Abstract
Detection of Brucella, causing brucellosis, is very challenging, since the applied techniques are mostly time-demanding and not standardized. While the common detection system relies on the cultivation of the bacteria, further classical typing up to the biotype level is mostly based on phenotypic or genotypic characteristics. The results of genotyping do not always fit the existing taxonomy, and misidentifications between genetically closely related genera cannot be avoided. This situation gets even worse, when detection from complex matrices, such as milk, is necessary. For these reasons, the availability of a method that allows early and reliable identification of possible Brucella isolates for both clinical and epidemiological reasons would be extremely useful. We evaluated micro-Raman spectroscopy in combination with chemometric analysis to identify Brucella from agar plates and directly from milk: prior to these studies, the samples were inactivated via formaldehyde treatment to ensure a higher working safety. The single-cell Raman spectra of different Brucella, Escherichia, Ochrobactrum, Pseudomonas, and Yersinia spp. were measured to create two independent databases for detection in media and milk. Identification accuracies of 92% for Brucella from medium and 94% for Brucella from milk were obtained while analyzing the single-cell Raman spectra via support vector machine. Even the identification of the other genera yielded sufficient results, with accuracies of >90%. In summary, micro-Raman spectroscopy is a promising alternative for detecting Brucella. The measurements we performed at the single-cell level thus allow fast identification within a few hours without a demanding process for sample preparation.
INTRODUCTION
In many respects, the detection of Brucella bacteria is very challenging. These small, coccoidal, and Gram-negative microorganisms are well-known biosafety level 3 (BSL-3) agents, which always bear a high risk for clinical and laboratory handling (4, 19, 58, 59), even if all safety measures are performed with the utmost care to avoid infections. This is supported by the fact that the health hazard of the bacteria causing brucellosis can be caused by small infection doses of 10 to 100 organisms as an aerosol to infect humans and by their potential as a biological warfare agent (13, 30, 58). Despite the low mortality rate of brucellosis, the zoonosis remains an important public health problem in wide areas of the world. The incidence of infection is especially high in regions such as the Middle East and countries bordering the Mediterranean Sea, as well as in China, Peru, and Mexico (31). In this context, humans were usually infected incidentally by contact with contagious animals or consumption of dairy foods, which were contaminated by Brucella species (13, 58).
Although all Brucella species are virulent, the major known pathogens for human brucellosis are Brucella melitensis, B. abortus, B. suis, and B. canis (51). The virulence is associated with changes in the composition of the lipopolysaccharides (LPS) in the outer cell membrane expressed in different colony morphologies: rough and smooth. Thereby, the smooth phenotype is regarded as fully virulent, whereas the species with rough surfaces seem to be less virulent (44). However, several exceptions from this rule exist: in contrast to the others, for example, B. canis appears to be naturally rough but causes occasional infections in humans (44).
Since brucellosis is a rare disease in industrialized countries, laboratories are often unfamiliar with the genus Brucella, and detection systems are often not reliable or sensitive enough. Even though the commonly used microbiological plating techniques for detection of Brucella are supplemented by more sophisticated and sensitive molecular genetic techniques such as PCR (27, 33, 51, 57, 60) or immunological assays such as the enzyme-linked immunosorbent assay (14), the handling and identification of this organism is still complicated. Bacterial culture techniques are hampered by a long cultivation time. Furthermore, the application of the cost-intensive PCR is limited when the detection of bacteria directly out of food is necessary, since an ability to detect them is influenced by PCR-inhibitory substances associated with the food matrix (37). In the case of Brucella, detection in milk and other dairy products is particularly complicated (2).
In addition, it has been reported that biochemical detection systems are not optimized for that agents, which causes unsafe identification or even misidentification of Brucella (3), most often as Ochrobactrum spp. (10, 50, 53) as result of PCR or as Yersinia spp. (49) in cross-reactions in serological analyses. In this context, the discrimination of the different Brucella species is even more problematic (60). These factors emphasize the need for a rapid and more efficient detection system.
Since vibrational spectroscopy, including infrared absorption (25, 26, 29, 55) and—in particular—all facets of Raman spectroscopy, has proved to be an appropriate tool for the rapid identification of microorganisms (35, 36), it could also be a promising approach for detecting BSL-3 agents (17), such as Brucella, by analyzing molecule-specific fingerprint information. A wide range of different techniques are applicable. Although the resonant excitation in the UV wavelength region probes taxonomic markers, e.g., DNA/RNA vibrations (28, 48), the application of visible or NIR Raman excitation wavelengths allows for a phenotypic characterization (20). By applying micro-Raman spectroscopy, even bacterial identification on the single-cell level can be performed (8, 15, 43, 54), and thus elaborate precultivation can be avoided. Thus, the detection process of microorganisms can be accelerated significantly, which is especially important in areas susceptible to bacterial contamination, e.g., clinical environments or the food-processing industry.
In order to fully exploit the potential of micro-Raman spectroscopy as a reliable identification tool, versatile spectral databases have to be created to evaluate all of the spectral information by statistical classifiers, such as linear discriminant analysis or a support vector machine (40).
To detect Brucella in milk via micro-Raman spectroscopy, we prepared a list of relevant pathogens to be considered in this context. In addition to Brucella spp., we chose genetically closely related and pathogenic Ochrobactrum species, as well as Yersinia species, to check whether misidentification with Brucella is also problematic. Pseudomonas and Escherichia spp. were used as Gram-negative representatives, which are well known to also be typical contaminants in dairy products (9, 56). In summary, 20 different species or subspecies (seven Brucella species, two strains of Escherichia, four strains of Ochrobactrum, five Pseudomonas species, and two strains of Yersinia) were selected.
With respect to the misidentification problem, we first took into account whether Brucella identification via Raman spectroscopy is workable when the bacteria have been cultivated on nutrient agar under standard conditions. We further focused on the identification of Brucella biovar samples, which have not been implemented in the database before. Beyond that, the discrimination potential of micro-Raman spectroscopy to the different Brucella species as a further issue was tested.
A second aim was to identify Brucella directly from an environmental sample: milk. Due to the changed nutrient and incubation conditions by adaptation to the new matrix, it is probable that the bacterial growth behavior was modified (16, 45). Changes caused by these factors become manifest as variations in the bacterial Raman spectra. Since the data are no longer comparable with the database of bacteria from medium, a second independent database has to be set up. Therefore, milk samples were spiked with the bacteria, processed and analyzed.
In addition, since handling BSL-3 agents is always dangerous, a suitable formalin inactivation procedure was applied to the bacterial samples prior to the analysis. By doing so, the microorganisms were concentrated and resuspended in distilled water; in that way, any further technique to isolate bacteria from milk was not necessary.
MATERIALS AND METHODS
Species and strains.
The bacteria were chosen according to their presence as known milk contaminants and their potential to be health hazards. The following species were examined in the present study for the databases: seven species of Brucella (B. abortus BA544, B. canis RM6/66, B. ceti NCTC 12891, B. melitensis BM16M, B. microti CCM 4915, B. pinnipedialis NCTC 12890, and B. suis BS1330), two strains of Escherichia coli (DSM 613 and DSM 1085), four strains of Ochrobactrum (O. anthropi ATCC 49188, O. anthropi field strain 10RB0263, O. intermedia DSM 17986, and O. intermedia field strain 06RB0416), five species of Pseudomonas (P. aeruginosa DSM 1117, P. aeruginosa DSM 50071, P. fluorescens DSM 50106, P. fragi DSM 3456, P. putida DSM 291, and P. stutzeri DSM 5190), and two Yersinia enterocolitica subsp. enterocolitica serovars (Y. enterocolitica DSM 4780 [O:3] and Y. enterocolitica DSM 11504 [O:5,27]). In addition, six Brucella biovars were considered as “unknown” samples: B. abortus biovar 3, B. abortus biovar 4, B. melitensis biovar 2, B. melitensis biovar 3, and two different isolates of B. suis biovar 2. In Table 1, the details about the source of each investigated species are given.
Table 1.
Sources of the investigated species
| Organism | Strain | Biovar | Sourcea |
|---|---|---|---|
| B. abortus | BA544 | 1 | ERL |
| 08RB1648 (field strain) | 3 | BfR | |
| 08RB1653 (field strain) | 4 | BfR | |
| B. canis | RM6/66 | ERL | |
| B. ceti | NCTC 12891 | SAC | |
| B. melitensis | BM16M | 1 | ERL |
| 09RB5269 (field strain) | 2 | FLI | |
| 09RB5696 (field strain) | 3 | FLI | |
| B. microti | CCM 4915 | IMB | |
| B. pinnipedialis | NCTC 12890 | SAC | |
| B. suis | BS1330 | 1 | ERL |
| 09RB5757 (field strain) | 2 | FLI | |
| 09RB5758 (field strain) | 2 | FLI | |
| E. coli | DSM 613 | DSMZ | |
| DSM 1085 | DSMZ | ||
| O. anthropi | ATCC 49188 | DSMZ | |
| 10RB0263 (field strain) | FLI | ||
| O. intermedia | DSM 17986 | DSMZ | |
| 06RB0416 (field strain) | FLI | ||
| P. aeruginosa | DSM 1117 | DSMZ | |
| DSM 50071 | DSMZ | ||
| P. fluorescens | DSM 50106 | DSMZ | |
| P. fragi | DSM 3456 | DSMZ | |
| P. putida | DSM 291 | DSMZ | |
| P. stutzeri | DSM 5190 | DSMZ | |
| Y. enterocolitica | DSM 4780, O:3 | DSMZ | |
| DSM 11504, O:5,27 | DSMZ |
ERL, European Reference Laboratory for Brucellosis, Maisons Alfort, France; IMB, Bundeswehr Institute of Microbiology, Munich, Germany; FLI, Institute of Bacterial Infections and Zoonoses, Friedrich Loeffler Institute, Federal Research Institute for Animal Health, Jena, Germany; DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany.
Sample preparation.
The Brucella spp. and Ochrobactrum spp. were grown according to the OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (www.oie.int). All other investigated species were cultivated on a standard nutrient agar for 24 h at an incubation temperature of 37°C.
The microorganisms from one plate were then suspended in 1 ml of 0.9% sodium chloride solution and cleaned three times by centrifugation at 12,100 × g for 1 min (MiniSpin; Eppendorf, Hamburg, Germany). Afterward, the bacterial pellet was resuspended in 0.9% sodium chloride solution. In this way, six independently cultivated batches of each species were prepared in order to take biological variability between the samples into account. Three of them were directly inactivated as described below, while each of the other batches was inoculated in 1 ml of commercially available ultra-high-temperature-treated milk with a 1.5% fat content to a concentration of ∼107 cells/ml. At room temperature, the bacteria were allowed to adapt to the new matrix over a time period of 24 h. Afterward, the spiked milk samples were also inactivated via a formaldehyde treatment, which is a useful Raman-compatible procedure (47). The samples were mixed with 37% formaldehyde solution (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) in a 10-ml tube to a final concentration of 10% formaldehyde. During a treatment time of 1 h, the tubes were shaken continuously at 150 rpm in a laboratory shaker (Gerhardt Shaker RO 15). The treatment was stopped by centrifugation at 2,200 × g for 10 min. After three additional washes, including centrifugation, the bacterial pellet was resuspended in 1 ml of 0.9% sodium chloride solution. To determine a successful inactivation, 50 μl of the bacterial solution was plated, followed by incubation at 37°C for a week. Only samples that showed no growth were used for Raman spectroscopic measurements. An inoculation loop with a volume of 1 μl was used to load the nickel foil with the sample. Before starting the measurement, the sample was air dried.
Spectroscopic instrumentation.
The Raman spectra were obtained with a micro-Raman setup (Bio-Particle Explorer; rap.ID Particle Systems GmbH, Berlin, Germany). A solid-state frequency-doubled Nd:YAG module (LCM-S-111-NNP25; Laser-Export Co., Ltd.) provides an excitation light with a wavelength of 532 nm. While an Olympus MPLFLN-BD 100× objective focuses the Raman excitation light onto the sample with a spot size of below 1 μm laterally, the samples are hit with ∼7 mW of laser power. After removal of the Rayleigh scattering, the scattered Raman light is diffracted with a single-stage monochromator (HE 532; Horiba Jobin Yvon, Munich, Germany) equipped with a 920-line/mm grating and collected with a thermoelectrically cooled charge-coupled device (CCD) camera (DV401-BV; Andor Technology, Belfast, Northern Ireland) with a spectral resolution of ∼10 cm−1. The integration time per single Raman spectrum (−113 to 3,186 cm−1) was 20 s, recorded as two 10-s measurements. These two spectra were compared afterward for spike removal and are summarized as one spectrum.
Databases.
After the collection of all of the spectra for each sample (∼50 single spectra per batch), two databases were created. All spectra for bacteria that were prepared on nutrient agar were centralized into a database named “medium” (3,761 spectra), while all spectra for the bacteria from milk were collected in a second database named “milk” (3,218 spectra). A more detailed breakdown of the data is given in Table SA1 in the supplemental material. In summary, two batches of each species were used to build the classification model, while an independent third batch was measured to identify them. This procedure is explained in the following section.
Chemometric methods.
The analysis of the Raman spectra took place in three steps: (i) preprocessing, (ii) training of the self-learning machine, and (iii) validation (6, 42, 52) and is performed with Gnu R (34). Each spectrum was preprocessed in the same way in order to reduce the contribution from noise and also the spectral variations due to the measuring procedure. First, the background and the cosmic spikes of the spectra were removed. The importance of the background correction was given, since the effects of various backgrounds, caused by fluorescence of the sample or thermal fluctuations on the CCD, were minimized (7). For this purpose the SNIP algorithm (for statistics-sensitive nonlinear iterative peak clipping algorithm), which is a composite of a low statistics filter and a peak clipping algorithm, was adopted (23, 38). Cosmic spikes could be easily localized by recording two Raman spectra of the same microbial cell, since their origin is neither correlated in time nor space. Intensity differences for each channel were calculated and spikes located in channels, where the intensity difference exceeded twice the standard deviation.
After the spike removal, a wavenumber calibration using acetaminophen as a standard was performed on the spectra. In addition, only significant wave number regions (650 to 1,750 cm−1 and 2,750 to 3,100 cm−1) were regarded to reduce data dimension. To ensure comparability between the spectra, vector normalization was conducted. Here, the spectra were divided by its 2-norm (Euclidean norm).
A principal component analysis (PCA) was performed prior to the linear discriminant analysis (LDA) to reduce the dimensionality of the problem. In addition, white noise can be removed by cutting off the scores after a particular channel in the new spectral space, and so the probability to find the best classification model representing the data can be increased (7). The exact number of the applied scores depends on the size of the data set and was separately determined for each calculation. The number of the chosen PCs was the best compromise between a minimal number of PCs and classification accuracy and practically never exceeded 10% of the whole spectral data to avoid overfitting. It should be mentioned that the PCA was only performed for the classification data set. To also convert the independent identification data in the same spectral space, the set was rotated by the loadings of the PCA of the preceding calculation.
After the data preprocessing was finished, the multivariate analyses were performed: LDA and support vector machine (SVM) were the classifiers of choice, since both strategies are widely used in pattern analysis of bacterial spectra (21, 47). Whereas LDA is performed with the scores from the PCA, the entire spectral information is used for SVM. A radial kernel function was chosen for SVM to map the data in a different space. The optimization of the values of cost and gamma allows some flexibility in separating the categories to create a more accurate classification model. A high cost value will force the SVM to create a complex enough prediction function to misclassify as few training points as possible, while a lower cost parameter will lead to a simpler prediction function (18).
A comparative study of both techniques was useful, since it is postulated that no specific classifier has the best fit for all problems and no classifier is always better than another one (39). This is especially true for experimental sciences dealing with real-world samples. Therefore, both classification algorithms were run on the same data set to compare their outputs for statistical significance. The accuracies given by these calculations were used as the accuracies of the classifiers. In addition, the sensitivity (the true-positive rate) and the specificity (the true-negative rate) for each genus or species was calculated. The spectra were smoothed by means of a Savitzky-Golay filter only for visualization.
RESULTS
In order to investigate the potential of micro-Raman spectroscopy for Brucella identification, spectral profiles of seven different Brucella spp. and genetically closely related pathogens (nine species), which are well known as typical microbial cross-reactants and milk contaminants, were obtained and analyzed more deeply by the chemometric methods LDA and SVM.
Initially, we took into account whether identification of Brucella prepared on nutrient agar plates was possible, whereas variances of the spectra between different genera should also be evaluated. Also, we tried to discriminate Brucella to the species level. Finally, the spectroscopic procedure was evaluated for the detection and differentiation of Brucella spp. in milk.
Identification of Brucella prepared on agar plates.
Since micro-Raman spectroscopy should be verified as a reliable identification tool for Brucella from the medium, we obtained Raman spectra for single microbial cells from seven Brucella spp., two strains of Escherichia, four strains of Ochrobactrum, five species of Pseudomonas, and two strains of Yersinia. A database was created, including 2,610 Raman spectra, to build up a classification model. Further, 1,151 Raman spectra of independently cultivated and measured bacterial samples were compiled as an identification set to be identified by the classification model.
On the basis of the spectral information, which needed to be analyzed, it is important to find a good classification algorithm. Since in the literature it has been reported that no single classifier is best for all problems and no classifier is always better than another one (39), we decided to perform a comparative study of two known algorithms practicable for bacterial discrimination (46, 54).
In order to validate the SVM performance, an LDA was also performed on the Raman spectra. A feature-reduction step was carried out using PCA before passing the data into the classification algorithm (LDA). The number of principal components used was 88. An overall accuracy of 92.9% was obtained, i.e., 2,424 of 2,610 Raman spectra were classified correctly. When the resulting classification model was applied to the identification set, 90.8% (1,045/1,151 spectra) of the spectra were assigned to the correct class. The outcomes are summarized in Table 2. The maximum sensitivity was obtained for the Raman spectra of Brucella spp. (96.8%), whereby the genetically closely related Ochrobactrum species showed the lowest accuracy (72.9%), since only 153 of 210 Raman spectra were identified accurately. Most of the incorrectly labeled spectra were put into the Brucella class (see Table SA2 in the supplemental material).
Table 2.
Results of LDA and SVM calculations: database “medium”a
| Genus | LDA |
SVM |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classification data |
Identification data |
Classification data |
Identification data |
|||||||||
| TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | |
| Brucella | 980/1048 | 93.5 | 94.8 | 368/380 | 96.8 | 92.4 | 1044/1048 | 99.6 | 98.5 | 369/380 | 97.1 | 92.8 |
| Escherichia | 212/215 | 98.6 | 99.6 | 101/106 | 95.3 | 99.8 | 214/215 | 99.5 | 99.9 | 94/106 | 88.8 | 99.8 |
| Ochrobactrum | 347/410 | 84.6 | 96.9 | 153/210 | 72.9 | 97.9 | 393/410 | 95.9 | 99.8 | 162/210 | 77.1 | 97.9 |
| Pseudomonas | 600/636 | 94.3 | 98.7 | 284/305 | 93.1 | 97.0 | 627/636 | 98.6 | 99.9 | 283/305 | 92.8 | 98.0 |
| Yersinia | 285/301 | 94.7 | 100 | 139/150 | 92.7 | 100 | 301/301 | 100 | 100 | 149/150 | 99.3 | 100 |
For the database “medium,” the values of true positives (TP) against all class members (CM) for classification and identification data are given as the results of LDA and SVM calculations (sensitivity, Sens; specificity, Spec).
For classification by means of SVM, a radial basis function was applied to the data. The results for this calculation are listed in Table 2 and also, in greater detail, in Table SA3 in the supplemental material. From 2,610 Raman spectra, 2,579 spectra were classified correctly (98.8%). The average recognition rate for the validation data was 91.8% (1,057/1,151 spectra). Although the sensitivity of Ochrobactrum was increased to 77.1% compared to the results of the LDA, misidentifications as Brucella and Pseudomonas were still not avoidable. Consequently, the specificity of the Brucella class (92.8%) is lower compared than that of the other classes. The Raman spectra of Escherichia were assigned with a correctness of 88.8%. In contrast to this, identification of Yersinia resulted in high sensitivities (99.3%) and specificities (100%).
To assess whether overfitting is problematic and to test whether a sample type yet unknown to the classification model of the database “medium” could be handled, six Brucella biovar samples (B. abortus biovar 3, B. abortus biovar 4, B. melitensis biovar 2, B. melitensis biovar 3, and two different isolates of B. suis biovar 2) were independently prepared in the same way as the other species. In summary, 324 Raman spectra on the single-cell level were obtained and labeled by the LDA classification model. In this way, 94.8% (307/324 Raman spectra) were correctly labeled as Brucella, while misclassified spectra were mainly labeled as Pseudomonas. In Table 3 the results are summarized on a subspecies level: the samplewise rates ranged from 90.0% (B. suis biovar 2) to 100% (B. abortus biovar 4). Applying SVM to these data generated similar results: 82.0% for B. suis biovar 2 to 100% for B. melitensis biovar 2 (data not shown).
Table 3.
LDA identification results for Brucella biovar samples not implemented in the database
| Strain | Spectra labeled as Brucella | % Sensa | Misidentification(s) (no. of spectra) |
|---|---|---|---|
| B. abortus biovar 3 | 63/66 | 95.5 | Escherichia (1), Pseudomonas (2) |
| B. abortus biovar 4 | 39/39 | 100 | |
| B. melitensis biovar 2 | 57/59 | 96.6 | Pseudomonas (2) |
| B. melitensis biovar 3 | 53/56 | 94.6 | Ochrobactrum (1), Pseudomonas (2) |
| B. suis biovar 2 | 45/50 | 90.0 | Pseudomonas (5) |
| 50/54 | 92.6 | Pseudomonas (4) |
Sens, sensitivity.
Significant spectral variations.
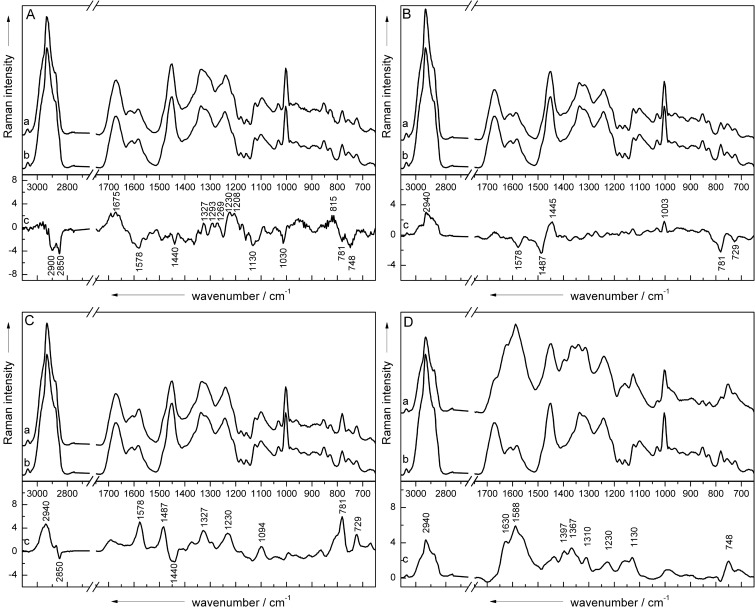
To analyze the spectral variations between the different genera, each of the four classes (Escherichia, Ochrobactrum, Pseudomonas, and Yersinia) was classified in a two-class approach against the Brucella class by means of LDA based on the classification data of the database “medium.” In this context, four classifications were performed using five principal components, since >98% of the spectral information of all samples was taken into account with this number of principal components. Afterward, the LDA factor loadings of the wavenumbers were calculated and compared to the mean spectra of the classification data (Fig. 1). Independent of each classification, bands with negative loading vector values were assigned as a marker band of the Brucella spectra, whereas peaks with positive values were associated with the mean spectra of the other class. The band assignment was performed as described previously (24, 32).
Fig 1.
The mean spectra of Brucella spp. (Ab, Bb, Cb, and Db) from media were compared to the mean spectra of Escherichia (Aa), Ochrobactrum (Ba), Pseudomonas (Ca), and Yersinia (Da) spp. isolated from medium by calculating the loading vector (Ac, Bc, Cc, and Dc) of a two-class LDA with five PCs.
While investigating spectral variations between Escherichia (line a) and Brucella (line b), shown in Fig. 1A, the loading vector (line c) indicates that some small signal variations were responsible for discrimination. Brucella spectra are differentiated by the higher signal intensities for DNA bands at 781 cm−1 (cytosine or uracil) and 1,578 cm−1 (guanine), cytochrome bands at 748, 1,130, and 1,578 cm−1, and peaks at 1,440 cm−1 (proteins), 2,850 cm−1 (lipids and fatty acids), and 2,900 cm−1 (lipids and proteins) assigned to the C—H vibrations. Escherichia, on the other hand, is separated by a number of bands, which can be predominantly assigned to protein signals (1,208, 1,230, 1,269, 1,293, and 1,675 cm−1). In addition, the signals for RNA structures at 815 cm−1 and for DNA at 1,327 cm−1 appear in the spectrum of Escherichia compared to the Brucella spectrum.
As a result of the loading vector (line c) in Fig. 1B, the discrimination of Brucella (line b) and Ochrobactrum (line a) is based on some significant protein and DNA signals. According to that the spectra of Ochrobactrum exhibit increased bands for phenylalanine (1,003 cm−1), the CH2 deformation vibration for proteins (1,448 cm−1), and the C—H stretching region at 2,940 cm−1, whereas typical DNA bands at 729 cm−1 (adenine), 781 cm−1 (cytosine or uracil), and 1,487 and 1,578 cm−1 (guanine) are stronger in the spectra of Brucella.
When considering the Pseudomonas (Fig. 1C, line a) versus the Brucella (line b) class, it is obvious that most of the spectral variations occurring in the loading vector (line c) are assignable to Pseudomonas. Thus far, almost all typical DNA signatures (729, 781, 1,094, 1,327, 1,487, and 1,578 cm−1) and, in addition, the amide III band (1,230 cm−1) and the C—H vibration (2,940 cm−1) appear to be enhanced compared to the mean spectrum of Brucella. Only lipid signals such as the CH2 vibration at 2,850 cm−1 and the CH2/CH3 deformation vibration at 1,440 cm−1 are slightly increased in the spectra of Brucella.
In Fig. 1D, the spectrum of Yersinia (line a) was compared to that of Brucella (line b). Recognizably, the mean spectra differ totally from each other. Thus, the separation criteria for these two classes are clearly given by the specific bands of Yersinia, which is also verified by the calculated loading vector (line c). The spectrum represents peaks from DNA, proteins, lipids, and possibly a cytochrome at 748 cm−1 (DNA, cytochrome), 1,130 cm−1 (fatty acids, cytochrome), 1,230 cm−1 (amide III), 1,310 cm−1 (lipids), 1,367 cm−1 (phospholipids), 1,397 cm−1 (CH2 deformation), 1,588 cm−1 (phenylalanine), 1,630 cm−1 (amide I), and 2,940 cm−1 (lipids and proteins).
Discrimination of Brucella species.
In Fig. 2, the mean spectra of B. abortus (line a), B. canis (line b), B. ceti (line c), B. melitensis (line d), B. microti (line e), B. pinnipedialis (line f), and B. suis (line g) are shown, where all essential microbial bands can be recognized. However, spectral variations between the different species are not obvious at first sight. The Raman spectra seem to be highly homogeneous, which is why chemometric analysis have to be applied to the data.
Fig 2.
Mean spectra were calculated from ∼100 background-corrected single-cell spectra of each investigated strain isolated from agar plates (B. abortus [line a], B. canis [line b], B. ceti [line c], B. melitensis [line d], B. microti [line e], B. pinnipedialis [line f], and B. suis [line g]) and are shown to highlight the close similarity of the spectra. The spectra are vertically shifted for clarity.
To investigate the discrimination and identification potential of Raman spectroscopy for Brucella on the species level, LDA and SVM were performed on the spectra of all investigated Brucella spp., which were cultivated on nutrient agar and inactivated by means of formalin treatment. Approximately 150 spectra of three batches of each species were used to set up a classification model, whereas a fourth independent batch was applied for validation.
Table 4 summarizes the results of the LDA. An overall accuracy of 85.7% was obtained for the classification using 86 principal components for dimension reduction. Although specificities >94% were observable for all species, only the sensitivity of B. microti was 94.4%. Among the other six species, the sensitivities varied from 75.0% for B. melitensis to 87.7% for B. abortus. B. melitensis was often misclassified as B. abortus and vice versa.
Table 4.
Discrimination results of the spectroscopic data from Brucella at the species levela
| Strain | LDA |
SVM |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classification data |
Identification data |
Classification data |
Identification data |
|||||||||
| TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | |
| B. abortus | 135/154 | 87.7 | 95.5 | 34/41 | 82.9 | 95.0 | 154/154 | 100 | 99.4 | 36/41 | 87.8 | 95.2 |
| B. canis | 121/141 | 85.8 | 98.8 | 67/73 | 91.8 | 95.8 | 141/142 | 100 | 100 | 70/73 | 95.9 | 96.3 |
| B. ceti | 72/88 | 81.8 | 99.8 | 36/60 | 60.0 | 97.6 | 88/88 | 100 | 100 | 53/60 | 88.3 | 96.9 |
| B. melitensis | 111/148 | 75.0 | 96.7 | 34/43 | 79.1 | 95.9 | 143/148 | 96.6 | 100 | 33/43 | 76.7 | 94.9 |
| B. microti | 202/214 | 94.4 | 97.4 | 63/78 | 80.8 | 98.1 | 214/214 | 100 | 99.9 | 58/78 | 74.4 | 99.6 |
| B. pinnipedialis | 115/132 | 87.1 | 94.9 | 43/53 | 81.1 | 95.5 | 131/132 | 99.2 | 100 | 43/53 | 81.1 | 98.6 |
| B. suis | 124/150 | 82.7 | 97.7 | 42/53 | 79.2 | 93.6 | 150/150 | 100 | 100 | 40/53 | 75.5 | 95.8 |
The results include all species-specific sensitivities (Sens) and specificities (Spec) obtained by performing an LDA and SVM on model and validation data.
By rotating the identification data in the same spectral space of the classification data and performing an LDA afterward, an overall accuracy of 79.6% was calculated. Again, specificities of >93% for each species were obtained, and the sensitivities were between 60.0% (B. ceti) and 91.8% (B. canis). Thus, B. ceti was often labeled as B. pinnipedialis (10/53 spectra) and B. suis (10/53 spectra), while B. microti was mainly misidentified as B. abortus (8/78 spectra). More-detailed confusion matrices are given in the Table SA6 in the supplemental material.
The same spectra were classified by means of a SVM using a radial kernel with an accuracy of 99.4% and an identification rate of 83.0%, as shown in Table 4. Although the classification was promising, since sensitivities of >96% and specificities of >99% for each species were calculated, the validation data were predicted only slightly better than with the LDA. Again, noteworthy interspecies misidentification was recognized similar to the analysis via the LDA. For example, some spectra of B. suis (8/53 spectra) and of B. microti (7/78 spectra) were often labeled as B. melitensis, whereas the sensitivities of both species (B. suis, 75.5%; B. microti, 74.4%) were considerably low. Also, the overlapping of B. pinnipedialis (9/53 spectra) and B. ceti (4/60 spectra) is worth mentioning. Instead, the identification rate of B. canis (95.9%) was increased in comparison to the LDA (see Table SA7 in the supplemental material).
Detection of Brucella from milk samples.
Since the results for identifying inactivated Brucella under standard laboratory conditions were promising, we used this approach for real-world samples. Hence, milk samples were spiked with the pathogens and analyzed after 1 day of storage. The prepared milk samples were inactivated by the described procedure. The resulting resuspended biomass was used to gather single-cell Raman spectra, which were collected in a second database “milk.” In summary, 3,218 Raman spectra were measured and divided in classification (2,140 Raman spectra) and identification data (1,078 Raman spectra). Again, the spectra were statistically analyzed: the results of the LDA and SVM are given in Table 5 and more detailed in Tables SA4 and SA5 in the supplemental material.
Table 5.
Results of LDA and SVM calculations: database “milk”a
| Genus | LDA |
SVM |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Classification data |
Identification data |
Classification data |
Identification data |
|||||||||
| TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | TP/CM | % Sens | % Spec | |
| Brucella | 583/624 | 93.4 | 96.3 | 325/351 | 92.6 | 96.3 | 615/624 | 98.6 | 98.3 | 330/351 | 94.0 | 98.4 |
| Escherichia | 199/205 | 97.1 | 99.7 | 100/102 | 98.0 | 99.1 | 204/205 | 99.5 | 99.7 | 102/102 | 100 | 99.6 |
| Ochrobactrum | 352/425 | 82.8 | 96.8 | 173/204 | 84.8 | 96.8 | 402/425 | 94.6 | 99.1 | 191/204 | 93.6 | 97.9 |
| Pseudomonas | 640/656 | 97.6 | 98.3 | 300/316 | 94.9 | 99.2 | 645/656 | 98.3 | 99.6 | 309/316 | 97.8 | 99.3 |
| Yersinia | 206/230 | 89.6 | 99.0 | 98/105 | 93.3 | 98.7 | 220/230 | 95.7 | 99.9 | 101/105 | 96.2 | 99.4 |
For the database “milk,” the values of true positives (TP) against all class members (CM) for model and validation data are given as results of LDA and SVM calculations (sensitivity, Sens; specificity, Spec).
The scores of 67 PCs were introduced in the data algorithm of the LDA. As a result, an overall accuracy of 92.5% was obtained for the classification data set, and the validation of the milk bacteria achieved almost the same rate (92.4%). Compared to the LDA results for the database “medium,” misidentification between Brucella (92.6% sensitivity) and Ochrobactrum (84.8% sensitivity) again appeared, whereas the discrimination of Yersinia (93.3% sensitivity) from Brucella was not problematic. Interestingly, the results could be improved when SVM was used. For the classification model data, the sensitivities varied from 94.6% (Ochrobactrum) to 99.5% (Escherichia), and the specificities were between 98.3% (Brucella) and 99.9% (Yersinia). Only 54 of 2,140 Raman spectra were misclassified (97.5%). Upon analyzing the results for the identification data, an overall accuracy of 95.8% (1,033/1,078 Raman spectra) was calculated. Although Brucella could be identified with a 94.0% sensitivity, even the spectra of Ochrobactrum were assigned correctly with a 93.6% sensitivity. The sensitivities of the other milk contaminants were up to 100% (Escherichia, 100%; Pseudomonas, 97.8%; and Yersinia, 96.2%).
Based on this data set, the 624 Raman spectra of Brucella in milk were used to evaluate the discrimination potential on the species level by performing both LDA and SVM (data not shown). Identification rates of 75.0% (LDA, 15 principal components; classification accuracy, 82.5%) and 70.2% (SVM; classification accuracy, 95.7%) were achieved here.
DISCUSSION
Although over 500,000 humans are infected by Brucella per year worldwide (57), the detection of the causative agents is still challenging. Diagnoses of brucellosis are currently based mainly on the cultivation of Brucella, on biochemical tests, or on classical serological methods, which often generate discordant results (4). In addition, these approaches bear a higher risk of infecting the personnel in the wake of their time-consuming and laborious character.
For this reason, more sophisticated techniques, such as PCR, are in development for clinical use that can be also performed directly on complex samples such as blood or milk (1, 41). However, the technique is not well established, since there is no standardization between the laboratories and sample preparation or the choice of target genes varies (60). As a consequence, PCR applications provide different sensitivities for Brucella identification. Furthermore, the detection is hampered by time-demanding precultivation or cost-intensive supplements. Therefore, we evaluated Raman spectroscopy as an identification tool for detecting Brucella.
Prior to the Raman spectroscopic measurements, the sample handling of pathogenic Brucella is simplified by a 1-h formalin inactivation step, which can be easily transferred to other organisms, such as Ochrobactrum or Yersinia, and can also be used directly for milk from which bacteria should be identified. This ensures more stable working safety, since bacteria are no longer infectious and so the risk of laboratory accidents is lowered. In addition, the inactivation procedure does not limit the subsequent analysis (47). Because we used milk spiked with 107 bacterial cells/ml of milk, no further isolation step has to be applied, since the bacterial biomass is concentrated during the inactivation process and afterward resuspended in distilled water to allow single cell measurements. Naturally, this high starting concentration was chosen to generate the high amount of the spectral data establishing a comprehensive database but is presumably orders of magnitude higher than the detection limit of the method.
To detect contaminations in the range of ≤103 cells/ml of milk, it is necessary to apply an additional isolation step for the Raman spectroscopic analysis. In this context, Raman-compatible extraction techniques based on buoyant density centrifugation or enzymatic milk clearing, found in the literature, can be used (12, 21). Although these isolation techniques are accompanied by a loss of one order of magnitude of the bacterial start concentration, the recovered biomass is still enough to detect single microbial cells by means of micro-Raman spectroscopy. Similar extraction steps are also essential for PCR, too, which is certainly limited when dealing with bacteria directly in milk due to inhibitory effects (5).
By means of a micro-Raman spectroscopic approach, a wealth of single-cell Raman spectra of all prepared samples were collected. On the basis of this amount of spectral information, which needed to be analyzed, it is important to find a good ongoing classification algorithm. Since it is postulated that no classifier is always better than another one (39), we decided to perform a comparative study of SVM and LDA, which are two algorithms known to be useful in pattern recognition of bacterial samples.
LDA is a common applied method for linear supervised dimensionality reduction, which is used to find a lower dimensional space that best discriminates the samples from different classes. Instead, SVM is a powerful supervised learning algorithm based on the principle of maximal margin bound. The application of both methods to the same data sets allowed us to compare the differences in their performances depending on the way looking at the problem. Since the performance of both classifiers is comparable, we are going to discuss only the results of the best classification model in each case.
Discrimination and identification of Brucella from media against Ochrobactrum or Yersinia, the most common cross-reactants in other biochemical detection systems, worked out with >92% sensitivity by analysis via Raman spectroscopy and SVM. Even further accompanying background organisms in milk, which are represented by Pseudomonas and Escherichia, can be separated from the rest.
It could also be shown that also a various number of signal variations, which are responsible for the discrimination, occur when analyzing the loading vectors between Brucella and the other genera. Escherichia and Brucella mostly differ in the cytochrome content, which is one possible reason why misidentification between these two classes is pretty rare. The spectra of Pseudomonas show significantly higher protein and DNA signals compared to Brucella. The changes in the spectra are provoked due to variations in the microbial composition and growth behavior of each genus. Yersinia shows a drastically changed Raman spectrum in comparison to the spectra of all other genera. Interestingly, the spectra of Ochrobactrum and Brucella show only few variations in DNA and protein bands. Conductively, misidentifications are more probable.
It has been reported that common biochemical identification routines are not reliable, since Ochrobactrum is often misidentified as Brucella and vice versa due to the high phenotypic and genotypic similarity between Ochrobactrum and Brucella, sharing 98.8% rRNA genes (10). Sensitivities in the range of 32 to 84% were reported for Ochrobactrum anthropi in APE 20NE systems, which encompassed also false identification of other genera such as Pseudomonas (50). We also obtained some misidentifications of Ochrobactrum as Brucella or Pseudomonas when using Raman spectroscopy. Thus, the identification rate of Ochrobactrum from culture media was reduced to 77%. Against this, the sensitivity for Ochrobactrum in milk is much higher (93.6%), which might be influenced by different adaptations and growth behaviors of the bacteria due to the different media.
Due to the high natural diversity of the genus Brucella, it was also imperative to test the classification model with several Brucella biovars, which were not implemented in the analysis before: the results ranging from 90 to 100% indicate that the method is stable enough to also identify reliably “unknown” Brucella subspecies at the genus level by using the LDA classifier.
By applying a combination of inactivation, Raman spectroscopy, and chemometrics to spiked milk samples, we can show that identification of Brucella in milk can be easily and rapidly ensured. Sensitivities in the range of 93.6 to 100% were obtained for all investigated genera applying SVM to the data.
Moreover, the discrimination potential of Raman spectroscopy for different Brucella species is a further concern in the present study. Although micro-Raman spectroscopy allows a satisfactory identification of the genus Brucella, the identification of independently prepared samples is less reliable for Brucella at the species level, since accuracies of only 83.0% from agar plates and 70.2% from milk were obtained, whereas the model operates with rates of >95.0%. One reason for this can be given by the fact that all Brucella spp. share >90% nucleic acid sequence homology (57). Also, other previously used techniques, such as PCR, have been partly hampered by these factors (60). In addition, the current internal taxonomy of Brucella is controversial, since it relies on a variety of biochemical, antigenic, and metabolic differences that are used simultaneously (22). Caused by this fact, detection of the single species could be limited, which is exemplified by an matrix-assisted laser desorption ionization–time of flight mass spectrometry study applied to six Brucella spp., with the result that 20.6% of the isolates were identified correctly (11).
Furthermore, the morphology of the species varies in the LPS composition due to spontaneous mutations, which directly influences the virulence of the bacteria. We took this fact into account by implementing only fully virulent and therefore smooth species, with the exception of B. canis, which appears naturally rough, but causes occasional infections in humans (44). Concluding, it is reasonable that B. canis can be identified with 95.6% sensitivity by means of SVM in comparison to the other species (sensitivities in the range of 74 to 88%). Even if the discrimination of Brucella spp. is still problematic, the availability of a method, which allows early identification of possible Brucella isolates independent of the LPS composition on genus-level both for clinical and epidemiological reasons, is extremely useful.
This report indicates that micro-Raman spectroscopy in combination with SVM may offer a promising alternative to detect Brucella both from agar-plates and directly from milk. Measurements on the single-cell level are enabled and so a faster and more accurate identification within 2 h can be achieved while avoiding a precultivation step.
Supplementary Material
ACKNOWLEDGMENTS
Funding of the research projects Pathosafe (FKZ 13N9547 and FKZ 13N9549) and RamaDek (FKZ 13N11168) from the Federal Ministry of Education and Research, Germany, and MikroPlex (PE 113-1) from the Thüringische Exzellenzinitiative is gratefully acknowledged.
We also thank Katja Fischer (Friedrich Loeffler Institute) for preparing the pathogenic organisms.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Al-Mariri A, Haj-Mahmoud N. 2010. Detection of Brucella abortus in bovine milk by polymerase chain reaction. Acta Vet. Brno 79:277–280 [Google Scholar]
- 2. Amoroso MG, et al. 2011. Validation of a real-time PCR assay for fast and sensitive quantification of Brucella spp. in water buffalo milk. Food Control 22:1466–1470 [Google Scholar]
- 3. Barham WB, Church P, Brown JE, Paparello S. 1993. Misidentification of Brucella species with use of rapid bacterial identification systems. Clin. Infect. Dis. 17:1068–1069 [DOI] [PubMed] [Google Scholar]
- 4. Batchelor BI, Brindle RJ, Gilks GF, Selkon JB. 1992. Biochemical misidentification of Brucella melitensis and subsequent laboratory-acquired infections. J. Hosp. Infect. 22:159–162 [DOI] [PubMed] [Google Scholar]
- 5. Bickley J, Short JK, McDowell DG, Parkes HC. 1996. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett. Appl. Microbiol. 22:153–158 [DOI] [PubMed] [Google Scholar]
- 6. Bocklitz T, et al. 2009. A comprehensive study of classification methods for medical diagnosis. J. Raman Spectrosc. 40:1759–1765 [Google Scholar]
- 7. Bocklitz T, Walter A, Hartmann K, Rösch P, Popp J. 2011. How to pre-process Raman spectra for reliable and stable models? Anal. Chim. Acta 704:47–56 [DOI] [PubMed] [Google Scholar]
- 8. Ciobotã V, et al. 2011. Influence of intracellular storage material on bacterial identification by means of Raman spectroscopy. Anal. Bioanal. Chem. 397:2929–2937 [DOI] [PubMed] [Google Scholar]
- 9. Daly P, Collier T, Doyle S. 2002. PCR-ELISA detection of Escherichia coli in milk. Lett. Appl. Microbiol. 34:222–226 [DOI] [PubMed] [Google Scholar]
- 10. Elsaghir AAF, James EA. 2003. Misidentification of Brucella melitensis as Ochrobactrum anthropi by API 20NE. J. Med. Microbiol. 52:441–442 [DOI] [PubMed] [Google Scholar]
- 11. Ferreira L, et al. 2010. Identification of Brucella by MALDI-TOF mass spectrometry: fast and reliable identification from agar plates and blood cultures. PLoS One 5:e14235 doi:10.1371/journal.pone.0014235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukushima H, Katsube K, Hata Y, Kishi R, Fujiwara S. 2007. Rapid separation and concentration of food-borne pathogens in food samples prior to quantification by viable-cell counting and real-time PCR. Appl. Environ. Microbiol. 73:92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guihot A, Bossi P, Bricaire F. 2004. Bioterrorism with brucellosis. Presse Med. 33:119–122 [DOI] [PubMed] [Google Scholar]
- 14. Gutierrez J, Liebana J. 1993. Immunological methods for the detection of structural components and metabolites of bacteria and fungi in blood. Ann. Biol. Clin. 51:83–90 [PubMed] [Google Scholar]
- 15. Harz M, et al. 2009. Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy. J. Biophoton. 2:70–80 [DOI] [PubMed] [Google Scholar]
- 16. Hutsebaut D, et al. 2004. Effect of culture conditions on the achievable taxonomic resolution of Raman spectroscopy disclosed by three Bacillus species. Anal. Chem. 76:6274–6281 [DOI] [PubMed] [Google Scholar]
- 17. Kalasinsky KS, et al. 2007. Raman chemical imaging spectroscopy reagentless detection and identification of pathogens: signature development and evaluation. Anal. Chem. 79:2658–2673 [DOI] [PubMed] [Google Scholar]
- 18. Karatzoglou A, Meyer D, Hornik K. 2006. Support vector machines in R. J. Stat. Software 15:1–28 [Google Scholar]
- 19. López-Goñi I, Moriyón I. 2004. Brucella: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 20. Maquelin K, et al. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255–271 [DOI] [PubMed] [Google Scholar]
- 21. Meisel S, Stöckel S, Elschner M, Rösch P, Popp J. 2011. Assessment of two isolation techniques for bacteria in milk toward their compatibility with Raman spectroscopy. Analyst 136:4997–5005 [DOI] [PubMed] [Google Scholar]
- 22. Moreno E, Cloeckaert A, Moriyon I. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90:209–227 [DOI] [PubMed] [Google Scholar]
- 23. Morhác M. 2009. An algorithm for determination of peak regions and baseline elimination in spectroscopic data. Nucl. Instruments Methods Phys. Res. Sect. A 600:478–487 [Google Scholar]
- 24. Movasaghi Z, Rehman S, Rehman IU. 2007. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 42:493–541 [Google Scholar]
- 25. Naumann D. 2000. Infrared spectroscopy in microbiology, p 102–131 In Meyers RA. (ed), Encyclopedia of analytical chemistry. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 26. Naumann D, Keller S, Helm D, Schultz C, Schrader B. 1995. FT-IR spectroscopy and FT-Raman spectroscopy are powerful analytical tools for the noninvasive characterization of intact microbial cells. J. Mol. Struct. 347:399–405 [Google Scholar]
- 27. Navarro E, Casao MA, Solera J. 2004. Diagnosis of human brucellosis using PCR. Expert Rev. Mol. Diagn. 4:115–123 [DOI] [PubMed] [Google Scholar]
- 28. Nelson WH, Manoharan R, Sperry JF. 1992. UV resonance Raman studies of bacteria. Appl. Spectrosc. Rev. 27:67–124 [Google Scholar]
- 29. Ngo-Thi NA, Kirschner C, Naumann D. 2003. Characterization and identification of microorganisms by FIF-IR micro-spectrometry. J. Mol. Struct. 661:371–380 [Google Scholar]
- 30. Pappas G, Panagopoulou P, Christou L, Akritidis N. 2006. Brucella as a biological weapon. Cell. Mol. Life Sci. 63:2229–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect. Dis. 6:91–99 [DOI] [PubMed] [Google Scholar]
- 32. Pätzold R, et al. 2008. In situ mapping of nitrifiers and anammox bacteria in microbial aggregates by means of confocal resonance Raman microscopy. J. Microbiol. Methods 72:241–248 [DOI] [PubMed] [Google Scholar]
- 33. Rees RK, Graves M, Caton N, Ely JM, Probert WS. 2009. Single tube identification and strain typing of Brucella melitensis by multiplex PCR. J. Microbiol. Methods 78:66–70 [DOI] [PubMed] [Google Scholar]
- 34. R Foundation for Statistical Computing 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 35. Rösch P, et al. 2006. Identification of single eukaryotic cells with micro-Raman spectroscopy. Biopolymers 82:312–316 [DOI] [PubMed] [Google Scholar]
- 36. Rösch P, et al. 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 71:1626–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossen L, Nørskov P, Holmstrøm K, Rasmussen OF. 1992. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 17:37–45 [DOI] [PubMed] [Google Scholar]
- 38. Ryan CG, Clayton E, Griffin WL, Sie SH, Cousens DR. 1988. SNIP, a statistics-sensitive background treatment for the quantitative-analysis of pixel spectra in geoscience applications. Nucl. Instruments Methods Phys. Res. Sect. B 34:396–402 [Google Scholar]
- 39. Salzberg SL. 1997. On comparing classifiers: pitfalls to avoid and a recommended approach. Data Mining Knowledge Disc. 1:317–328 [Google Scholar]
- 40. Schmid U, et al. 2009. Gaussian mixture discriminant analysis for the single-cell differentiation of bacteria using micro-Raman spectroscopy. Chemometrics Intell. Lab. Systems 96:159–171 [Google Scholar]
- 41. Scholz HC, et al. 2008. Specific detection and differentiation of Ochrobactrum anthropi, Ochrobactrum intermedium, and Brucella spp. by a multi-primer PCR that targets the recA gene. J. Med. Microbiol. 57:64–71 [DOI] [PubMed] [Google Scholar]
- 42. Schumacher W, Kühnert M, Rösch P, Popp J. 2011. Identification and classification of organic and inorganic components of particulate matter via Raman spectroscopy and chemometric approaches. J. Raman Spectrosc. 42:383–392 [Google Scholar]
- 43. Schuster KC, Urlaub E, Gapes JR. 2000. Single-cell analysis of bacteria by Raman microscopy: spectral information on the chemical composition of cells and on the heterogeneity in a culture. J. Microbiol. Methods 42:29–38 [DOI] [PubMed] [Google Scholar]
- 44. Smith LD, Ficht TA. 1990. Pathogenesis of Brucella. Crit. Rev. Microbiol. 17:209–230 [DOI] [PubMed] [Google Scholar]
- 45. Stöckel S, et al. 2009. Effect of supplementary manganese on the sporulation of Bacillus endospores analyzed by Raman spectroscopy. J. Raman Spectrosc. 40:1469–1477 [Google Scholar]
- 46. Stöckel S, Meisel S, Elschner M, Rösch P, Popp J. 2012. Raman spectroscopic detection of anthrax endospores in powder samples. Angew. Chem. Int. Ed. Engl. 51:5339–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stöckel S, et al. 2010. Raman spectroscopy-compatible inactivation method for pathogenic endospores. Appl. Environ. Microbiol. 76:2895–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tarcea N, et al. 2007. UV Raman spectroscopy: a technique for biological and mineralogical in situ planetary studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 68:1029–1035 [DOI] [PubMed] [Google Scholar]
- 49. Thirlwall RE, et al. 2008. Improving the specificity of immunodiagnosis for porcine brucellosis. Vet. Res. Commun. 32:209–213 [DOI] [PubMed] [Google Scholar]
- 50. Thoma B, et al. 2009. Identification and antimicrobial susceptibilities of Ochrobactrum spp. Int. J. Med. Microbiol. 299:209–220 [DOI] [PubMed] [Google Scholar]
- 51. Tomaso H, et al. 2007. Einsatzrelevanz der PCR als diagnostisches Verfahren. Wiener Klin. Wochen. 119:26–32 [DOI] [PubMed] [Google Scholar]
- 52. Vapnik V. 1995. The nature of statistical learning theory. Springer-Verlag, Inc, New York, NY [Google Scholar]
- 53. Velasco J, et al. 1998. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 48:759–768 [DOI] [PubMed] [Google Scholar]
- 54. Walter A, et al. 2011. Raman spectroscopic detection of physiology changes in plasmid-bearing Escherichia coli with or without antibiotic treatment. Anal. Bioanal. Chem. 400:2763–2773 [DOI] [PubMed] [Google Scholar]
- 55. Wenning M, Buechl NR, Scherer S. 2010. Species and strain identification of lactic acid bacteria using FTIR spectroscopy and artificial neural networks. J. Biophoton. 3:493–505 [DOI] [PubMed] [Google Scholar]
- 56. Wiedmann M, Weilmeier D, Dineen SS, Ralyea R, Boor KJ. 2000. Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk. Appl. Environ. Microbiol. 66:2085–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Winchell JM, Wolff BJ, Tiller R, Bowen MD, Hoffmaster AR. 2010. Rapid identification and discrimination of Brucella isolates by use of real-time PCR and high-resolution melt analysis. J. Clin. Microbiol. 48:697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yagupsky P, Baron EJ. 2005. Laboratory exposures to brucellae and implications for bioterrorism. Emerg. Infect. Dis. 11:1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Young EJ, Corbel MJ. 1989. Brucellosis: clinical and laboratory aspects. CRC Press, Inc, Boca Raton, FL [Google Scholar]
- 60. Yu WL, Nielsen K. 2010. Review of detection of Brucella spp. by polymerase chain reaction. Croatian Med. J. 51:306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.