Abstract
Soil pH is one of the most influential factors for the composition of bacterial and fungal communities, but the influence of soil pH on the distribution and composition of soil archaeal communities has yet to be systematically addressed. The primary aim of this study was to determine how total archaeal abundance (quantitative PCR [qPCR]-based estimates of 16S rRNA gene copy numbers) is related to soil pH across a pH gradient (pH 4.0 to 8.3). Secondarily, we wanted to assess how archaeal abundance related to bacterial and fungal growth rates across the same pH gradient. We identified two distinct and opposite effects of pH on the archaeal abundance. In the lowest pH range (pH 4.0 to 4.7), the abundance of archaea did not seem to correspond to pH. Above this pH range, there was a sharp, almost 4-fold decrease in archaeal abundance, reaching a minimum at pH 5.1 to 5.2. The low abundance of archaeal 16S rRNA gene copy numbers at this pH range then sharply increased almost 150-fold with pH, resulting in an increase in the ratio between archaeal and bacterial copy numbers from a minimum of 0.002 to more than 0.07 at pH 8. The nonuniform archaeal response to pH could reflect variation in the archaeal community composition along the gradient, with some archaea adapted to acidic conditions and others to neutral to slightly alkaline conditions. This suggestion is reinforced by observations of contrasting outcomes of the (competitive) interactions between archaea, bacteria, and fungi toward the lower and higher ends of the examined pH gradient.
INTRODUCTION
Ever since Archaea was recognized as a separate domain (albeit initially termed a “kingdom”) 35 years ago, it had been believed that the organisms in the domain mainly thrive in extreme environments (48). However, with the advent of DNA-based molecular tools to analyze environmental samples, it grew increasingly evident that archaea are ubiquitous in both terrestrial and aquatic environments (15, 45). For example, archaea have been found to make up 20% to 30% of the total prokaryotes in pelagic marine environments (42), while studies of soil environments have found archaeal abundance in ranges from between 0% and 10% (5, 45) all the way to 38% (25).
The causes for the variable abundance of archaea in terrestrial environments remain largely unknown. One factor that may have contributed to an incomplete assessment of archaeal distribution patterns in the environment is that, ever since it was discovered that archaea may be the dominant group of microorganisms governing NH4+ oxidation (29), soil archaeal research has primarily focused on understanding the biogeography and functioning of the subgroup of ammonium-oxidizing archaea (AOA). In many soils, the archaeal community seems to almost exclusively consist of AOA (5). However, there are also reports contradicting those results, and it now seems clear that the AOA (Crenarchaeota groups 1.1a and 1.1b) rarely represent the most abundant group of archaea in, e.g., acidic soils rich in organic matter (11, 14, 25, 28, 45). Furthermore, an archaeal abundance of up to 38% of the prokaryotic community (25) is unlikely to be sustained by purely autotrophic growth. Accordingly, studies from marine environments suggest that archaea are capable of heterotrophic growth on low-molecular-weight organic acids (35, 43, 44). In fact, there are reports that archaea, rather than bacteria, represent the major group of microorganisms responsible for the turnover of, e.g., amino acids in marine environments (6, 43, 44).
If archaea and bacteria use the same resources, this niche overlap, combined with phenotypic similarities, suggests a high potential for competition between the two groups. During the course of evolution, this might have led to niche differentiation. Accordingly, it has been suggested that archaea are adapted to chronic energy-deficiency stress, while bacteria are adapted to higher substrate concentrations and fast growth and might outcompete archaea when resources are plentiful (24, 47). In support of this proposal, it has also been suggested that archaea are more abundant in the habitats with low occurrence of currently cultivable bacteria (45). Studies on the few isolated strains of AOA also seem to back this theory, as AOA have a much higher affinity for NH4+ compared to most ammonium-oxidizing bacteria (AOB) (23, 27, 30, 46) and have even been found to be inhibited at high substrate concentrations (27, 46). Observations that NH4+ oxidation is dominated by archaea in soil environments under acidic conditions (29), while bacteria dominate when conditions are neutral to alkaline (16), seem to suggest that AOA and AOB have indeed differentiated their niches, adding support to the idea that competition partly determines the environmental distribution of these groups. Further reinforcing this conjecture, there are some indications that archaea and bacteria could potentially compete for ammonium at intermediate substrate concentrations (16, 46).
Currently, there is no evidence to suggest that fungi and archaea compete for the same nutrient and energy sources, but this may well be a result of limited scientific study addressing this particular issue. Karlsson et al. (24) noted that the ratio between archaeal abundance and bacterial abundance generally decreased when the organisms were in direct proximity to mycorrhized roots and mycorrhizal hyphae. However, there are also indications to the contrary. For example, the diversity of organisms belonging to group 1.1c Crenarchaeota, commonly found in acidic forest soils, has been found to be higher in proximity to mycorrhizal hyphae and mycorrhized roots than in proximity to bulk soil (10, 11). In fact, this pattern seems to be a common feature not restricted to group 1.1c Crenarchaeota, as a variety of crenarchaeotes and also euryarchaeotes are found in the mycorrhizosphere and the rhizosphere of a wide variety of plants (11, 41, 45).
The findings described above emphasize a need to improve our understanding of the factors regulating the distribution and functioning of archaea and bacteria in soil environments. Soil pH is one of the most influential factors for the composition of bacterial (4, 18, 38) and fungal (38) communities. While the influence of soil pH on the distribution and composition of soil archaeal communities has yet to be systematically addressed, there are indications that the composition of AOA communities is affected by pH (20). Thus, the primary aim of this study was to determine how total archaeal abundance is related to soil pH. As AOA have been proposed to make up the majority of soil archaea (5) and seem relatively more abundant at low pH (16, 29), we hypothesized that total archaeal abundance is negatively related to soil pH. Secondarily, we wanted to assess how archaeal abundance related to bacterial abundance and bacterial and fungal growth rates across the pH gradient.
MATERIALS AND METHODS
Sample collection and soil characterization.
As previously described (38, 39), the Hoosfield acid strip is the outcome of a one-time application of chalk in the mid-19th century. Since the application was uneven, a pH gradient ranging from 4.0 to 8.3 within 200 m emerged over the years. The field has not been treated with fertilizers, and since the chalk application, winter wheat has been continuously grown. Environmental factors other than pH vary minimally over the strip; for example, the organic carbon (C) content of the soil is nearly constant at 0.9% ± 0.01% and the C:N ratio is stable at about 9 between pH 4.5 and 8.3 (39), while below pH 4.5 the organic C decreases to about 0.8% due to lower plant growth rates at these pHs. The soil is classified as Typic Paleudalf (USDA, 1992), and for a more complete description, see references 1 and 2.
The pH gradient was sampled, taking 5-cm-diameter, 0-to-23-cm-depth cores at each sampling position, in April 2008. Every 10 m was sampled in stretches between 0 and 40 m and between 120 and 180 m, while every 5 m was sampled between 40 and 120 m, due to the higher rate of pH change in that interval (1). The soil samples were sieved (pore size, <2.8 mm) in the laboratory and soil characteristics determined (as reported in reference 39). The soil samples were then stored frozen until molecular analyses were performed in January 2009.
DNA extraction and quantitative PCR analyses.
DNA was extracted from moist field subsamples by the use of a PowerSoil DNA Isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) following the manufacturer's instructions, and the abundances of archaeal and bacterial small-subunit rRNA gene copy numbers were measured by quantitative PCR (qPCR). Archaeal 16S rRNA gene fragments (ca. 140 bp) were amplified using universal primers Arch 967F (5′-AATTGGCGGGGGAGCAC-3′) and Arch-1060R (5′-GGCCATGCACCWCCTCTC-3′) (reference 13 and references therein). The abundance of bacterial 16S rRNA gene copies in the same DNA extracts has been previously reported in reference 38 and was assessed as detailed in that report, using forward primer Eub338 and reverse primer Eub518 (17). The primers were checked for specificity using the Ribosomal Database Project (http://rdp.cme.msu.edu/) and amplicon sequencing in independent soil samples (24), and both primer pairs were found to be domain specific. To estimate archaeal small-subunit rRNA gene abundances, standard curves were generated using a 10-fold serial dilution of Halobacterium salinarum DNA, while bacterial 16S rRNA gene abundance was quantified against a standard curve generated from a plasmid containing a full-length copy of the Escherichia coli 16S rRNA gene, again using a 10-fold serial dilution (as previously reported in reference 38). The 25-μl qPCR mixtures contained 12.5 μl of ABgene SYBR MasterMix (ABgene, Rochester, NY), 0.5 μl each of the 10 μM forward and reverse primers, and 9.5 μl of sterile, DNA-free water. Standard and environmental (ca. 0.5 ng) DNA samples were added at 2.0 μl per reaction. The reaction was carried out using an Eppendorf Mastercycler ep realplex thermocycler (Eppendorf North America, Westbury, NY) and a program of 94°C for 15 min followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. Melting curve and gel electrophoresis analyses were performed to confirm that the amplified products were the appropriate size. Gene copy numbers were determined using a regression equation for each assay and relating the cycle threshold (CT) value to the known numbers of copies in the standards. The efficiencies of the qPCR were 90% to 95% (R2 > 0.991). All qPCRs were run in quadruplicate with the DNA extracted from each soil sample.
Fungal and bacterial growth.
We compared the qPCR data on archaeal and bacterial abundances to estimates of fungal and bacterial growth in the same soil samples (previously published in reference 39). These estimates were obtained using the acetate-into-ergosterol-incorporation method to determine fungal growth and the leucine (Leu)-and-thymidine (TdR)-incorporation technique to determine bacterial growth. Briefly, bacterial growth was estimated using leucine (26) or thymidine (19) with a homogenization/centrifugation step (3) to adapt it for use in soil (37). Radiolabeled leucine ([3H]leucine; Amersham) (37 MBq ml−1, 5.74 TBq mmol−1), combined with nonradioactive leucine for a final concentration of 275 nM or with thymidine ([3H]TdR; Amersham) (37 MBq ml−1, 925 GBq mmol−1) at a final concentration of 130 nM, was added to the bacterial suspension. After a 2-h incubation step at 22°C, growth was terminated with trichloroacetic acid, and samples were washed (3), after which the level of incorporated radioactivity was determined using liquid scintillation. Fungal growth was assessed by measuring acetate incorporation into ergosterol (32, 37) by the use of [1-14C]acetate (sodium salt; Amersham) (7.4 MBq ml−1, 2.04 GBq mmol−1) combined with unlabeled sodium acetate to create a final concentration of 220 μM in a soil slurry and an incubation time of 5 h at 22°C, after which growth was terminated by addition of formalin. Ergosterol was extracted, separated, and quantified using high-performance liquid chromatography (HPLC) with a UV detector (39), and the ergosterol eluent was collected to determine its radioactivity by the use of liquid scintillation.
Calculations and statistics.
All statistical analyses were performed using linear regression (Statistica 9.1; StatSoft, Inc., Tulsa, OK). If necessary, the input values were log-transformed in order to ensure normal distribution of the residuals.
RESULTS AND DISCUSSION
There was consistently a higher abundance of bacterial than archaeal 16S rRNA gene copy numbers across the pH gradient, as shown by a ratio between archaeal and bacterial copy numbers that ranged between 0.002 and 0.07. We identified two distinct and opposite effects of pH on the abundance of archaeal 16S rRNA gene copies across the pH interval covered by the gradient. Between pH 4.0 and 4.7 (referred to here as the “low-pH” range), the abundance of archaeal 16S rRNA gene copies was on average 8.7 × 103 ± 0.92 × 103 copies g−1 of soil (mean ± standard error [SE]) (Fig. 1). Above this pH range, there was an almost 4-fold decrease in archaeal abundance, with a minimum of only 2.5 × 103 to 2.7 × 103 copies g−1 of soil at pH 5.1 to 5.2. The archaeal abundance then sharply increased with pH between pH 5.1 and 8.3 (referred to here as the “high-pH” range) (P < 0.001), from a minimum of 2.5 × 103 copies g−1 to a maximum of 3.7 × 105 at pH 8 (Fig. 1), resulting in an almost 150-fold increase in archaeal copy numbers. Our further analyses were carried out in the light of this finding by analyzing the archaeal response at pH 4.0 to 4.7 separately from that of archaea found at pH 5.1 to 8.3.
Fig 1.
The relationship between soil pH and archaeal abundance. Open circles represent samples with pH 5.1 to 8.3 and filled squares samples with pH 4.0 to 4.7. Only samples in the pH range 5.1 to 8.3 were included in the regression analysis.
The two disparate responses of the archaeal community at low and high pHs are consistent with the recent proposal that some ammonium-oxidizing archaea (AOA) are specialized to the conditions of high-pH environments and others to low pH (20, 33). Even so, the strong positive relationship between soil pH and archaeal abundance at pH 5.1 to 8.3 was surprising. Soil pH is considered one of the major factors regulating the abundance of AOA, and their abundance relative to bacterial ammonium oxidizers is generally higher at low pH (16, 33). Our findings seem to contrast with this. In the low-pH range (pH 4.0 to 4.7), the ratio between archaeal and bacterial copy numbers was on average 0.009 ± 0.0006 (mean ± SE), which decreased to a minimum of 0.002 at pH 5.2. However, above this pH the ratio steadily increased by about 40-fold with higher pH (P < 0.001), reaching a maximum of more than 0.07 at pH 8 (Fig. 2), consistent with other reports of decreasing AOA abundance with decreasing pH (12, 20, 22). A possible explanation for the contradictory results in different reports may be harbored in our results obtained here: the archaeal response to pH is possibly dependent on, and variable with, the span and resolution of the pH range that is examined. We present here one of the most highly resolved and comprehensive ranges of soil pHs so far used within the same study to assess archaeal abundances, and our findings suggest that archaeal abundance responded to pH in two disparate ways along the gradient, probably as a result of some archaea being specialized to the conditions of high-pH environments and others to low pH (20, 33).
Fig 2.
The relationship between soil pH and the ratio between archaeal and bacterial copy numbers [i.e., log10 16S rRNA archaeal gene copy numbers (log10 16S rRNA bacterial gene copy numbers−1)]. The lowest ratio between logarithmically transformed copy numbers (0.55 at pH 5.2) corresponds to a ratio of archeal/bacterial copy numbers of 0.002 and the highest value for the ratio between logarithmically transformed counts (0.85 at pH 8.0) to a ratio of archeal/bacterial copy numbers of 0.07. Open circles represent samples with pH 5.1 to 8.3 and filled squares samples with pH 4.0 to 4.7. Only samples in the pH range of 5.1 to 8.3 were included in the regression analysis. The data on bacterial 16S rRNA gene copy numbers has been previously reported in reference 38.
Another possible reason for the nonuniform response of archaea to soil pH along the gradient is that we quantified the total archaeal community, not constraining our analysis to only the AOA subgroup. A previous study (28) observed a massive decline of the abundance of group 1.1c Crenarchaeota with increased pH (between pH 4.5 and 6.0). Crenarchaeota group 1.1c is also commonly the dominant group of archaea in acidic forest soils (11, 34, 45). It is possible that our observation of decreases in archaeal abundance from pH 4.0 to 4.7 to pH 5.2 (Fig. 1) reflects a reduced abundance of this particular group of archaea, and to our knowledge there is no evidence that these archaea are capable of ammonia oxidation. On the other hand, Crenarchaeota group 1.1c archaea were recently found to have the ability to grow on methanol and methane (9), and observations of archaeal growth in the presence of acetylene (an inhibitor of ammonia oxidation) provides further evidence that archaea in upland soils might not be sustained by NH4+ oxidation alone (22).
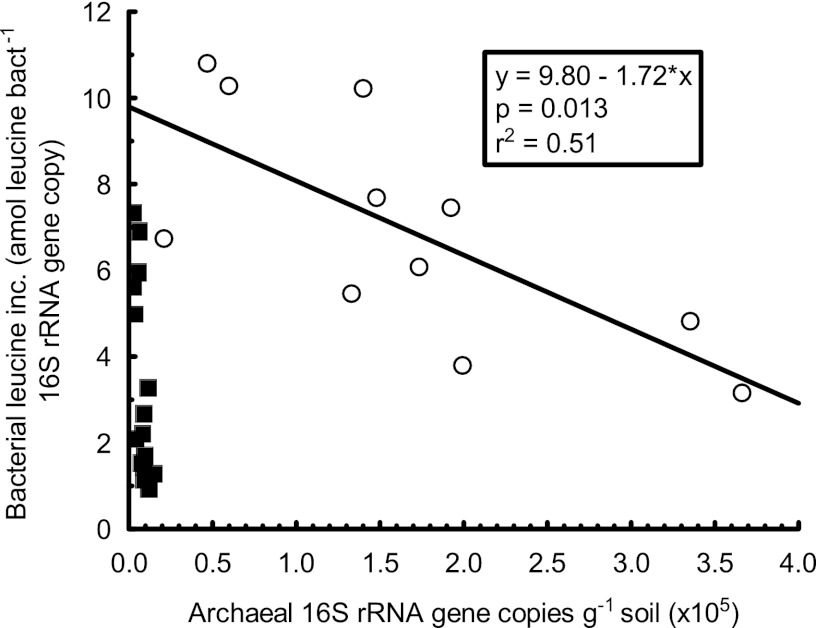
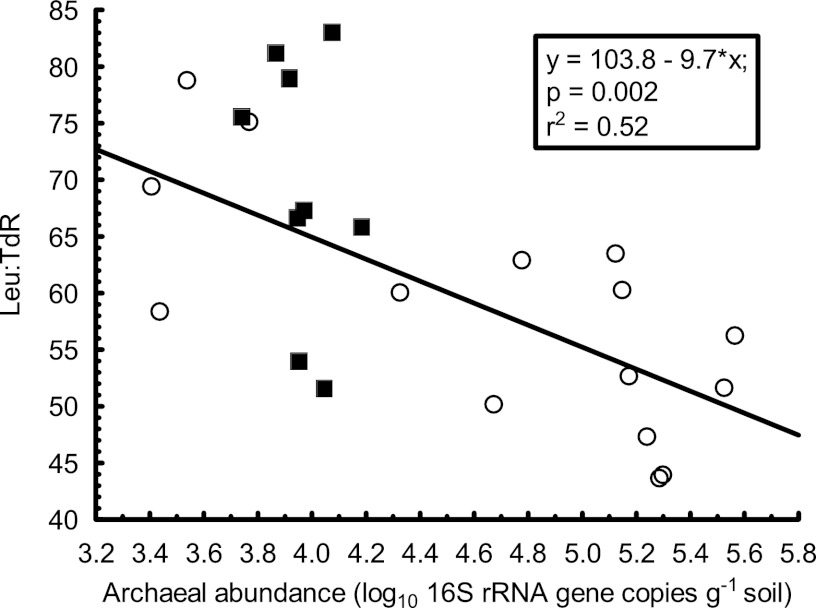
The highest pH values in our study correspond well to the pH of seawater (∼pH 8), where archaea are abundant (42) and are at least partly sustained by heterotrophic growth on organic acids (6, 35, 43). In agreement with these findings, the strong positive relationship between the abundance of archaea and pH in the high-pH range (pH 5.1 to 8.3) in this study (Fig. 1) coincided with a slightly reduced leucine uptake per bacterial cell, as suggested by a weak but nonsignificant (P = 0.09) negative relationship between archaeal abundance and specific bacterial growth (ratio between leucine incorporation and bacterial 16S rRNA gene copy numbers). The same relationship became highly significant (P = 0.013) when only samples with pH between 6.0 and 8.3 were included in the analysis (Fig. 3), i.e., the pH range where the increase in the abundance of archaeal 16S rRNA gene copies was most notable (Fig. 1). The same effect was not observed to the same extent for bacterial thymidine incorporation, as the ratio between bacterial leucine uptake and thymidine uptake decreased slightly when archaea became more abundant (P = 0.002; Fig. 4). Thymidine uptake is generally considered to be specific to bacteria (3, 31), while the capacity for uptake of exogenous leucine is a more ubiquitous trait among microorganisms (7, 36). Thus, it cannot be ruled out that archaeal leucine utilization contributed to the observed decrease in bacterial leucine incorporation versus thymidine incorporation (Fig. 4). However, as our study was not designed to elucidate the potential for archaeal amino acid uptake or heterotrophy, we have no direct evidence for this conjecture. Nevertheless, the observations warrant further investigation of the possibility that archaea contribute to the rapid cycling of organic acids not only in marine environments (6) but also in soils of neutral to alkaline pHs. Reinforcing this argument is evidence that the recently isolated archaeal ammonium oxidizer Nitrososphaera viennensis grows more vigorously when the growth medium is enriched with the organic acid pyruvate (46). On a separate note, the potential archaeal uptake of leucine suggests a path forward to estimate archaeal growth rates in soil (21).
Fig 3.
The relationship between archaeal abundance and “specific” bacterial leucine incorporation (attomoles leucine incorporated per bacterial 16S rRNA gene copy). There was a weak but nonsignificant (P = 0.09) negative relationship at pH 5.1 to 8.3 which became highly significant (P = 0.013) when only samples with pH between 6.0 and 8.3 were included in the analysis. Open circles represent samples with pH 6.0 to 8.3 and filled squares samples with pH 4.0 to 5.6. The bacterial qPCR data and the bacterial growth rate here used to evaluate the archaeal results were previously reported in reference 38 and reference 39, respectively.
Fig 4.
The relationship between archaeal abundance and the ratio of bacterial leucine (Leu) incorporation to thymidine (TdR) incorporation. Open circles represent samples with pH 5.1 to 8.3 and filled squares samples with pH 4.0 to 4.7. Only samples in the pH range of 5.1 to 8.3 were included in the regression analysis. The bacterial growth rates here used to evaluate the archaeal results have been previously reported in a different context in reference 39.
If there is overlap in the use of resources by archaea and bacteria, we might expect to find low archaeal abundance where conditions promote high bacterial growth rates (24, 47). This was not the case. On the contrary, archaea became more abundant at high pH (Fig. 1), where bacterial growth rates were also the highest (39). However, these findings alone do not allow us to exclude the possibility that there are competitive interactions between archaea and bacteria, as the sharp increase in archaeal abundance in the high-pH range might have been even more pronounced in the absence of bacteria.
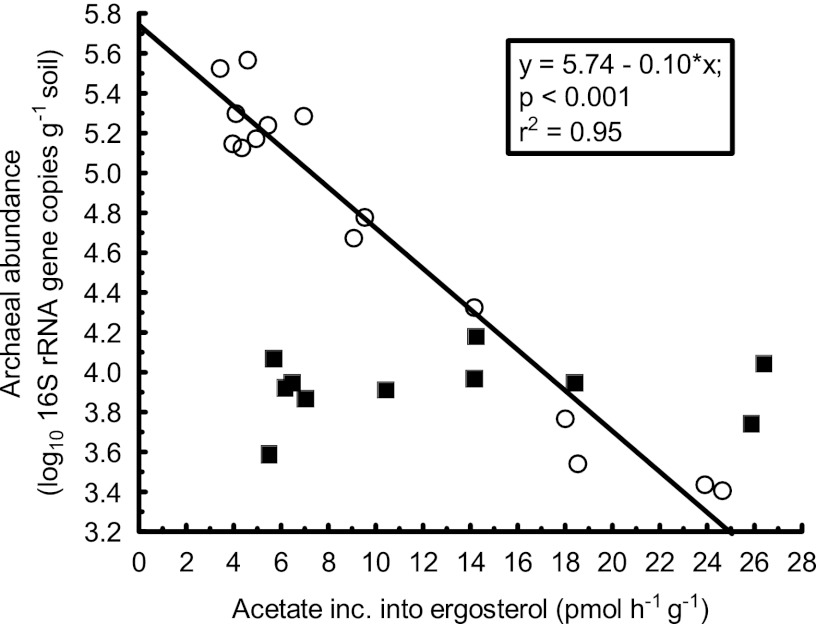
In contrast, there was a strong negative relationship between fungal growth and archaeal abundance (Fig. 5). That is, when conditions favored high fungal growth rates, archaeal abundance decreased. However, the response was obvious only in the high-pH range (pH 5.1 to 8.3; P < 0.001) whereas there was no correlation between fungal growth and archaeal abundance below pH 5.1. The results contradict observations suggesting that mycorrhizal fungi might actually promote growth of archaea (8, 9, 11). However, those observations are derived from experiments using boreal pine forest soils with pH matching the low-pH range in our study (pH 4.0 to 4.7), i.e., the range where archaeal abundance was not negatively affected by fungal growth (Fig. 5). In contrast, another study (24) found a pH-independent decrease in archaeal abundance, as well as in the ratio between archaea and bacteria, in direct proximity to mycorrhizal roots and mycorrhizal hyphae in a forest soil with pH 5.5. Our observation of decreased archaeal abundance at high fungal growth rates is consistent with those findings.
Fig 5.
The relationship between fungal growth and archaeal abundance. Open circles represent samples with pH 5.1 to 8.3 and filled squares samples with pH 4.0 to 4.7. Only samples in the pH range of 5.1 to 8.3 were included in the regression analysis. The fungal growth rates here used to evaluate the archaeal results have been previously reported in a different context (39).
The ratio between archaeal and bacterial 16S rRNA gene copy numbers also decreased considerably with increasing fungal growth rates in the high-pH range (P < 0.001, Fig. 6), suggesting that archaea rather than bacteria are the prokaryotes most strongly affected by interactions with fungi. However, the results should be interpreted with caution, as autocorrelation between the measured biological and environmental factors makes the observed correlative relationships between microbial groups difficult to assign to causal factors. Moreover, that competitive interaction between fungi and bacteria appears to explain their pH dependencies (40) complicates the picture further. It should also be noted that the acetate incorporation assay used to estimate fungal growth rates measures only the growth of saprotrophic fungi and not that of mycorrhizal fungi. It can be speculated that mycorrhizal fungi provide growth factors or an environment that favors archaeal growth, while the same is not true for saprotrophic fungi. Furthermore, the correlative measures of archaeal abundance and fungal and bacterial growth should be interpreted with some caution, since we do not have any information on archaeal growth rates in the same samples. However, lacking a better measure of the archaeal presence, this is a valuable initial assessment.
Fig 6.
The relationship between fungal growth and the ratio between archaeal and bacterial abundance (log10 16S rRNA gene copy numbers g−1 soil). Open circles represent samples with pH 5.1 to 8.3 and filled squares samples with pH 4.0 to 4.7. Only samples in the pH range of 5.1 to 8.3 were included in the regression analysis. The bacterial qPCR data and the fungal growth rates here used to evaluate the archaeal results were previously reported in reference 38 and reference 39, respectively.
In conclusion, our results show that soil pH is an important factor that regulates archaeal abundance in the terrestrial environment. Interestingly, however, the archaeal response to pH was not uniform and our results implicitly suggest that two disparate influences of pH on the archaeal community were present. This might possibly be a reflection of variations in the archaeal community composition along the gradient, with some archaea being adapted to acidic conditions and others to neutral to slightly alkaline conditions. This conclusion is reinforced by observations of contrasting outcomes of the (competitive) interactions between archaea, bacteria, and fungi toward the lower and higher ends of the examined pH gradient. Our assessment of the influence of soil pH on the distribution of archaeal abundance was devised to factor in the whole archaeal domain, and our results contrast with what is growing to be a consensus regarding the relationship between AOA and soil pH. This, combined with recent finding of alternative archaeal nutritional strategies, emphasizes a need to reconsider the ecology and resource use of soil archaea—they may be more diverse than is currently recognized.
ACKNOWLEDGMENTS
We thank Noah Fierer for enabling the archaeal qPCR measurements and Phil C. Brookes for endorsing and facilitating our work on the Hoosfield acid strip at the Biotechnological and Biological Sciences Research Council-funded Rothamsted Research.
P.B. and J.R. were supported by grants from the Swedish Research Council (grants 621-2007-3740, 623-2009-7343, and 621-2011-5719) and the Crafoord foundation (grant number 20100996).
Footnotes
Published ahead of print 15 June 2012
REFERENCES
- 1. Aciego Pietri JC, Brookes PC. 2008. Nitrogen mineralisation along a pH gradient of a silty loam UK soil. Soil Biol. Biochem. 40:797–802 [Google Scholar]
- 2. Aciego Pietri JC, Brookes PC. 2008. Relationships between soil pH and microbial properties in a UK arable soil. Soil Biol. Biochem. 40:1856–1861 [Google Scholar]
- 3. Bååth E. 1990. Thymidine incorporation into soil bacteria. Soil Biol. Biochem. 22:803–810 [Google Scholar]
- 4. Bååth E, Anderson TH. 2003. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35:955–963 [Google Scholar]
- 5. Bates ST, et al. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biddle JF, et al. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U. S. A. 103:3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bloem J, Bolhuis PR. 2006. Thymidine and leucine incorporation to assess bacterial growth rate, p 142–149 In Bloem J, Hopkins DW, Benedetti A. (ed), Microbial methods for assessing soil quality. CABI, Wallingford, United Kingdom [Google Scholar]
- 8. Bomberg M, Jurgens G, Saano A, Sen R, Timonen S. 2003. Nested PCR detection of archaea in defined compartments of pine mycorrhizo spheres developed in boreal forest humus microcosms. FEMS Microbiol. Ecol. 43:163–171 [DOI] [PubMed] [Google Scholar]
- 9. Bomberg M, Montonen L, Timonen S. 2010. Anaerobic Eury- and Crenarchaeota inhabit ectomycorrhizas of boreal forest Scots pine. Eur. J. Soil Biol. 46:356–364 [Google Scholar]
- 10. Bomberg M, Timonen S. 2007. Distribution of cren- and euryarchaeota in scots pine mycorrhizospheres and boreal forest humus. Microb. Ecol. 54:406–416 [DOI] [PubMed] [Google Scholar]
- 11. Bomberg M, Timonen S. 2009. Effect of tree species and mycorrhizal colonization on the archaeal population of boreal forest rhizospheres. Appl. Environ. Microbiol. 75:308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bru D, et al. 2011. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 5:532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cadillo-Quiroz H, et al. 2006. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, U. S. A. Environ. Microbiol. 8:1428–1440 [DOI] [PubMed] [Google Scholar]
- 14. Chu HY, Neufeld JD, Walker VK, Grogan P. 2011. The influence of vegetation type on the dominant soil bacteria, archaea, and fungi in a low Arctic tundra landscape. Soil Sci. Soc. Am. J. 75:1756–1765 [Google Scholar]
- 15. DeLong EE, Pace NR. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470–478 [PubMed] [Google Scholar]
- 16. Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. 2009. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 33:855–869 [DOI] [PubMed] [Google Scholar]
- 17. Fierer N, Jackson JA, Vilgalys R, Jackson RB. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuhrman JA, Azam F. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters—evaluation and field results. Mar. Biol. 66:109–120 [Google Scholar]
- 20. Gubry-Rangin C, et al. 2011. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:21206–21211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ionescu D, et al. 2009. Archaea in the Gulf of Aqaba. FEMS Microbiol. Ecol. 69:425–438 [DOI] [PubMed] [Google Scholar]
- 22. Jia ZJ, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671 [DOI] [PubMed] [Google Scholar]
- 23. Jung MY, et al. 2011. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl. Environ. Microbiol. 77:8635–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karlsson AE, Johansson T, Bengtson P. 2012. Archaeal abundance in relation to root and fungal exudation rates. FEMS Microbiol. Ecol. 80:305–311 [DOI] [PubMed] [Google Scholar]
- 25. Kemnitz D, Kolb S, Conrad R. 2007. High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol. Ecol. 60:442–448 [DOI] [PubMed] [Google Scholar]
- 26. Kirchman D, Knees E, Hodson R. 1985. Leucine incorporation and its potential as a measure of protein-synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Könneke M, et al. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 28. Lehtovirta LE, Prosser JI, Nicol GW. 2009. Soil pH regulates the abundance and diversity of Group 1.1c Crenarchaeota. FEMS Microbiol. Ecol. 70:367–376 [DOI] [PubMed] [Google Scholar]
- 29. Leininger S, et al. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809 [DOI] [PubMed] [Google Scholar]
- 30. Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:U976–U234 [DOI] [PubMed] [Google Scholar]
- 31. Martinez J, Riera M, Lalucat J, Vives-Rego J. 1989. Thymidine incorporation into algal DNA from axenic cultures of Synechococcus, Chlorella and Tetraselmis. Lett. Appl. Microbiol. 8:135–138 [Google Scholar]
- 32. Newell SY, Fallon RD. 1991. Toward a method for measuring instantaneous fungal growth rates in field samples. Ecology 72:1547–1559 [Google Scholar]
- 33. Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978 [DOI] [PubMed] [Google Scholar]
- 34. Oline DK, Schmidt SK, Grant MC. 2006. Biogeography and landscape-scale diversity of the dominant crenarchaeota of soil. Microb. Ecol. 52:480–490 [DOI] [PubMed] [Google Scholar]
- 35. Ouverney CC, Fuhrman JA. 2000. Marine planktonic Archaea take up amino acids. Appl. Environ. Microbiol. 66:4829–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pérez MT, Hörtnagl P, Sommaruga R. 2010. Contrasting ability to take up leucine and thymidine among freshwater bacterial groups: implications for bacterial production measurements. Environ. Microbiol. 12:74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rousk J, Bååth E. 2011. Growth of saprotrophic fungi and bacteria in soil. FEMS Microbiol. Ecol. 78:17–30 [DOI] [PubMed] [Google Scholar]
- 38. Rousk J, et al. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351 [DOI] [PubMed] [Google Scholar]
- 39. Rousk J, Brookes PC, Bååth E. 2009. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 75:1589–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rousk J, Brookes PC, Bååth E. 2010. Investigating the mechanisms for the opposing pH relationships of fungal and bacterial growth in soil. Soil Biol. Biochem. 42:926–934 [Google Scholar]
- 41. Sliwinski MK, Goodman RM. 2004. Comparison of crenarchaeal consortia inhabiting the rhizosphere of diverse terrestrial plants with those in bulk soil in native environments. Appl. Environ. Microbiol. 70:1821–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stoica E, Herndl GJ. 2007. Contribution of Crenarchaeota and Euryarchaeota to the prokaryotic plankton in the coastal northwestern Black Sea. J. Plankton Res. 29:699–706 [Google Scholar]
- 43. Teira E, Hv. Aken Veth C, Herndl GJ. 2006. Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol. Oceanogr. 51:60–69 [Google Scholar]
- 44. Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol. 70:4411–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Timonen S, Bomberg M. 2009. Archaea in dry soil environments. Phytochem. Rev. 8:505–518 [Google Scholar]
- 46. Tourna M, et al. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108:8420–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valentine DL. 2007. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nature Rev. Microbiol. 5:316–323 [DOI] [PubMed] [Google Scholar]
- 48. Woese CR, Fox GE. 1977. Phylogenetic structure of prokaryotic domain—primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 74:5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]