Abstract
The aim of this study was to determine the relationship between the composition and function of gut microbiota. Here, we compared the bacterial compositions and fermentation metabolites of human and chicken gut microbiotas. Results generated by quantitative PCR (qPCR) and 454 pyrosequencing of the 16S rRNA gene V3 region showed the compositions of human and chicken microbiotas to be markedly different, with chicken cecal microbiotas displaying more diversity than human fecal microbiotas. The nutrient requirements of each microbiota growing under batch and chemostat conditions were analyzed. The results showed that chicken cecal microbiotas required simple sugars and peptides to maintain balanced growth in vitro but that human fecal microbiotas preferred polysaccharides and proteins. Chicken microbiotas also produced higher concentrations of volatile fatty acids than did human microbiotas. Our data suggest that the availability of different fermentable substrates in the chicken cecum, which exist due to the unique anatomical structure of the cecum, may provide an environment favorable to the nourishment of microbiotas suited to the production of the higher-energy metabolites required by the bird. Therefore, gut structure, nutrition, immunity, and life-style all contribute to the selection of an exclusive bacterial community that produces types of metabolites beneficial to the host.
INTRODUCTION
In monogastric animals, food passes through the esophagus and stomach to enter the small intestine in a one-way fashion. The upper digestive tract not only harvests the majority of the energy from the diet to meet the energy requirements of the host but also delivers fermentable materials to the lower digestive tract to nourish the bacterial community inside (10). The bacterial communities in the large intestines of hindgut fermenters can be highly complex. For example, over 1,000 bacterial phylotypes inhabit the human gut (4). Approximately 15 to 20 g protein and 40 to 50 g carbohydrates enter the large intestine daily to maintain highly condensed bacterial populations (11, 18). The type and amount of substrates available for fermentation in the colon play a significant role in determining the diversity and metabolic functions of the colonic gut microbiota. Analyses of 16S rRNA gene sequences of the fecal microbiotas from 60 mammalian species have led scientists to recognize both diet and host phylogeny as two of the most important driving forces affecting the diversity of the colonic microbiota (25).
Although both human and chicken are classified as the monogastric animals, the relative intensity of bacterial fermentation in compartments of the gastrointestinal tract varies. For humans, the primary fermentation site in the lower digestive tract is the colon. However, in chickens, bacterial fermentation occurs mainly in the cecum. The development of different digestion strategies in animals has been found to be related to each animal's life-style (2). The manner in which the environment of the chicken cecum selects a unique microbial community beneficial to the host, however, remains unknown. The fermentable substrates in chicken ceca are more water soluble and contain more uric acid than the fermentable substrates available in the human colon, which receives digesta directly from the terminal ileum (23). In this way, it is reasonable to hypothesize that the composition of the microbiota and its derived fermentation profiles in the chicken cecum differ from those of the human colon. However, because the host physiological background has significant influences on the composition of the gut microbiota, the lack of cross-species comparisons of microbiota compositions with a normalized in vitro system hinders an understanding of the fermentation characteristics associated with each gut microbiota.
In vitro gut modeling systems, also known as chemostat systems, are some of the in vitro systems suitable for the study of colonic bacterial interactions and metabolisms (28). With appropriate operation parameters, such systems can simulate the physiological conditions of different regions within the gut, and the major groups of colonic bacteria can be maintained in numbers similar to those observed in vivo (1, 42). This system has been successfully applied to the production of competitive-exclusion products used in chicken and pig farming that prevent the colonization of pathogenic bacteria during early life (35, 39). The media used in the chemostat system to mimic the physiochemical conditions of human colonic microbiota are completely different from those used in chicken and pig chemostats. The former is modified veal infusion broth (VI medium) containing mainly polymerized carbon and protein (9, 29). The latter, Viande Levure (VL) medium, contains mainly hydrolyzed nitrogen and simple sugars (19). From this, it can be seen that medium preferences can reflect the metabolic characteristics of microbial communities of different origins. Therefore, the in vitro gut modeling system is an effective tool for the study of the contribution of non-host-associated factors to the shaping of the vertebrate gut microbiota community. The chemostat system allows direct measurements of fermentation products derived from each microbiota, providing an easy way to compare the functions of gut microbiotas from different species.
In the current study, which is based on a survey of the fermentability of human and chicken gut microbiotas in batch cultures, we compared the fermentation characteristics of microbial populations of chicken and human gut microbiotas using chemostat systems and two growth media, VI and VL media. The main chemical compositions of chicken ileal digesta and feces were determined, and the ability of chicken cecal microbiotas to digest carbohydrates and proteins was compared to that of human colon microbiotas. Based on these results, we conclude that the composition of the gut bacterial community and the corresponding fermentation profiles coevolved with the host life-style to meet the specific physiological requirements of each animal species.
MATERIALS AND METHODS
Origin of samples.
All chickens used in the experiment (commercial strain of the Hubbard strain layer) were obtained from a local chicken farm (Zhejiang Zhenda Broilers Co., Ltd., Hangzhou, China). All birds were free from coccidian infection and had received commercial vaccinations. Throughout the experiment, they were fed antibiotic-free maize and a soybean-based diet, according to the Feeding Standards of Chickens in China (NY-T 33-2004) (43). The chicks were brooded in steel-wire cages and grown under housing conditions described previously by Wang et al. (44). A plastic sheet was placed under the cage to collect the excreta from the birds, and the sheet was changed daily. All the animals used in the current experiment were handled in strict compliance with the current regulations and guidelines concerning the use of laboratory animals in China. Procedures were approved by the Laboratory Animal Care and Usage Committee of the Zhejiang Academy of Agricultural Sciences.
A total of six healthy human volunteers, aged 20 to 30 years old, were recruited for these experiments. All volunteers gave informed, written consent, and the study was approved by the Ethics Committee of the Zhejiang Academy of Agricultural Sciences.
Batch culture fermentations.
To compare the fermentation metabolites of microbiotas derived from chicken cecum samples and human colon samples, batch culture fermentations were first conducted according to procedures described previously by Pieper et al. (34). Briefly, 100-ml serum bottles with rubber stoppers were filled with 40 ml of VL or VI medium. VL medium was comprised of 2.4 g liter−1 beef extract, 5.0 g liter−1 yeast extract, 2.5 g liter−1 glucose, 10.0 g liter−1 tryptose, 0.6 g liter−1 l-cysteine hydrochloride, and 5.0 g liter−1 NaCl (19), and VI medium contained 8.0 g liter−1 starch, 3.0 g liter−1 tryptone, 3.0 g liter−1 peptone, 4.5 g liter−1 yeast extract, 0.4 g liter−1 bile salts no. 3, 0.8 g liter−1 l-cysteine hydrochloride, 0.05 g liter−1 hemin, 4.5 g liter−1 NaCl, 2.5 g liter−1 KCl, 0.45 g liter−1 MgCl2 · 6H2O, 0.2 g liter−1 CaCl2 · 6H2O, 0.4 g liter−1 KH2PO4, 1 ml Tween 80, and 2 ml of a solution of trace elements (9). To test the effect of carbohydrates on volatile fatty acid (VFA) production, 8 g of wheat bran (which had been thoroughly washed twice with boiling water to remove soluble materials) or maltose replaced the starch in VI medium. Because uric acid is a source of nitrogen for the chicken cecal microbiota, 0.7 g liter−1 uric acid was also included (7). All media were adjusted to pH 6.4, and 2 ml of 0.025% (wt/vol) resazurin were added before autoclaving.
Six adult hens (approximately 120 days old; commercial strain of the Hubbard strain) were anesthetized by using ether and were killed by cervical dislocation. The contents of the ceca were weighed, mixed with anaerobic phosphate-buffered saline (PBS) (0.1 M, pH 6.8, and degassed with N2), and homogenized in a stomacher bag in a stomacher (Huifeng, Shanghai, China) to yield 10% (wt/vol) slurries. The fecal samples collected from these six human volunteers were homogenized in stomacher bags with 0.1 M anaerobic PBS to make 10% (wt/vol) slurries. Large food residues were removed by passing the mixture through a 0.4-mm sieve twice. Ten milliliters of human fecal slurry or chicken cecal slurry was inoculated into the serum bottle and then incubated at 37°C for 24 h in an anaerobic chamber (miniMACS Anaerobic Workstation; Don Whitley, United Kingdom). Two milliliters of samples was removed at the end of the experiment for analyses of VFA concentrations.
Single-stage chemostat fermentation.
A parallel chemostat system containing two single-stage chemostat systems (330-ml working volume) was set up as described previously by Yin et al. (48). The pH (6.2) was automatically controlled by using a pH controller (Baoxing, Shanghai, China), and the temperature (37°C) was maintained by using a circulating water bath. The systems were kept anaerobic by continuous sparging with O2-free N2 and operated at dilution rate of 0.04 h−1, as described below.
Three chicken cecal slurries and three human fecal slurries were prepared as described above and inoculated into the parallel chemostat systems containing VL or VI medium. After overnight equilibration, fresh medium was supplied to the system by a peristaltic pump. The system was equilibrated for at least 168 h before the samples (15 ml) were removed from the chemostats.
DNA extraction and PCR-DGGE analysis.
Genomic DNA was extracted from each sample by using a QIAamp DNA stool minikit (Qiagen, Dusseldorf, Germany) according to the manufacturer's instructions. The concentration of extracted DNA was determined by the use of a NanoDrop ND-2000 instrument (Thermo, Waltham, MA), and its integrity and size were checked by using 1.0% agarose gel electrophoresis. For analyses of the microbial communities, the V3 region of the 16S rRNA gene (positions 341 to 534 of the Escherichia coli gene) was analyzed by using PCR-denaturing gradient gel electrophoresis (DGGE), as described previously (20, 26, 32). DGGE was performed by using a DCode universal mutation detection system (Bio-Rad, Hercules, CA) in an 8% (wt/vol) polyacrylamide gel containing a linear 30%-to-60% denaturant gradient with a constant voltage of 200 V at 60°C for 4 h (26). The gels were then visualized by staining with SYBR green I nucleic acid (Sigma, St. Louis, MO) for 45 min and washed twice with deionized water. The similarities of the DGGE profiles were analyzed by using Quantity One software (version 4.6.1; Bio-Rad, Hercules, CA) with a match tolerance of 4%.
Enumeration of the major bacterial populations by quantitative PCR.
The major bacterial groups in human and chicken microbiotas and chemostat samples were assessed by quantitative PCR (qPCR) using a 7500 Fast real-time PCR system (Applied Biosystems, Carlsbad, CA). All primers used are listed in Table S1 in the supplemental material. Each reaction was performed in duplicate with a volume of 20 μl in 96-well optical-grade PCR plates. The reaction mixture comprised 50 pmol each primer, 1× SYBR green Realtime PCR Master Mix (Toyobo, Japan), and 40 ng template DNA. Amplifications were performed with the following temperature profiles: 1 cycle of 95°C for 3 min, 40 cycles of 95°C for 15 s, primer-specific annealing temperature for 25 s, and then 72°C for 30 s. In order to determine the specificity of the PCRs, melt-curve analysis was carried out after amplification by heating the PCR mixtures slowly from 60°C to 95°C. Fluorescence was assessed at 0.5°C intervals with a hold of 10 s at each decrement. A quantitative analysis of unknowns was achieved by using standard curves, which were made from known concentrations of plasmid DNA containing the corresponding amplicon for each set of primers. Copy numbers of 16S rRNA genes per mg of sample were transformed into logarithms, and the major bacterial groups were detected by using the abundance relative to the total bacterial population.
Pyrosequencing and sequencing process.
PCR amplification of the V3 region of the 16S rRNA gene was performed by using 8-bp key-tagged primers, 341F (5′-CCT ACG GGA GGC AGC AG-3′) and 533R (5′-ATT ACC GCG GCT GCT-3′), as described previously (49). PCR mixtures (50 μl) contained 0.2 μM each primer (Integrated DNA Technologies), 1.8 mM MgSO4, 0.2 mM 2′-deoxynucleoside 5′-triphosphate, 1 unit of Platinum Pfx DNA polymerase (Invitrogen), and 10 ng of DNA template. The PCR program consisted of a 3-min initial denaturation step at 94°C and 30 cycles of 94°C for 35 s, 55°C for 45 s, and 72°C for 1 min, followed by a final 3-min extension step at 72°C. For each sample, amplicons of three replicated PCRs were recovered by using a QIAquick gel extraction kit followed by a QIAquick PCR purification kit (Qiagen, Dusseldorf, Germany). Equimolar amplicons were combined and submitted to pyrosequencing by using the Genome Sequencer GS-FLX system (454 Life Sciences, Branford, CT) at the Chinese National Human Genome Center in Shanghai.
Raw sequence reads were trimmed for quality according to an in-house-developed program with the following criteria: (i) a perfect match to at least one end of the barcode and 16S rRNA gene primer, (ii) a length of at least 50 nucleotides (nt), and (iii) no ambiguous bases in the sequence read (8). A detailed analysis is provided in Fig. S1 in the supplemental material. A total of 8,659 of 11,394 sequences were then confirmed to be high-quality sequences with an average length of 183 bp. Library information, including operational taxonomic units (OTUs) (98% identity as the cutoff value), Shannon index, evenness, and Chao estimator of OTUs, was obtained by using Mothur software (version 1.11.0) (38). The taxonomy assignments were performed by RDP Classifier (RDP release 10.25 [http://rdp.cme.msu.edu/classifier/classifier.jsp]) with 50% bootstrap support. Sequences in each OTU were then blasted with the BLASTN algorithm of the NCBI database with all sequences except the those from environmental samples, and the closest relatives were considered the most discriminatory species (see Fig. S1 in the supplemental material). Uncultured bacteria were defined according to criteria established previously by Eckburg et al. (17). Any sequence in each OTU that failed to match any sequences already present in the public database (preferably from named organisms) was designated a novel phylotype, and any novel phylotypes that matched only the uncultured sequences in GenBank were designated uncultured bacteria. The proportion of the uncultured bacteria was calculated as the number of uncultured phylotypes in the total numbers of sequences in each genus assigned by RDP Classifier.
VFA analysis.
Fermentation samples were centrifuged at 13,000 × g for 10 min at 4°C to collect the supernatants. After filtration through a 0.22-μm membrane filter (GLScience, Tokyo, Japan), 0.5-ml samples were acidified with 0.5 ml of 100 mM H2SO4, and 4 μl of tert-butyl acetate was added as the internal standard to give a final concentration of 30 mM (27). After extraction with diethyl ether, a quantitative analysis of acetic acid, propionic acid, butyric acid, valeric acid, and isovaleric acid was performed with a gas chromatograph (6820GE; Agilent, Santa Clara, CA) equipped with a glass column (HP-innovax, 30 m by 0.320 mm). The temperatures of the detector, injection port, and column were 225°C, 200°C, and 200°C, respectively. The concentrations of VFAs were determined by a comparison of sample peak heights with those of authentic standards.
Chemical analysis.
Eighteen female chickens (approximately 60 days old; commercial strain of the Hubbard strain) were used for chemical analysis. A plastic sheet was placed under the cage to collect the excreta from birds, and the fecal samples were collected and rapidly frozen at −20°C for chemical analysis. After the chickens were anesthetized by ether and killed by cervical dislocation, the contents of the terminal ileum (from 5 cm of the upper cecum) were squeezed out and rapidly frozen at −20°C for chemical analysis. All samples were dried and then analyzed for total nitrogen (N), starch, sugar, and nonstarch polysaccharide (NSP) contents.
The total nitrogen content was determined by using a 2300 Kjeltec Auto Distillation instrument (Foss Benelux, Amersfoort, Netherlands). The concentration of crude protein was calculated by using an N-to-protein conversion factor of 6.25. Because chicken excreta include nonprotein nitrogen (NPN) such as that found in urea and ammonia, the true level of protein in the feces was determined by using the Stutzer method (5). The total sugar content was determined by using the phenol sulfuric colorimetric method (15). The determination of the total starch content was performed by the hydrochloric acid hydrolysis method. NSP levels in each sample were determined by gas chromatography (6980N; Agilent, Santa Clara, CA) using a 30-m- by 0.53-mm-internal-diameter (ID) capillary column (Restek Corp., Canada) with alditol acetate derivatives. The chicken cecum digestibility was calculated as the difference between the terminal ileal output (g day−1) and feces output (g day−1) relative to the terminal ileal output (g day−1).
Correlation coefficients and statistical analysis.
For DGGE analysis, Dice coefficients were used to evaluate the similarity between the chemostat products. The DGGE gels were analyzed by using Quantity One software (version 4.6.1; Bio-Rad, Hercules, CA) with a match tolerance of 4%. After matching all bands in each lane, the similarity matrix under the reports menu was exported by the Dice coefficient method by using the unweighted-pair group method using average linkages (UPGMA) clustering algorithm to create dendrograms of DGGE profiles from the stabilization period, taking into account both the band position and band density.
For comparisons of pyrosequencing data, correlation coefficients were analyzed by using SPSS software (version 13.0; SPSS Inc., Chicago, IL) and the Pearson correlation. The correlation coefficients of pyrosequencing data between samples were calculated based on the percentage of each bacterial classification unit at a different classified level.
The VFAs of each fermentation sample were measured in technical triplicates. Means and standard deviations (SD) were calculated. The differences between means were assessed by using an analysis of variance (ANOVA) with a (post hoc) Bonferroni test by using SPSS software. P values of <0.05 were considered statistically significant. To compare the similarity of VFA productions between fermentation samples, correlation coefficients were calculated by using SPSS based on the values of VFAs for different fermentation samples.
Nucleotide sequence accession number.
All sequences from pyrosequencing were submitted to the NCBI SRA database under accession number SRA045744.1.
RESULTS
Production of VFAs by chicken cecal microbiotas and human fecal microbiotas.
To analyze the function of the gut microbiota, we first conducted batch fermentations with microbiota derived from human fecal and chicken cecal contents. VI and VL media, which were previously used for the growth of human colonic microbiota and chicken cecal microbiota, respectively, were chosen as the basic medium formulas. Results showed the overall VFA production from the chicken cecal microbiota to be significantly higher than that of human feces in both VI and VL media. Levels of VFAs were 76.6 mM and 75.7 mM for chicken cecal microbiota and 63.6 mM and 48.4 mM for human fecal microbiota, respectively (Table 1). In addition to the acetate, propionate, and butyrate produced by both human and chicken microbiotas, valerate and isovalerate were also detected in the chicken cecal microbiota under all medium conditions. However, they were detected in the human fecal microbiota under only some medium conditions. Decreased VFA levels were detected when insoluble polysaccharides in the form of wheat bran were substituted for soluble starch in VI medium incubated with chicken cecal contents. In contrast, wheat bran increased the level of VFA production of human fecal microbiotas. In addition, neither growth medium had much effect on VFA production by the chicken or human gut microbiota after 24 h of fermentation when supplemented with uric acid (0.7 g liter−1).
Table 1.
Comparative analysis of VFA concentrations in batch fermentations of chicken cecal microbiota and human fecal microbiota in different growth mediaa
| Specimen | Growth medium | Mean VFA concn (mM) ± SD |
|||||
|---|---|---|---|---|---|---|---|
| Acetate | Propionate | Butyrate | Isovalerate | Valerate | Total | ||
| Chicken | VIb | 39.5 ± 7.1B | 22.0 ± 1.5A | 7.9 ± 1.2B | 2.7 ± 0.2B,C | 4.5 ± 0.4A | 76.6 ± 6.2A |
| VI-wheat bran | 28.4 ± 4.3C | 8.5 ± 1.3C | 3.5 ± 0.2C | 5.1 ± 0.4A | 3.2 ± 0.3A,B | 48.7 ± 1.7B | |
| VI-maltose | 58.0 ± 2.7A | 10.7 ± 0.5C | 6.9 ± 0.3B | 2.0 ± 0.2C,D | 4.1 ± 0.4A | 81.7 ± 1.1A | |
| VL | 36.8 ± 1.4B | 17.5 ± 1.7B | 12.0 ± 0.7A | 3.9 ± 0.4A,B | 5.6 ± 1.7A | 75.7 ± 3.5A | |
| VI + uric acidc | 43.6 ± 9.3A,B | 24.9 ± 0.9A | 9.5 ± 1.5A,B | 0.4 ± 0.4D | 1.1 ± 0.3B | 79.5 ± 6.9A | |
| VL + uric acidc | 40.6 ± 7.5B | 17.8 ± 2.7B | 11.5 ± 1.6A | 4.8 ± 1.4A | 5.2 ± 1.3A | 79.9 ± 6.0A | |
| Human | VI | 35.9 ± 3.0A | 15.0 ± 0.8C | 12.6 ± 0.9B,C | ND | ND | 63.6 ± 2.7B,C |
| VI-wheat bran | 34.1 ± 1.9A | 22.2 ± 1.3B | 10.7 ± 0.7C | 0.6 ± 0.1B | 0.5 ± 0.2C | 68.1 ± 1.2B | |
| VI-maltose | 40.9 ± 1.2A | 29.9 ± 0.7A | 23.5 ± 1.4A | 1.0 ± 0.1A | 3.1 ± 0.4A | 98.4 ± 1.0A | |
| VL | 25.3 ± 2.7B | 11.3 ± 0.9C,D | 11.8 ± 1.5C | ND | ND | 48.4 ± 2.7D | |
| VI + uric acidc | 36.7 ± 0.3A | 14.9 ± 2.1C | 15.1 ± 0.5B | 1.1 ± 0.1A | 2.1 ± 0.1B | 69.9 ± 1.4B | |
| VL + uric acidc | 36.6 ± 1.1A | 10.4 ± 1.1C,D | 9.5 ± 0.7C | ND | ND | 56.5 ± 1.1C,D | |
Data are the means of data from six individual human fecal or chicken cecal samples ± SD (n = 6). Each sample was measured in triplicate. Mean values with unlike superscript roman letters are significantly different (P < 0.05), as determined by an ANOVA with a (post hoc) Bonferroni test. ND, none detected.
VI medium contained 8 g liter−1 starch, VI-wheat bran medium contained 8 g liter−1 wheat bran instead of starch, and VI-maltose contained 8 g liter−1 maltose instead of starch.
Uric acid (0.7 g liter−1) was added to VI and VL media.
Chemostat system as a model for analysis of gut microbiota.
Based on the results generated from batch fermentations, we propose that the microbiota from chicken cecum may generate different fermentation patterns than the human fecal microbiota. To better simulate the growth dynamics of the gut microbiota, we used chemostat systems to investigate the fermentation characteristics possessed by the two microbiota communities. We first evaluated the stability and reproducibility of the chemostat system by performing two experiments with the same operation parameters with two fecal inocula taken from the same donor 10 months apart (inocula HU08 and HU09). A parallel system containing two growth media was constructed. VL and VI media were inoculated into the fecal slurry. The structures of the bacterial communities were checked daily by using PCR-DGGE profiles. The PCR-DGGE profiles fluctuated significantly in the first 96 h and became stable at 120 h (data not shown), suggesting that the chemostat had reached the steady state. Samples were then taken at 168 h, and the microbiota community was assessed by using PCR-DGGE profiles, VFA production, and 454 pyrosequencing. As shown in Fig. S2 in the supplemental material, the PCR-DGGE profile patterns of the two inocula demonstrated a high level of similarity. Correlation coefficient indexes of VI and VL chemostats between the two experiments were 0.977 and 0.867, respectively, based on a phylogenetic analysis of 454 pyrosequencing data at the genus level; 96.2 and 92.1, respectively, based on PCR-DGGE profile analysis; and 0.990 and 0.983, respectively, based on VFA production levels (Table 2). The distributions of bacterial genera from inocula HU08 and HU09 as well as the corresponding chemostat samples are presented in Fig. S3 in the supplemental material.
Table 2.
Evaluation of the reproducibility of duplicated chemostat products at the steady state based on PCR-DGGE profiles, pyrosequencing data, and VFA concentrations
| Samplea | Correlation coefficient |
||||
|---|---|---|---|---|---|
| PCR-DGGE profileb | 454 Pyrosequencingc |
Volatile fatty acidc | |||
| Phylum level | Family level | Genus level | |||
| HF08VI/HF09VI | 96.2 | 0.995 | 0.991. | 0.977 | 0.990 |
| HF08VL/HF09VL | 92.1 | 0.939 | 0.896 | 0.867 | 0.983 |
HF08VI/HF09VI represents duplicate experiments conducted in chemostats containing VI medium with the same operation parameters with two fecal inocula collected from the same donor in 2008 and 2009, respectively. HF08VL/HF09VL represents the same experiment conducted in chemostats containing VL medium.
Correlation coefficients of PCR-DGGE profiles were calculated with Quantity One software based on the similarity matrix of matched bands.
For pyrosequencing and VFA analyses, correlation coefficients were calculated with SPSS 13.0 software based on the percentage of bacterial classification units at different phylogenetic levels and the concentrations of VFAs in fermentation samples.
Because the numbers of pyrosequencing reads may influence the diversity and coverage of the microbiota community, we compared the distributions and compositions of the microbiota community of inoculum HU08 produced by 129,934 reads to those produced by 1,751 reads. There were a total of 116 genera generated from the 129,934 reads. Among them, 27 genera had an abundance higher than 0.05%. From the 1,751 reads, there were 34 genera detected with a relative abundance higher than 0.05%. After we combined the low-abundance genera from 129,934 reads (designated “others”), a similar distribution of bacterial genera was observed (see Fig. S4 in the supplemental material). Previous studies indicated that at least 1,000 reads are required to describe the structure of a microbial community (31, 46). For this reason, approximately 1,000 reads were used in the following analysis of the fermentation samples.
Effects of the composition of growth media on the structure of the bacterial community and metabolic profiles.
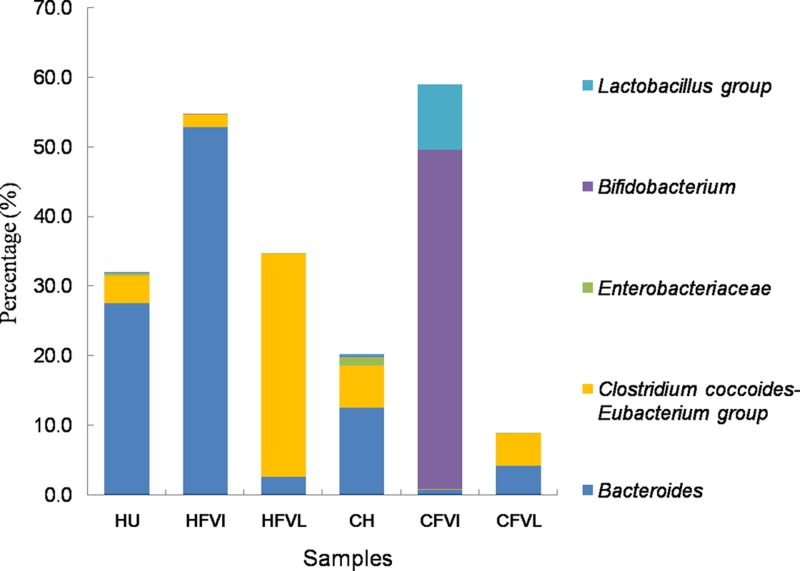
The effects of VI and VL growth media on the structures of the microbial communities in the chemostat systems were evaluated by using gut microbial communities derived from three individual human fecal samples and three chicken cecal samples. The populations of major bacterial groups in VI and VL chemostats were measured by using qPCR and are presented as the proportion of total bacteria in Fig. 1. All tested samples showed statistically similar total bacterial cell densities (Table 3). However, the medium composition significantly affected the proportions of major bacterial groups. VI medium increased the number of Bacteroides bacteria in the human fecal chemostat, and the same medium enhanced the growth of Bifidobacterium bacteria in the chicken cecal chemostat. VFA productions from VI and VL media in chemostats were measured and are expressed as specific products of VFA per unit of microbial biomass (Table 3). Consistent with the results described above, the overall VFA production level in the chemostat system inoculated with chicken cecal microbiotas was higher in both VI and VL media than that of those inoculated with human colonic microbiotas. Under the same conditions, the total amounts of VFAs produced by chicken cecal microbiotas in VL and VI media were 1.6 mM and 1.7 mM per 1010 cells, respectively, but were only 1.1 mM and 1.1 mM per 1010 cells, respectively, in the chemostats inoculated with human fecal microbiotas.
Fig 1.
Relative abundances of major bacterial groups in human and chicken microbiotas and their corresponding chemostat samples. Data are presented as the percentages of the total bacterial population made up by each bacterial group, as assessed by qPCR. The results are given as the means of measurements from three individual human (HU) and three chicken (CH) gut microbiotas and their chemostat products in VI (HFVI and CFVI) and VL (HFVL and CFVL) media.
Table 3.
Concentrations of VFAs and the ability for production of VFAs per 1010 bacterial cells in chemostat fermentation samples of chicken and human microbiotas with VI and VL media
| Sampleb | Mean concn (mM) ± SDa |
Total bacterial population (16 rRNA gene copies g−1)c | VFA concn (mM per 1010 cells) | |||||
|---|---|---|---|---|---|---|---|---|
| Acetate | Propionate | Butyrate | Isovalerate | Valerate | Total | |||
| CHVI | 51.7 ± 1.3A | 21.7 ± 2.6A | 18.3 ± 2.7B | ND | ND | 91.6 ± 2.4B | 5.8 × 1011 | 1.6 |
| CHVL | 46.5 ± 0.8A | 11.5 ± 2.4B | 24.4 ± 2.1A | 16.8 ± 1.6 | 15.2 ± 2.5 | 114.2 ± 2.6A | 6.6 × 1011 | 1.7 |
| HUVI | 34.1 ± 0.8B | 25.8 ± 3.9A | 13.9 ± 1.5B | ND | ND | 73.9 ± 2.2B,C | 7.0 × 1011 | 1.1 |
| HUVL | 23.9 ± 1.67B | 21.8 ± 3.5A | 17.3 ± 1.5B | ND | ND | 62.9 ± 2.3C | 5.9 × 1011 | 1.1 |
Data are shown as means ± SD (n = 3). Mean values with unlike superscript roman letters are significantly different (P < 0.05). ND, none detected.
Samples were obtained from hemostats containing VI or VI medium inoculated with chicken (CH) cecal microbiota or human (HU) fecal microbiota.
Detected by qPCR.
Analysis of bacterial community structures in chemostats inoculated with human fecal microbiota and chicken cecal microbiota.
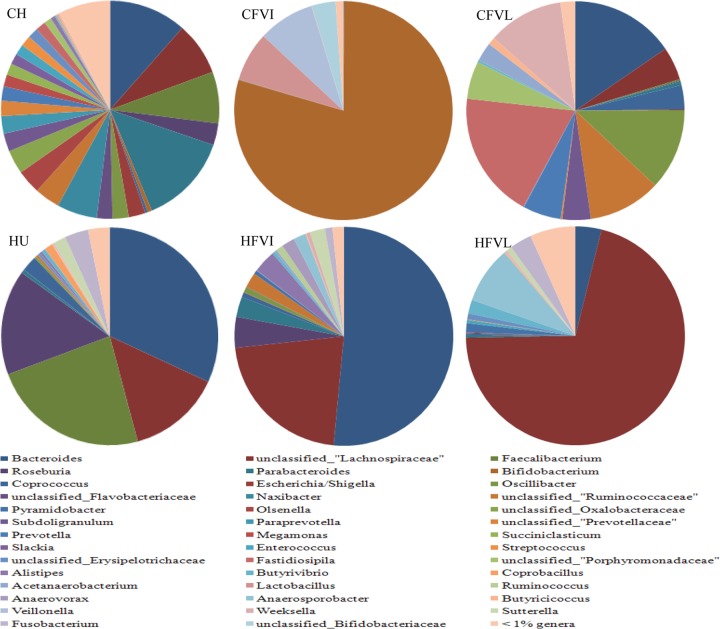
The bacterial communities of the human fecal inoculum (no. 3) and chicken cecal inoculum (no. 3) at the equilibrium stage in chemostats were further analyzed by pyrosequencing. A total of 8,659 reads were generated, and the diversity of each microbiota community is presented in Table S2 in the supplemental material as OTU numbers (98% cutoff), ACE (abundance-based coverage estimator), Shannon index, evenness, and Chao estimator of OTUs. In general, chicken cecum showed a higher level of bacterial diversity than did human feces. However, the OTU numbers in both VI and VL media with the human fecal inoculum were higher than those with chicken cecal contents, suggesting that the operation parameters applied in the current study, such as growth medium composition, dilution rate, temperature, and pH, closely resembled the conditions of the human colon. The microbiota communities of the initial inocula and the stabilized chemostat samples were compared to the relative abundance at the genus level using RDP Classifier (Fig. 2), and 25 genera with a relative abundance of over 1% were detected in the chicken cecal microbiota, but only 8 genera were found in the human fecal microbiota. Relative numbers of genera were more evenly represented in chickens than in humans. Bacteroides, Roseburia, Faecalibacterium, Parabacteroides, and unclassified Lachnospiraceae were the major organisms found in both human and chicken samples. The genus Bacteroides was the most predominant genus in both species. As shown in Table S3 in the supplemental material, the Bacteroides organism in the human fecal sample was classified as Bacteroides vulgatus, but in chicken cecal samples, it included five phylotypes, B. plebeius, B. uniformis, B. clarus, B. faecis, and B. barnesiae. Similar observations were also made for Roseburia, Parabacteroides, and unclassified Lachnospiraceae.
Fig 2.
Compositional comparison of human and chicken gut microbiotas grown in chemostats with two media. The percentage of sequences in each of the bacterial genera is shown according to pyrosequencing data assessed by RDP Classifier. The chicken microbiota (CH) and chemostat products in VI (CFVI) and VL (CFVL) media were compared to human microbiotas (HU) and their chemostat products in VI (HFVI) and VL (HFVL) media.
The enrichment effect of the composition of the growth medium on the microbial communities in the chemostat was obvious at the genus level (Fig. 2). In general, the similarity of microbial communities produced from VI medium and the human inoculum was higher than that of the microbial communities produced from VL medium. Conversely, in chemostats inoculated with chicken cecal microbiotas, the microbial community in VL medium resembled the inoculum relatively closely. VI medium inoculated with chicken cecal contents maintained only four genera, of which Bifidobacterium was the most predominant, taking up to 79.37% of the total. The remaining organisms were Lactobacillus (7.30%), Veillonella (8.34%), and unclassified Bifidobacteriaceae (3.53%). Of the 25 genera in the original chicken cecal microbiota inoculum, 19 were detected in VL medium. Among them, the populations of Weeksella, Oscillibacter, Fastidiosipila, and unclassified Ruminococcaceae increased, while the populations of Parabacteroides, Faecalibacterium, and Naxibacter decreased. The effects of the growth medium on changes in the structures of the microbial community were also found in chemostat systems inoculated with human feces. For example, numbers of Anaerosporobacter and classified Lachnospiraceae bacteria increased from negligible and 13.94%, respectively, in human feces to 8.44% and 70.76%, respectively, in VL medium. The populations of Faecalibacterium and Roseburia decreased from 23.42% and 15.59%, respectively, to undetectable and 4.46%, respectively, although the bacterial community structure in VI medium inoculated with human fecal samples resembled that of the original inoculum, and a remarkable decline in numbers of Faecalibacterium bacteria was detected in the chemostat. All bacterial genera that were abundant in the inocula and corresponding chemostat samples are listed in Table S4 in the supplemental material.
Estimation of levels of uncultured bacteria in the chemostat systems.
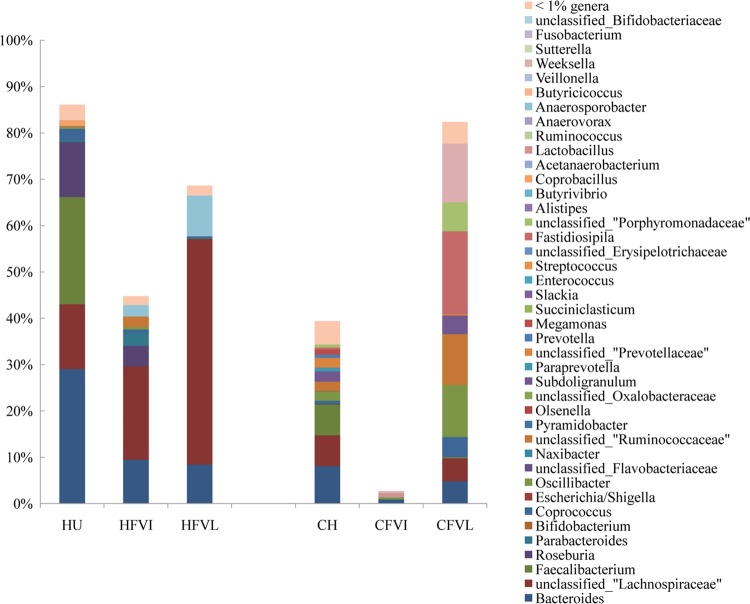
Uncultured bacteria are defined as sequences that do not match any named organisms in any public databases but that match only uncultured sequences in GenBank neighbors (17). In the present study, if the first 100 sequences to appear after the blasting of an unknown sequence in the NCBI database were all uncultured sequences, that unknown sequence was defined as an uncultured phylotype. According to this criterion, 86.12% of phylotypes in human fecal samples belonged to uncultured bacteria, which is consistent with data from a previous study (17). In contrast, only 39.41% of phylotypes in chicken cecal samples belonged to uncultured bacteria (Fig. 3). The composition of the medium also significantly influenced the percentage of uncultured bacteria in chemostats. VL medium grown with human fecal microbiota changed the proportion of unclassified Lachnospiraceae markedly, from 13.88% in inocula to 48.74%, indicating that the medium may favor the Lachnospiraceae family of human gut microbiota. The proportion of a single phylotype of unclassified Lachnospiraceae increased from 0.01% in the human inoculum to 39.40% in the VL chemostat (see Appendix A1 in the supplemental material). For chicken cecal microbiota, 82.41% of the population in VL medium was made up of uncultured bacteria. In contrast, the total percentage of uncultured bacteria was only 2.70% in VI medium. The major communities were made up of lactobacilli and bifidobacteria.
Fig 3.
Compositions of uncultured bacteria in human and chicken gut microbiotas grown in chemostats with two media. The uncultured bacteria were defined according to criteria set up previously by Eckburg et al. (17). The percentage of uncultured bacteria in each of the bacterial genera was calculated according to the pyrosequencing data assessed by RDP Classifier. The chicken microbiota (CH) and chemostat products in VI (CFVI) and VL (CFVL) media were compared to the human microbiota (HU) and chemostat products in VI (HFVI) and VL (HFVL) media.
Digestibility of crude protein, starch, sugars, and nonstarch polysaccharides in the lower digestive tracts of chickens.
The data generated from the in vitro modeling systems suggested that the microbiota communities derived from human feces and chicken cecum required different nutrients to maintain balanced growth. We assumed that this difference in nutritional preferences could be ascribed to the communities' unique types of metabolic activity, which were determined by the availability of substrates in the lower gastrointestinal tract. We then collected the terminal ileal contents and feces from 18 60-day-old chickens to analyze the levels of crude protein, total sugars, starch, and nonstarch polysaccharides (NSPs). Due to the unique anatomical structure of bird cecum, in which urine flows in a retrograde fashion from the duodenum to the ceca, the true protein level, excluding nonprotein nitrogen (NPN), of chicken excreta was also analyzed. Data from a previous study indicated a difference of approximately 2.5 g dry matter between intact and cecectomized adult chickens (36). We predicted that the daily ileal output in dry matter for adult chickens would be the daily total dry matter concentration in the excreta plus 2.5 g. Based on the predicted dry matter concentration, concentrations of crude protein, starch, total sugars, and NSP were calculated and expressed as grams per chicken per day. As shown in Table S5 in the supplemental material, 2.51 g day−1 of crude protein, 1.37 g day−1 of starch, 0.17 g day−1 of NSP, and 4.02 g day−1 of sugars were detected in the contents of terminal ilea. The postileal digestibility was then calculated as the difference between the terminal ileal output (g day−1) and feces output (g day−1) relative to the terminal ileal output (g day−1). Hence, the digestibility of crude protein was 23.51%. The digestibilities of starch, NSP, and sugars were 35.77%, 47.06%, and 88.06%, respectively.
DISCUSSION
The correlation between the gut physiologies and dietary compositions within the structure of the microbiota community and of the fermentation properties has generated perennial interest (25, 33). For mammals, food quality and abundance are the fundamental drivers of the evolution of the gut anatomy and its bacterial residents (24, 47). Because both humans and chickens are omnivores, and the diversity of the microbial communities in the gastroenterological tracts of different animal species is related to the host life-style, we initially proposed that the different nutritional requirements for maintaining the homeostasis of the microbiota communities in the chemostats would be ascribable to the different fermentable substrates available in the terminal ilea. However, due to the difficulty in obtaining human ileostomates, which would have permitted direct measurements of nutrient levels, we compared the concentrations of nutrients in the terminal ilea of chickens and humans by using data reported previously by Coles et al. (10). The study by Coles et al. evaluated four types of experimental diets, but we chose to use the data from the high-fiber-diet group because the nutrient composition of this diet is similar to the Chinese daily dietary formula. As shown in Table S5 in the supplemental material, a relatively high level of starch residues and a low level of NSP were detected in the chicken terminal ileum samples. In addition, there was no obvious difference in the proportions of crude protein between human terminal ileal samples and chicken terminal ileal samples. However, the overall amounts of carbohydrates outweighed total protein amounts in the terminal ilea of chickens (see Table S5 in the supplemental material), suggesting that there is no remarkable difference between chickens and humans with respect to the types and amounts of substrate excreted from the small intestine.
The fermentable substrates entering the lower digestive tract are of both dietary (exogenous) and endogenous origins. Endogenous substrates include pancreatic enzymes, mucins, and other secretions from epithelial cells, and they are species specific. Food residues are the major sources of carbon and nitrogen for colonic bacterial fermentation, but glycoproteins and mucins with host-specific structures also have an impact on the compositions and metabolic patterns of colonic microbiotas (12). On the other hand, anatomical differences in the lower digestive tracts of chickens and humans may affect the availability of fermentable substrates. Unlike the human colon, the cecum has villi at its entrance that act as a mesh, allowing only fluid and fine particles smaller than 200 nm in diameter to enter (16). The substrates available in the chicken cecum consist mainly of water-soluble carbohydrates and peptides and extra uric acid refluxed from the cloaca to the cecum. The morphology of the chicken gut microbiota, assessed by using scanning electron microscopy (SEM), confirmed that large particles of food residue existed in the digesta of the terminal ileum and feces but not in the ceca (data not shown). In contrast, water-insoluble plant materials constituted approximately 17% of the total human digesta in the colon (27). Therefore, we propose that the differences in anatomical structures between the chicken and human lower intestinal tracts may affect the types and amounts of fermentable substrates and that these differences cause the different compositions of colonized microbiotas and eventually the different nutritional requirements for maintaining the homeostasis of the microbiota community in vitro. In the present study, chemostat systems showed that chicken cecal microbiotas require media containing simple carbohydrates and hydrolyzed peptides but that human fecal microbiotas prefer polysaccharides and proteins.
VFAs are the major end metabolites for gut bacterial fermentation. The amount of acetate produced in vitro by the human gut microbiota in current experiments seems to be relatively low. After comparing our VFA data to those reported previously for other chemostat modeling studies, we noticed that the origin of the fecal inocula may influence the molar ratio of VFA production (see Table S6 in the supplemental material) (30). The small amount of acetate in current experiments could be ascribable to the typical Asian food eaten by the volunteers. Fermentation by chicken cecal microbiotas in tested media in vitro generates a significantly higher yield of VFA than that of human microbiotas, suggesting that the efficient production of VFA is one of the metabolic characteristics associated with the chicken cecal microbiota community. This is consistent with the in vivo situation. Cummings et al. previously estimated the total concentration of short-chain fatty acids in the human cecum to be around 131 ± 9 mM (13). However, 179.5 mM VFA was detected in cecal samples of 21-day-old chickens (6). The highly efficient production of VFAs is particularly important for the maintenance of avian health because it not only satisfies the intense energy demands of birds but also participates in the recycling of inorganic ions via an enhancement of the ion transport processes within the ceca (37). The rapid utilization of uric acid by chicken cecal microbiota was reported previously by Braun and Campbell (7). However, in the current study, fermentation supplemented with 0.7 g liter−1 uric acid in both VI and VL media resulted in a slight increase in the level of VFA production. Consistent with SEM observations, chicken cecal microbiotas were found to produce less VFA when wheat bran was the sole carbon source rather than soluble starch or maltose. Considering birds' special energy requirements, the ability to quickly discharge large, slowly fermenting, high-fiber particles through the excreta and recruit more easily fermentable substrates to efficient cecal microbiotas, which would then produce higher levels of VFA, would be extremely advantageous.
Our results revealed that human fecal microbiotas share many major phylogenetic groups with chicken cecal microbiotas at the genus level. However, the composition of chicken cecal microbiotas is more complex than that of human fecal microbiotas, and there are significant differences at the species level. For example, Bacteroides is the predominant genus in both human feces and chicken cecum, but at the species level, the phylotype detected in human feces was B. vulgatus, and those obtained from chicken ceca were classified as B. plebeius and B. clarus. B. vulgatus is the most common type of bacteria in the human large intestine. It is well known for its ability to produce a wide range of polysaccharide hydrolases (14, 41). In contrast, B. plebeius and B. clarus were more recently isolated and identified in colonic samples, and little metabolic information was available (22, 45). Among the five common bacterial genera shared by chicken cecum and human feces (see Table S3 in the supplemental material), Faecalibacterium prausnitzii was the only common species identified. The other species showed exclusive associations with host specificity. Ochman et al. reported recently that among great ape species, common components of the fecal microbiotas could be found only at lower taxonomic levels (33). The significant difference at the species level could be attributed to the metabolic characteristics associated with each species.
The current study demonstrates that the chemostat system is a reliable in vitro gut modeling system with good reproducibility. However, because the transit times of the gastrointestinal tracts of humans and chickens differ significantly, the converted retention time, 24 h, used in the current experiments more closely resembled the human gut transit time than the chicken gut transit time. For example, the overall gastrointestinal transit time for humans is 36.2 ± 5.1 h, but it is only 3.5 h for chickens (21, 40). The dilution rate (0.04 h−1, converted to a retention time of 24 h) used in the current experiment is more close to the human gut transit time than that of chickens. In addition, because we used the same circulating water bath to control the temperature of both chemostats, the temperature set for the chemostat inoculated with chicken ceca was below the natural chicken body temperature. Altogether, the operation parameters applied in the current study, such as the composition of the growth medium, dilution rate, temperature, and pH, all more closely resembled the conditions of the human colon, which may have contributed to the overall similarity of the structures of the microbiotas in the chemostat inoculated with the human sample to those in the chemostat inoculated with the chicken cecal microbiota. Further study is needed to improve the degree of microbiota community simulation in chicken chemostats by using the operational parameters that more closely resemble the physiological conditions of the chicken cecum.
One previous study demonstrated that 80% of the phylotypes in human fecal samples belonged to uncultured bacteria, which is consistent with current data (17). Based on the same criteria, only 39.41% of the phylotypes for chicken cecal samples were classified as uncultured bacteria. Apajalahti et al. reported previously that 90% of the bacteria in the chicken gastrointestinal tract are of unknown species and that 55% are of unknown genera (3). The discrepancy in terms of the proportions of the uncultured bacteria between the current experiments and the previous data could be due to the different criteria used for classification: Apajalahti and colleagues identified only unknown species, whereas we also counted uncultured bacteria from unknown species. It is always desirable to enrich human uncultured colonic bacteria with low abundance in vitro. The in vitro model system demonstrates the advantage of the enrichment of bacterial species with a relatively low abundance. By using unbalanced medium, for example, VL medium, in the current study, the prevalence of a single phylotype of unclassified Lachnospiraceae from the Firmicutes increased from 0.01% in the human inoculum to 39.40% in the chemostat (see Appendix A1 in the supplemental material). Therefore, our results demonstrate that it is possible to use a chemostat system to increase the number of individuals from rare species using medium selection and then study the metabolic activity.
The goal of the current study was to identify differences in the compositions of the microbiotas of the lower digestive tracts of chickens and humans and to use these data to better understand the functional differences. Although the current study has certain limitations, a comparative analysis of the nutrient requirements and fermentation metabolites of human and chicken gut microbiotas allowed us to find evidence to show that the composition of the bacterial community colonizing the host colon could be determined by using the nutritional substrates available in the host colon. These nutritional substrates allow the production of a distinct fermentation profile beneficial to the respective animal species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Nature Science Foundation of China (grant 30970108), the Zhejiang Nature Science Foundation (grant Y3080102), and the National Basic Research Program of China (973) (grant 2009CB522605). The cooperation with Hongwei David Yu is supported by the International S&T cooperation project of China (2008DFA32080).
We also thank Junying Li at Zhejiang University, who provided technical help with the SEM analysis.
Footnotes
Published ahead of print 8 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allison C, McFarlan C, MacFarlane GT. 1989. Studies on mixed populations of human intestinal bacteria grown in single-stage and multistage continuous culture systems. Appl. Environ. Microbiol. 55:672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Apajalahti J. 2005. Comparative gut microflora, metabolic challenges, and potential opportunities. J. Appl. Poult. Res. 14:444–453 [Google Scholar]
- 3. Apajalahti J, Kettunen A, Graham H. 2004. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. Worlds Poult. Sci. J. 60:223–232 [Google Scholar]
- 4. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920 [DOI] [PubMed] [Google Scholar]
- 5. Bhattacharya AN, Sleiman FT. 1971. Beet pulp as a grain replacement for dairy cows and sheep. J. Dairy Sci. 54:89–94 [Google Scholar]
- 6. Biggs P, Parsons CM. 2009. The effects of whole grains on nutrient digestibilities, growth performance, and cecal short-chain fatty acid concentrations in young chicks fed ground corn-soybean meal diets. Poult. Sci. 88:1893–1905 [DOI] [PubMed] [Google Scholar]
- 7. Braun EJ, Campbell CE. 1989. Uric acid decomposition in the lower gastrointestinal tract. J. Exp. Zool. Suppl. 3:70–74 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y, et al. 2011. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54:562–572 [DOI] [PubMed] [Google Scholar]
- 9. Child MW, et al. 2006. Studies on the effect of system retention time on bacterial populations colonizing a three-stage continuous culture model of the human large gut using FISH techniques. FEMS Microbiol. Ecol. 55:299–310 [DOI] [PubMed] [Google Scholar]
- 10. Coles LT, Moughan PJ, Awati A, Darragh AJ, Zou ML. 2010. Predicted apparent digestion of energy-yielding nutrients differs between the upper and lower digestive tracts in rats and humans. J. Nutr. 140:469–476 [DOI] [PubMed] [Google Scholar]
- 11. Cummings JH. 1997. Carbohydrate and protein digestion: the substrates available for fermentation, p 16 In Cummings JH. (ed), The large intestine in nutrition and disease. Institut Danone, Brussels, Belgium [Google Scholar]
- 12. Cummings JH, Englyst HN. 1987. Fermentation in the human large-intestine and the available substrates. Am. J. Clin. Nutr. 45:1243–1255 [DOI] [PubMed] [Google Scholar]
- 13. Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Degnan BA, Macfarlane S, Macfarlane GT. 1997. Utilization of starch and synthesis of a combined amylase/alpha-glucosidase by the human colonic anaerobe Bacteroides ovatus. J. Appl. Microbiol. 83:359–366 [DOI] [PubMed] [Google Scholar]
- 15. Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. 1951. A colorimetric method for the determination of sugars. Nature 168:167. [DOI] [PubMed] [Google Scholar]
- 16. Duke GE. 1986. Alimentary canal: anatomy, regulation of feeding, and motility, p 269–288 In Sturkie PD. (ed), Avian physiology. Springer-Verlag, New York, NY [Google Scholar]
- 17. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elia M, Cummings JH. 2007. Physiological aspects of energy metabolism and gastrointestinal effects of carbohydrates. Eur. J. Clin. Nutr. 61(Suppl 1):S40–S74 [DOI] [PubMed] [Google Scholar]
- 19. Genovese KJ, et al. 2003. Competitive exclusion of Salmonella from the gut of neonatal and weaned pigs. J. Food Prot. 66:1353–1359 [DOI] [PubMed] [Google Scholar]
- 20. Holben WE, Feris KP, Kettunen A, Apajalahti JH. 2004. GC fractionation enhances microbial community diversity assessment and detection of minority populations of bacteria by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hughes RJ. 2008. Relationship between digesta transit time and apparent metabolisable energy value of wheat in chickens. Br. Poult. Sci. 49:716–720 [DOI] [PubMed] [Google Scholar]
- 22. Kitahara M, Sakamoto M, Ike M, Sakata S, Benno Y. 2005. Bacteroides plebeius sp. nov. and Bacteroides coprocola sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 55:2143–2147 [DOI] [PubMed] [Google Scholar]
- 23. Lai HC, Duke GE. 1978. Colonic motility in domestic turkeys. Dig. Dis. Sci. 23:673–681 [DOI] [PubMed] [Google Scholar]
- 24. Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848 [DOI] [PubMed] [Google Scholar]
- 26. Li M, et al. 2008. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. U. S. A. 105:2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macfarlane GT, Gibson GR, Cummings JH. 1992. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72:57–64 [DOI] [PubMed] [Google Scholar]
- 28. Macfarlane GT, Macfarlane S. 2007. Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr. Opin. Biotechnol. 18:156–162 [DOI] [PubMed] [Google Scholar]
- 29. Macfarlane S, Woodmansey EJ, Macfarlane GT. 2005. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl. Environ. Microbiol. 71:7483–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuura Y. 1998. Degradation of konjac glucomannan by enzymes in human feces and formation of short-chain fatty acids by intestinal anaerobic bacteria. J. Nutr. Sci. Vitaminol. (Tokyo) 44:423–436 [DOI] [PubMed] [Google Scholar]
- 31. Momozawa Y, Deffontaine V, Louis E, Medrano JF. 2011. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS One 6:e16952 doi:10.1371/journal.pone.0016952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ochman H, et al. 2010. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 8:e1000546 doi:10.1371/journal.pbio.1000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pieper R, Bindelle J, Rossnagel B, Van Kessel A, Leterme P. 2009. Effect of carbohydrate composition in barley and oat cultivars on microbial ecophysiology and proliferation of Salmonella enterica in an in vitro model of the porcine gastrointestinal tract. Appl. Environ. Microbiol. 75:7006–7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pivnick H, Nurmi E. 1982. The Nurmi concept and its role in the control of Salmonella in poultry, p 41–70 In Davies R. (ed), Developments in food microbiology. Applied Science Publishers, Barking, Essex, England [Google Scholar]
- 36. Rezvani MR. 2007. Standardisation of precaecal and total tract amino acid digestibility measurement in laying hens. Ph.D. thesis Martin Luther University, Halle-Wittenberg, Germany [Google Scholar]
- 37. Rice GE, Skadhauge E. 1982. Caecal water and electrolyte absorption and the effects of acetate and glucose, in dehydrated, low-NaCl diet hens. J. Comp. Physiol. B 147:61–64 [Google Scholar]
- 38. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schneitz C. 2005. Competitive exclusion in poultry—30 years of research. Food Control 16:657–667 [Google Scholar]
- 40. Tomita R, et al. 2011. Study of segmental colonic transit time in healthy men. Hepatogastroenterology 58:1519–1522 [DOI] [PubMed] [Google Scholar]
- 41. Tozaki H, et al. 1997. Chitosan capsules for colon-specific drug delivery: improvement of insulin absorption from the rat colon. J. Pharm. Sci. 86:1016–1021 [DOI] [PubMed] [Google Scholar]
- 42. Van den Abbeele P, et al. 2010. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 76:5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang ML, et al. 2008. Influence of grape seed proanthocyanidin extract in broiler chickens: effect on chicken coccidiosis and antioxidant status. Poult. Sci. 87:2273–2280 [DOI] [PubMed] [Google Scholar]
- 44. Wang XL, et al. 2009. Laying performance and egg quality of blue-shelled layers as affected by different housing systems. Poult. Sci. 88:1485–1492 [DOI] [PubMed] [Google Scholar]
- 45. Watanabe Y, Nagai F, Morotomi M, Sakon H, Tanaka R. 2010. Bacteroides clarus sp. nov., Bacteroides fluxus sp. nov. and Bacteroides oleiciplenus sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 60:1864–1869 [DOI] [PubMed] [Google Scholar]
- 46. Wommack KE, Bhavsar J, Ravel J. 2008. Metagenomics: read length matters. Appl. Environ. Microbiol. 74:1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yildirim S, et al. 2010. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One 5:e13963 doi:10.1371/journal.pone.0013963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yin Y, et al. 2010. Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J. 4:367–376 [DOI] [PubMed] [Google Scholar]
- 49. Zhang C, et al. 2010. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 4:232–241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.