa
| Strain (carbon source) | Sp act (U/mg protein) |
|
|---|---|---|
| Xylose isomerase | Xylulokinase | |
| R. jostii RHA1 (glucose) | NDb | 0.16 ± 0.02 |
| R. jostii XYLA (glucose) | 0.27 ± 0.01 | 0.17 ± 0.04 |
| R. jostii XYLA (xylose) | 0.39 ± 0.02 | 0.53 ± 0.10 |
| R. jostii XYLAB (xylose) | 0.40 ± 0.05 | 4.89 ± 0.61 |
| R. opacus PD630 (glucose) | ND | 0.21 ± 0.01 |
| R. opacus XYLA (glucose) | 0.20 ± 0.01 | 0.23 ± 0.02 |
| R. opacus XYLA (xylose) | 0.42 ± 0.05 | 0.46 ± 0.03 |
| R. opacus XYLAB (xylose) | 0.37 ± 0.02 | 2.96 ± 0.30 |
The data were calculated from triplicate measurements, and data are represented as the average ± standard deviation. Glucose or xylose at 20 g/liter was used as the carbon source in this study.
ND, not detected.
Fig 2.
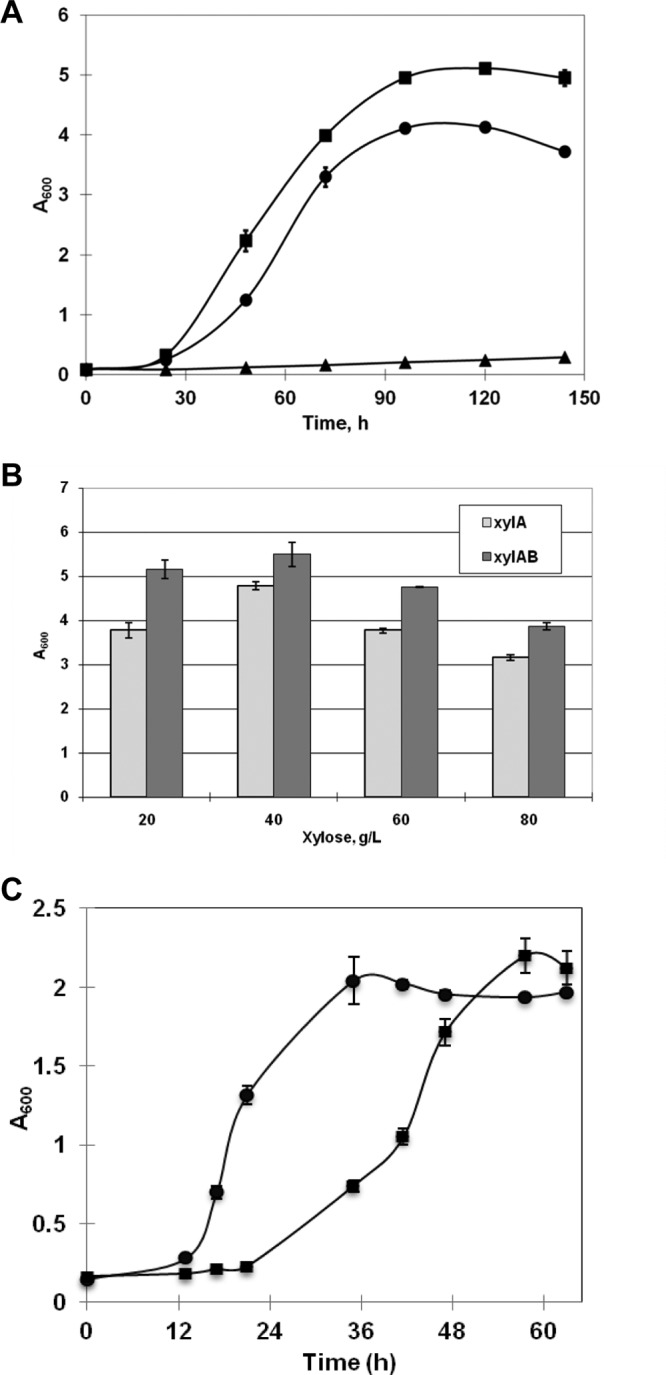
Growth of recombinant cells of R. jostii RHA1 in W medium at 30°C and 200 rpm. (A) The initial A600 of the culture was adjusted to about 0.1, and the cells were grown in medium containing 30 g/liter d-xylose. Samples were taken every 12 h, and the A600 was determined. ■, R. jostii XYLAB; •, R. jostii XYLA; ▲, R. jostii/pNV18. (B) Cells were grown in medium containing different contents from 20 g/liter to 80 g/liter of d-xylose. The cell biomass yield, indicated as the A600, was detected at 120 h. xylA, R. jostii XYLA; xylAB, R. jostii XYLAB. (C) Recombinant R. jostii XYLAB bearing xylA and xylB from S. lividans TK23 was incubated on 4 g/liter d-xylose (■), and R. jostii/pNV18 harboring the empty plasmid pNV18 was grown in W medium containing 4 g/liter glucose (•).
Endogenous xylB genes from R. jostii RHA1.
The growth profiles of R. jostii RHA1 recombinants suggested that some endogenous enzyme with the activity of xylulokinase was present in wild-type strain R. jostii RHA1. The enzyme assay of xylulokinase further confirmed this result (Table 2). Additionally, as shown in Table 2, the activity of the enzyme xylose isomerase for conversion of d-xylose into xylulose was not detectable in the wild-type strain. Two possible genes, RHA1_ro02812 (xylB1) and RHA1_ro02901 (xylB2), encoding xylulokinase were found in R. jostii RHA1 by homology analysis. As shown in Fig. 3, the xylB2-disrupted strain R. jostii ΔxylB1_xylA grew more efficiently than strain R. jostii ΔxylB2_xylA with the xylB1 deletion when xylA was coordinately expressed in these two strains. The strain with the double knockout of xylB1 and xylB2 carrying the expression vector pXYLA showed minimal growth in medium containing 10 g/liter of d-xylose (Fig. 3).
Fig 3.
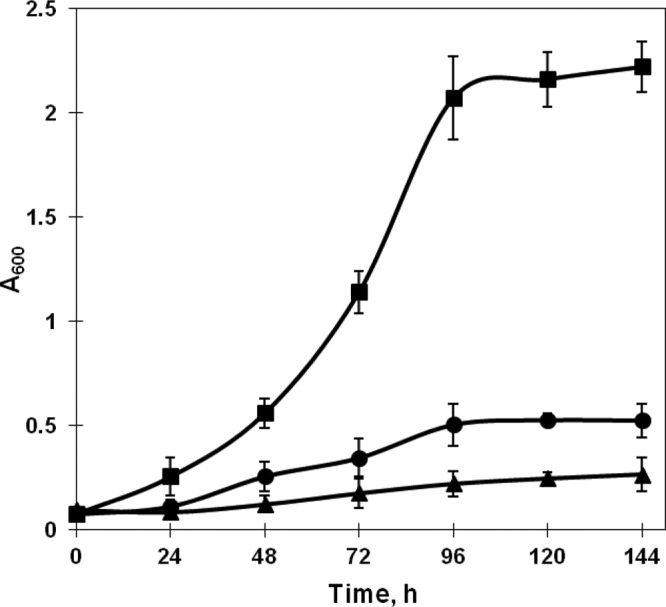
Growth of xylB-disrupted R. jostii RHA1 bearing xylA on 10 g/liter d-xylose at 200 rpm and 30°C. The samples were taken every 24 h, and the A600 was determined. ■, R. jostii ΔxylB2_xylA; •, R. jostii ΔxylB1_xylA; ▲, R. jostii ΔxylB1B2_xylA.
Consumption of a mixture of sugars consisting of glucose and d-xylose.
The results showed that both glucose and d-xylose could be consumed by the cells of R. jostii XYLAB bearing plasmid pXYLAB when the two sugars were present in the medium as a mixture at 4 g/liter each (Fig. 4), and there was no apparent diauxic shift during the cultivation. However, the results still showed that the consumption of d-xylose did not begin until the glucose was almost depleted, at about 36 h. The specific growth rate of strain R. jostii XYLAB was about 0.18 h−1, which was comparable to that of the wild type grown on 4 g/liter glucose, 0.19 h−1. However, the biomass yield was remarkably improved on the mixture containing both C5 and C6 sugars (Fig. 2 and Fig. 4).
Fig 4.
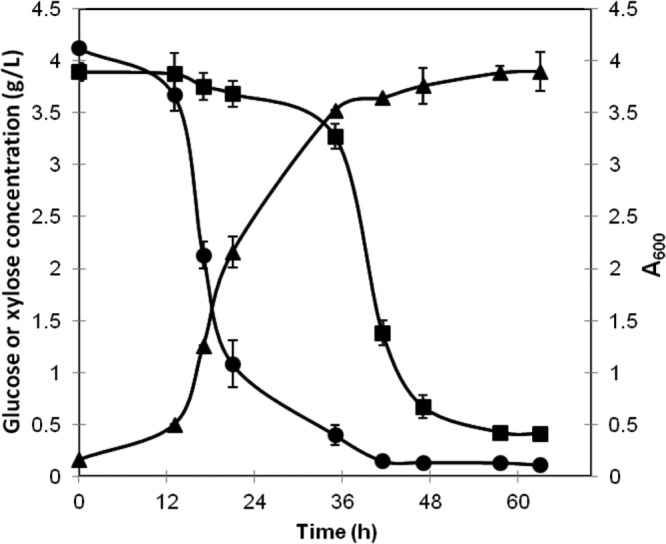
Growth of R. jostii XYLAB bearing plasmid pXYLAB and time function of sugar consumption in medium containing a sugar mixture consisting of about 4 g/liter of glucose and d-xylose. The incubation was carried out in a 250-ml flask at 200 rpm and 30°C with an initial A600 of 0.15. ▲, A600 of the culture; •, residue glucose content (g/liter); ■, residue xylose content (g/liter).
Construction and characterization of R. opacus PD630 recombinants.
Two recombinants, R. opacus XYLA and R. opacus XYLAB, were developed by electroporation of strain R. opacus PD630 with plasmids pXYLA and pXYLAB, respectively (Table 1). As shown in Table 2, the enzyme activity of xylulokinase could be detected in wild-type strain R. opacus PD630. It also shows that the specific activity of xylose isomerase was observed in the recombinant strains bearing xylA, whereas little activity was detected with wild-type strain R. opacus PD630 (Table 2). The result showed that both strains could utilize xylose as the sole carbon source with similar growth rates and biomass yields (Fig. 5). The specific growth rate of the strains cultivated on 30 g/liter xylose was about 0.05 h−1, which was similar to the growth rate of the recombinant of R. jostii RHA1 (Fig. 2 and Fig. 5). However, the final A600 of R. opacus XYLAB was approximately 4.3, which was lower than that of R. jostii XYLAB. It also showed that unlike the growth of the R. jostii RHA1 recombinants, expression of xylB from S. lividans TK23 could enhance neither the cell growth rate nor the yield of biomass on xylose.
Fig 5.
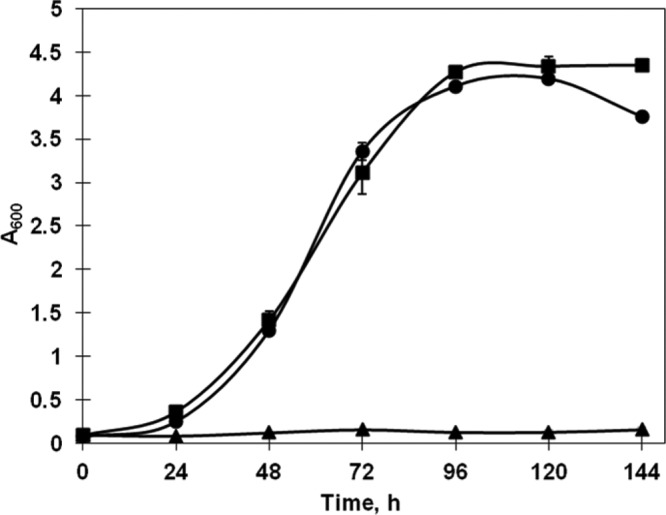
Growth of recombinant cells of R. opacus PD630 in W medium containing 30 g/liter xylose at 30°C and 200 rpm. The initial A600 of the culture was adjusted to about 0.1. After inoculation, samples were taken every 12 h and the A600 was determined. ■, R. opacus XYLAB; •, R. opacus XYLA; ▲, R. opacus/pNV18.
Lipid production by Rhodococcus recombinants from d-xylose.
In this study, the content of lipid, indicated as the proportion of total fatty acids, was detected. To allow cells to accumulate lipids, the cells were cultured on glucose or d-xylose under nitrogen-limited conditions (22). As shown in Table 3, the lipids present in the cells consisted of saturated and unsaturated fatty acids, and other than fatty acids with even numbers of carbons, there was a certain amount of fatty acids with odd numbers of carbons, which, peculiarly, existed in some bacteria (14). Like most of the organisms, fatty acids with chain lengths of more than 14 predominantly existed in the cells. Although all of the strains grown in the presence of different carbon sources contained the same prominent fatty acids, including pentadecenoic acid (C15:0), palmitic acid (C16:0), heptadecanoic acid (C17:0), heptadecenoic acid (C17:1n7), and oleic acid (C18:1n9), the relative proportions of the fatty acids were different. Furthermore, the lipid content of the cells grown in d-xylose was a little higher than the lipid content of cells grown in glucose. As also demonstrated by previous studies, the results showed that the lipid content of R. opacus PD630 was higher than that of R. jostii RHA1 (22, 24, 55). The recombinants R. opacus PD630 and R. opacus XYLAB grown in d-xylose could accumulate lipid to up to 68.3% and 52.5% of CDW, respectively, whereas the lipid contents of the natural strain cultivated in glucose were about 61.2% and 42.63% of CDW, respectively (Table 3). However, the recombinants R. jostii XYLAB and R. opacus XYLAB grown on W medium containing 5 g/liter of xylose and 1 g/liter of (NH4)2SO4 with a carbon-to-nitrogen ratio of 5:1 produced only 5.2% and 8.8% lipid, respectively. The results of this study verified that culture conditions consisting of a high carbon-to-nitrogen ratio like 40:1 could lead to accumulation of high contents of lipid in the recombinants R. jostii RHA1 and R. opacus PD630 (32).
Table 3.
Lipid content of R. jostii RHA1, R. opacus PD630, and their recombinants grown in W medium containing glucose or xylose under nitrogen-limited conditions
| Fatty acid or lipid | % lipid contenta |
|||
|---|---|---|---|---|
| PD630 (glucose) | PD630 XYLAB (xylose) | RHA1 (glucose) | RHA1 XYLAB (xylose) | |
| Fatty acid (total) | 61.78 ± 1.2 | 68.30 ± 2.9 | 42.63 ± 1.5 | 52.5 ± 3.0 |
| Myristic acid (C14:0) | 1.68 ± 0.2 | 0.92 ± 0.1 | 1.66 ± 0.1 | 1.38 ± 0 |
| Pentadecanoic acid (C15:0) | 6.63 ± 0.4 | 11.61 ± 1.2 | 6.89 ± 0.1 | 9.79 ± 0.2 |
| Palmitic acid (C16:0) | 25.92 ± 1.0 | 21.54 ± 1.7 | 25.33 ± 2.3 | 21.2 ± 1.7 |
| Palmitoleic acid (C16:1n7) | 5.50 ± 0.9 | 4.37 ± 1.3 | 4.91 ± 1.6 | 4.71 ± 1.1 |
| Heptadecanoic acid (C17:0) | 14.82 ± 2.3 | 19.62 ± 1.8 | 19.38 ± 0.7 | 22.39 ± 0.8 |
| Heptadecenoic acid (C17:1n7) | 14.32 ± 0.3 | 18.31 ± 0.6 | 14.37 ± 1.9 | 19.37 ± 0.9 |
| Stearic acid (C18:0) | 5.24 ± 0.1 | 3.38 ± 0.1 | 6.05 ± 0.8 | 3.33 ± 0.2 |
| Oleic acid (C18:1n9) | 17.85 ± 1.5 | 10.60 ± 0.9 | 13.8 ± 1.0 | 7.89 ± 0.7 |
| Linoleic acid (C18:2n6) | 1.78 ± 0.1 | 1.65 ± 0 | 1.47 ± 0.3 | 1.17 ± 0.1 |
Total fatty acids are expressed as a percentage of cell dry weight (CDW), and the fatty acid composition profile is shown as percent (wt/wt). The data were calculated from triplicate measurements, and data are represented as the average ± standard deviation. The carbon source is indicated in parentheses after the strain name.
DISCUSSION
Biomass, including crop residues, woody biomass, and organic waste, can serve as a renewable source for biofuel production (48). Therefore, to utilize these various feedstocks, including sugars derived from lignocellulosic biomass, a strain with the ability to grow on a broad range of substrates is desirable (1). In this study, the utilization of various carbon substrates by the two oleaginous bacteria R. jostii RHA1 and R. opacus PD630 for their growth was investigated. The further genome-scale survey of the genes for carbohydrate transportation and catabolism will provide the foundation for a better understanding of the metabolic pathways and regulatory networks of utilization of diverse substrates in these organisms (8, 24, 37). The results showed that these two strains could not initially utilize the pentose d-xylose. For utilization of d-xylose by bacteria, the enzymes d-xylose isomerase and xylulokinase are generally required to transform the transported intracellular d-xylose into d-xylulose 5-phosphate, which can subsequently be converted into d-ribulose-5-phosphate and further metabolized through the pentose phosphate pathway (PPP) (6, 20, 27). To develop the xylose-utilizing strains, most of the effort has been toward expression of the genes xylA and xylB encoding these two enzymes from E. coli in different hosts, including Corynebacterium glutamicum, Pseudomonas putida S12, Zymobacter palmae, and Zymomonas mobilis (29, 38, 60, 62). It was found that the first enzyme for xylose catabolism, xylose isomerase from Streptomyces strains, possessed high thermostability and a broad optimum pH (20). Furthermore, the genes from Streptomyces will be more readily expressed in rhodococci because both genera belong to the high-GC-content actinomyces and thus possess similar codon usage bias and regulation for gene expression (21). Therefore, in this study, the genes xylA and xylB, encoding enzymes for catabolism of d-xylose, were cloned from S. lividans TK23. After expression of xylA from S. lividans TK23 under the tac promoter in R. jostii RHA1, the recombinant could grow in medium containing d-xylose as the sole carbon source. The growth of recombinant R. jostii RHA1 on d-xylose was further improved by additional expression of xylB (Fig. 2A). The functional expression of xylA and xylB was also confirmed by the enzyme assay. In addition, it seems that there is an efficient xylose uptake system in the wild-type strain, as R. jostii RHA1 required the expression of only xylAB for xylose utilization (38). Several xylose transporters have been found in bacteria, including both the ABC-type transporter and xylose-proton symporter (58). However, only two sugar transporters, the major facilitator superfamily (MFS) glucose transporter GlcP and phosphotransferase system (PTS) PtsH, responsible for the uptake of fructose, were characterized in strain R. jostii RHA1 (5). In the genome of R. jostii RHA1, there was a putative protein, Ro05189, annotated as the possible xylose transporter (37), but its function needs to be further identified.
Assay of the enzyme xylulokinase of wild-type strains R. jostii RHA1 and R. opacus PD630 showed that the strains possessed an intrinsic enzyme(s) giving xylulokinase activity (Table 2). To study its function, the two candidates of the native xylB in R. jostii RHA1, RHA1_ro02812 (xylB1) and RHA1_ro02901 (xylB2), were cloned and disrupted. After transformation of the expression vector pXYLA, only one of them, the xylB2-disrupted strain R. jostii ΔxylB2_xylA, showed adequate growth on d-xylose (Fig. 3). It revealed that the first one, xylB1, played the major role in d-xylose metabolism, according to the growth performance on d-xylose. Furthermore, in wild-type strain R. jostii RHA1, the enzyme activity of xylulokinase was induced by d-xylose and repressed by glucose (Table 2). This phenomenon was observed in many bacteria, including E. coli, Streptomyces sp. strain TK24, and Bacillus subtilis (31). There was a regulatory protein, XylR, present in some microorganisms, including natural d-xylose-utilizing and non-d-xylose-utilizing strains (20, 31, 49). Generally, expression of the related genes was induced by d-xylose but strictly repressed by glucose due to the control of this regulator (20, 49). However, the existence and function of the potential protein used for regulation of xylose metabolism in Rhodococcus strains are unknown. The gene xylA was also expressed alone and coexpressed with xylB in the high-lipid-producing strain R. opacus PD630. The recombinants were able to utilize d-xylose as the carbon source for their growth (Fig. 5). However, unlike the recombinants of R. jostii RHA1, the two recombinants of R. opacus PD630 showed similar growth characteristics. This implied that the rate-limiting step for xylose utilization in this strain was not controlled by the xylulokinase encoded by xylB after expression of the xylA gene in strain R. opacus PD630.
In this study, the recombinants of R. jostii RHA1 demonstrated sequential consumption of glucose and xylose, and even the genes xylA and xylB were expressed constitutively by Ptac (15). It was also found that many other wild-type and engineered bacteria, such as E. coli, Pseudomonas spp., and Zymomonas spp., could not coutilize d-xylose and glucose either (28, 38). This drawback prevents the strains from efficient utilization of complex sugar mixtures while extending the cultivation time in the fermentor (30). Therefore, the development of strains with the capability of simultaneous utilization of both C5 and C6 sugars is critical for biofuel production from lignocellulosic biomass. Previous studies suggested that this sequential carbohydrate utilization in microorganisms resulted from carbon catabolite repression or allosteric competition of the sugars during sugar transport (16, 29). In a recombinant C. glutamicum strain, the results showed that additional expression of the l-arabinose transporter gene araE enhanced its ability to utilize pentose, and simultaneous consumption of pentose and glucose was mainly observed under oxygen-deprived conditions (28). E. coli mutants devoid of the phosphotransferase system (PTS) were developed to avoid this regulatory system, but problems still remain to be resolved (23), although it was recently reported that overexpression of the xylose transporter xylT and other catabolism genes in the glucose PTS-deficient organism Clostridium acetobutylicum allowed the strain to utilize glucose and xylose simultaneously (58). Our previous studies showed that oleaginous yeast and fungi were capable of using all forms of sugars and even other degradation products of lignocellulosic biomass from pretreatment for cell biomass and lipid production (61). However, most of them also preferred the glucose in the sugar mixtures as their carbon source (25). The ethanol-producing yeast Saccharomyces cerevisiae was intensively engineered to use d-xylose, and most of the resulting strains could not efficiently use the C5 sugar in the sugar mixture either (27). This bottleneck was overcome by expression of an intracellular β-glucosidase and cellodextrin transporter to bypass glucose repression for d-xylose fermentation, and the recombinant strain coutilized the transported cellobiose and d-xylose (19). Although the metabolic pathways of d-xylose utilization among bacteria, fungi, and bacteria are diverse (26), the results are instructive for the further genetic engineering of oleaginous strains for pentose utilization.
Some SCO-producing microorganisms are capable of accumulating substantial amounts of oil, sometimes up to 70% of their dry weight biomass (34). The lipid accumulation in such microorganisms was triggered by imbalanced culture conditions, such as starvation for nitrogen or other limiting nutrients. The profiles of fatty acids occurring in the lipids of the strains were dependent on cultivation factors such as temperature and carbon source (34). In this study, the recombinants of R. opacus PD630 and R. jostii RHA1 grown in d-xylose could accumulate up to 68.3% and 52.5% lipid (CDW) under nitrogen-limited conditions. Actually, the highest contents of the lipid for these two strains, 76% and 56.9%, respectively, were achieved by using the same carbon source, gluconic acid (3, 22). By combination of the results with those of this study, it is suggested that carbon sources such as d-xylose and gluconic acid metabolized by the pentose phosphate pathway are desirable for lipid accumulation. This is also supported by the calculation of the maximum theoretical yield of SCO produced per xylose molecule consumed, which is about 0.34 g/g (40). The culture conditions for biomass yield and lipid accumulation need to be optimized to achieve higher yields, and genetic modification of the strains can further improve productivity (7, 32, 63). The profiles of the fatty acid of the lipid were studied, and the straight-chain odd-carbon-number fatty acids were detected in the strains due to both type 1a fatty acid synthetase (FAS) and FAS II, responsible for fatty acid synthesis (24). The results confirmed that the d-xylose could serve as one of the desirable carbon sources for lipid production by the recombinants developed in this study.
Rhodococcus has been serving as an important platform organism for biodegradation, biotransformation, and biocatalysis, such as xenobiotic degradation, and for biosynthesis of fuels, pharmaceuticals, and valuable chemicals (51). The results in this study provide insight into a means of improvement of the performance of Rhodococcus strains by enabling them to utilize sugars, including xylose, from renewable lignocellulosic biomass. However, our research presented in this study is still preliminary. Future work will include further improvement of the stains for substrate utilization, optimization of the cultivation conditions for biofuel production, and evaluation and enhancement of the tolerance of the strains to inhibitors derived from the lignocellulosic biomass pretreatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lindsay D. Eltis (Department of Microbiology & Immunology, University of British Columbia) for the gift of R. jostii RHA1, Anthony Sinskey (Department of Biology, Massachusetts Institute of Technology) for the gift of R. opacus PD630, and Jun Ishikawa (Department of Bioactive Molecules, National Institute of Infectious Diseases, Japan) for the gift of plasmid pNV18. We also thank Jim O'Fallon for his assistance with the lipid analysis.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alper H, Stephanopoulos G. 2009. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7:715–723 [DOI] [PubMed] [Google Scholar]
- 2. Alvarez AF, Alvarez HM, Kalscheuer R, Wältermann M, Steinbüchel A. 2008. Cloning and characterization of a gene involved in triacylglycerol biosynthesis and identification of additional homologous genes in the oleaginous bacterium Rhodococcus opacus PD630. Microbiology 154:2327. [DOI] [PubMed] [Google Scholar]
- 3. Alvarez HM, Mayer F, Fabritius D, Steinbüchel A. 1996. Formation of intracytoplasmic lipid inclusions by Rhodococcus opacus strain PD630. Arch. Microbiol. 165:377–386 [DOI] [PubMed] [Google Scholar]
- 4. Arabolaza A, Rodriguez E, Altabe S, Alvarez H, Gramajo H. 2008. Multiple pathways for triacylglycerol biosynthesis in Streptomyces coelicolor. Appl. Environ. Microbiol. 74:2573–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Araki N, et al. 2011. Identification and characterization of uptake systems for glucose and fructose in Rhodococcus jostii RHA1. J. Mol. Microbiol. Biotechnol. 20:125–136 [DOI] [PubMed] [Google Scholar]
- 6. Aristidou A, Penttila M. 2000. Metabolic engineering applications to renewable resource utilization. Curr. Opin. Biotechnol. 11:187–198 [DOI] [PubMed] [Google Scholar]
- 7. Beopoulos A, et al. 2008. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 74:7779–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertram R, et al. 2004. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol. 186:1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen HP, et al. 2012. Vanillin catabolism in Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 78:586–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiba K, et al. 2007. Construction of a pair of practical Nocardia-Escherichia coli shuttle vectors. Jpn. J. Infect. Dis. 60:45–47 [PubMed] [Google Scholar]
- 11. Chisti Y. 2007. Biodiesel from microalgae. Biotechnol. Adv. 25:294–306 [DOI] [PubMed] [Google Scholar]
- 12. Choi MH, Yoon SC, Lenz RW. 1999. Production of poly (3-hydroxybutyric acid-co-4-hydroxybutyric acid) and poly (4-hydroxybutyric acid) without subsequent degradation by Hydrogenophaga pseudoflava. Appl. Environ. Microbiol. 65:1570–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chundawat SPS, Beckham GT, Himmel ME, Dale BE. 2012. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. 2:121–145 [DOI] [PubMed] [Google Scholar]
- 14. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 15. de Boer HA, Comstock LJ, Vasser M. 1983. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. U. S. A. 80:21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desai TA, Rao CV. 2010. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Microbiol. 76:1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dische Z, Borenfreund E. 1951. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J. Biol. Chem. 192:583–587 [PubMed] [Google Scholar]
- 18. Freer SN, Skory CD, Bothast RJ. 1997. d-Xylose metabolism in Rhodosporidium toruloides. Biotechnol. Lett. 19:1119–1122 [Google Scholar]
- 19. Ha SJ, et al. 2011. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl. Acad. Sci. U. S. A. 108:504–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heo GY, et al. 2008. Deletion of xylR gene enhances expression of xylose isomerase in Streptomyces lividans TK24. J. Microbiol. Biotechnol. 18:837–844 [PubMed] [Google Scholar]
- 21. Herai S, et al. 2004. Hyper-inducible expression system for streptomycetes. Proc. Natl. Acad. Sci. U. S. A. 101:14031–14035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hernández M, et al. 2008. Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics 9:600–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hernandez-Montalvo V, Valle F, Bolivar F, Gosset G. 2001. Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl. Microbiol. Biotechnol. 57:186–191 [DOI] [PubMed] [Google Scholar]
- 24. Holder JW, et al. 2011. Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development. PLoS Genet. 7:e1002219 doi:10.1371/journal.pgen.1002219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsiao HY, Chiang LC, Ueng PP, Tsao GT. 1982. Sequential utilization of mixed monosaccharides by yeasts. Appl. Environ. Microbiol. 43:840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeffries T. 1983. Utilization of xylose by bacteria, yeasts, and fungi. Adv. Biochem. Eng. Biotechnol. 27:1–32 [DOI] [PubMed] [Google Scholar]
- 27. Jeffries TW. 2006. Engineering yeasts for xylose metabolism. Curr. Opin. Biotechnol. 17:320–326 [DOI] [PubMed] [Google Scholar]
- 28. Kawaguchi H, Sasaki M, Vertès AA, Inui M, Yukawa H. 2008. Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 77:1053–1062 [DOI] [PubMed] [Google Scholar]
- 29. Kawaguchi H, Vertes AA, Okino S, Inui M, Yukawa H. 2006. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72:3418–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim SR, Ha SJ, Wei N, Oh EJ, Jin YS. 2012. Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends Biotechnol. 30:274–282 [DOI] [PubMed] [Google Scholar]
- 31. Kreuzer P, Gartner D, Allmansberger R, Hillen W. 1989. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J. Bacteriol. 171:3840–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurosawa K, Boccazzi P, de Almeida NM, Sinskey AJ. 2010. High-cell-density batch fermentation of Rhodococcus opacus PD630 using a high glucose concentration for triacylglycerol production. J. Biotechnol. 147:212–218 [DOI] [PubMed] [Google Scholar]
- 33. Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. 2010. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol. Bioeng. 106:193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Q, Du W, Liu D. 2008. Perspectives of microbial oils for biodiesel production. Appl. Microbiol. Biotechnol. 80:749–756 [DOI] [PubMed] [Google Scholar]
- 35. MacEachran DP, Prophete ME, Sinskey AJ. 2010. The Rhodococcus opacus PD630 heparin-binding hemagglutinin homolog TadA mediates lipid body formation. Appl. Environ. Microbiol. 76:7217–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20–W25 doi:10.1093/nar/gkh435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McLeod MP, et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meijnen JP, De Winde JH, Ruijssenaars HJ. 2008. Engineering Pseudomonas putida S12 for efficient utilization of d-xylose and l-arabinose. Appl. Environ. Microbiol. 74:5031–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogata H, et al. 1999. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27:29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Papanikolaou S, Aggelis G. 2011. Lipids of oleaginous yeasts. Part I. Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 13:1031–1051 [Google Scholar]
- 41. Patrauchan MA, et al. 2005. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J. Bacteriol. 187:4050–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rehm BHA, Mitsky TA, Steinbuchel A. 2001. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 67:3102–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rude MA, Schirmer A. 2009. New microbial fuels: a biotech perspective. Curr. Opin. Microbiol. 12:274–281 [DOI] [PubMed] [Google Scholar]
- 44. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Schafer A, et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 46. Sergeeva YE, Galanina LA, Andrianova DA, Feofilova EP. 2008. Lipids of filamentous fungi as a material for producing biodiesel fuel. Appl. Biochem. Microbiol. 44:523–527 [PubMed] [Google Scholar]
- 47. Shao Z, Dick WA, Behki RM. 1995. An improved Eschehchia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. using electroporation. Lett. Appl. Microbiol. 21:261–266 [DOI] [PubMed] [Google Scholar]
- 48. Somerville C, Youngs H, Taylor C, Davis SC, Long SP. 2010. Feedstocks for lignocellulosic biofuels. Science 329:790–792 [DOI] [PubMed] [Google Scholar]
- 49. Song S, Park C. 1997. Organization and regulation of the d-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 179:7025–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Steen EJ, et al. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562 [DOI] [PubMed] [Google Scholar]
- 51. van der Geize R, Dijkhuizen L. 2004. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7:255–261 [DOI] [PubMed] [Google Scholar]
- 52. van der Geize R, Hessels GI, Van Gerwen R, Van der Meijden P, Dijkhuizen L. 2002. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 45:1007–1018 [DOI] [PubMed] [Google Scholar]
- 53. Voss I, Steinbüchel A. 2001. High cell density cultivation of Rhodococcus opacus for lipid production at a pilot-plant scale. Appl. Microbiol. Biotechnol. 55:547–555 [DOI] [PubMed] [Google Scholar]
- 54. Wältermann M, Steinbüchel A. 2005. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J. Bacteriol. 187:3607–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wältermann M, Luftmann H, Baumeister D, Kalscheuer R, Steinbüchel A. 2000. Rhodococcus opacus strain PD630 as a new source of high-value single-cell oil? Isolation and characterization of triacylglycerols and other storage lipids. Microbiology 146:1143–1149 [DOI] [PubMed] [Google Scholar]
- 56. Wang J, Zhang B, Chen S. 2011. Oleaginous yeast Yarrowia lipolytica mutants with a disrupted fatty acyl-CoA synthetase gene accumulate saturated fatty acid. Process Biochem. 46:1436–1441 [Google Scholar]
- 57. Wendisch VF, Bott M, Eikmanns BJ. 2006. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr. Opin. Microbiol. 9:268–274 [DOI] [PubMed] [Google Scholar]
- 58. Xiao H, et al. 2011. Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Appl. Environ. Microbiol. 77:7886–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiong X, et al. 2007. Enhancement of biodesulfurization in two-liquid systems by heterogeneous expression of Vitreoscilla hemoglobin. Appl. Environ. Microbiol. 73:2394–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yanase H, et al. 2007. Genetic engineering of Zymobacter palmae for production of ethanol from xylose. Appl. Environ. Microbiol. 73:2592–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu X, Zheng Y, Dorgan KM, Chen S. 2011. Oil production by oleaginous yeasts using the hydrolysate from pretreatment of wheat straw with dilute sulfuric acid. Bioresour. Technol. 102:6134–6140 [DOI] [PubMed] [Google Scholar]
- 62. Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240. [DOI] [PubMed] [Google Scholar]
- 63. Zhang Y, Adams IP, Ratledge C. 2007. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153:2013–2015 [DOI] [PubMed] [Google Scholar]
- 64. Zheng Y, Chi Z, Lucker B, Chen S. 2012. Two-stage heterotrophic and phototrophic culture strategy for algal biomass and lipid production. Bioresour. Technol. 103:484–488 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.