Abstract
Environmental sampling for microbiological contaminants is a key component of hygiene monitoring and risk characterization practices utilized across diverse fields of application. However, confidence in surface sampling results, both in the field and in controlled laboratory studies, has been undermined by large variation in sampling performance results. Sources of variation include controlled parameters, such as sampling materials and processing methods, which often differ among studies, as well as random and systematic errors; however, the relative contributions of these factors remain unclear. The objective of this study was to determine the relative impacts of sample processing methods, including extraction solution and physical dissociation method (vortexing and sonication), on recovery of Gram-positive (Bacillus cereus) and Gram-negative (Burkholderia thailandensis and Escherichia coli) bacteria from directly inoculated wipes. This work showed that target organism had the largest impact on extraction efficiency and recovery precision, as measured by traditional colony counts. The physical dissociation method (PDM) had negligible impact, while the effect of the extraction solution was organism dependent. Overall, however, extraction of organisms from wipes using phosphate-buffered saline with 0.04% Tween 80 (PBST) resulted in the highest mean recovery across all three organisms. The results from this study contribute to a better understanding of the factors that influence sampling performance, which is critical to the development of efficient and reliable sampling methodologies relevant to public health and biodefense.
INTRODUCTION
Effective surface sampling for biological contaminants is critical to the development of hygiene and decontamination plans to ensure public health and product integrity. Recovery, enumeration, and identification of microorganisms on surfaces have been a focus of scientific inquiry for over a century, with initial applications primarily related to food safety (2), the integrity of the National Aeronautics and Space Administration (NASA) programs (35), and hygiene in domestic (22, 50) and clinical (1, 6, 19) settings. However, the reliability of traditional surface sampling methods came under intense scrutiny in 2001 following the widespread contamination of multiple large building complexes with spores identified as Bacillus anthracis, the causative agent of anthrax. Confidence in the qualitative results primarily reported during that event was undermined in part by the use of sample collection and analytical methods that had not been previously validated for B. anthracis (47). Since that event, quantitative sampling and analysis methods have been recognized as essential components of a biothreat investigation for determination of the extent of contamination as well as decontamination effectiveness (27). As a result, over the last decade needs for quantitative assessment capabilities have driven research on the relative effectiveness of different sampling processes as well as the development of validated surface sampling and sample processing methods (27, 49). Notably, the vast majority of these studies have utilized B. anthracis spores (13, 20, 23, 27, 28, 48) or spores of a surrogate organism (4, 5, 8, 9, 18, 58). Comparatively little effort has been applied in recent years toward improved surface sampling of viruses (31) and vegetative bacterial cells that represent likely biothreat (BT) agents (or surrogates thereof) (7, 29, 39, 60).
In addition to biodefense-related surface sampling needs, emerging hygiene requirements in food production and clinical settings are also fueling demand for improved reliability and efficiency of traditional surface sampling methods. In response to the establishment of a zero tolerance policy for Listeria monocytogenes in ready-to-eat foods (57), food industry stakeholders have implemented microbiological testing programs for food contact and environmental surfaces to assess effectiveness of sanitation standard operating procedures. Although traditional culture enrichment methods for Listeria provide only qualitative (i.e., presence/absence) results, there is increasing interest in quantitative measurements for use in risk assessments (59). The rising prevalence of certain hospital-acquired infections, such as methicillin-resistant Staphylococcus aureus (MRSA), has spurred a proposal for microbiological hygiene standards in hospitals (12), which also necessitates the capability to quantitatively assess bacterial loads on surfaces. In the absence of standardized and validated surface sampling procedures that could be incorporated into environmental monitoring programs, efforts to optimize methods for recovery of these food-borne and hospital-associated microorganisms from contaminated surfaces have been reported (36, 44, 45, 59). However, most have focused on the relative effectiveness of different sampling materials, and few studies have examined the impact of processing method, including extraction solution and physical dissociation methods (e.g., sonication and vortexing) on recovery efficiency (42).
Although vegetative bacterial cells lack the stability and resistance to environmental degradation characteristic of bacterial endospores, the ability of some bacteria to persist on surfaces in a vegetative state for as long as several months has been clearly demonstrated (14, 37, 61). Consequently, fomites, environmental surfaces, and even clothing (62) may act as reservoirs of bacterial pathogens and can play an important role in disease transmission (3, 56). While this is of particular concern for health care settings and food contact surfaces, it is also relevant to clean room and biodefense applications, necessitating sample collection methods that facilitate monitoring for a wide range of surface-associated microbial contaminants. A clear understanding of the factors that impact sampling performance is prerequisite to intelligent design and optimization of robust methods that meet multidisciplinary surface sampling needs.
Traditional surface sampling methods utilizing swabs and wipes have been plagued by poor recovery and highly variable results (17), with reported recovery efficiencies ranging from less than 1% (7) to greater than 90% (18), depending on the experimental conditions. Comparison among studies has been hindered by the wide variation in experimental conditions and the lack of standardized methods for characterization of sampling performance. Performance is generally measured by depositing a known quantity of cells or spores onto a surface, removing the adherent microorganisms using an absorptive sampling device (e.g., swab or wipe), and extracting the particles into solution. Recovered cells or spores can then be quantified by culture or quantitative PCR (8, 38). The recovery efficiency is dependent on numerous experimental parameters, including the method for depositing biocontaminants on surfaces, surface characteristics, wetting agents, extraction solutions, the physical dissociation method, the sampling material, variation in sampling technique, and the biological agent. These parameters may impact the removal of microorganisms from the surface (i.e., removal efficiency) and/or release of the particles from the absorptive material during the extraction step (i.e., extraction efficiency), both of which affect the overall recovery. However, removal and extraction efficiency are rarely measured independently, and most studies report only the overall recovery efficiency. Consideration of each step separately facilitates elucidation of the mechanism by which experimental parameters impact the recovery process and thus how sampling performance can be optimized. For example, Rose et al. (48) found that recovery efficiency for B. anthracis spores sampled from stainless steel coupons was significantly lower for rayon and polyester swabs as compared to cotton and macrofoam. However, in that study, the extraction efficiency for directly inoculated rayon swabs was not statistically different from those of cotton and macrofoam swabs, indicating that the lower recovery efficiency for rayon can likely be attributed to comparatively poorer removal of spores from the surface.
Together the extraction solution and physical dissociation method (PDM) make up the extraction method. Extraction methods for wipes reported in the literature vary considerably. Sonication, stomaching (generally used for sponge-type wipes), and vortexing are the most commonly reported PDMs; however, processing times can range from 30 s to 15 min. Similarly, a range of extraction solutions, including Butterfield's buffer (BB), potassium phosphate buffer, phosphate-buffered saline (PBS), and water, all with and without surfactant like Tween 80 or Triton X-100, as well as Ringer-based solutions and maximum recovery diluent (MRD), have been reported in the sampling literature. The objective of the present study was to characterize the impact of wipe processing method on recovery of vegetative bacterial cells from directly inoculated wipe materials. Specifically, the effects of different extraction solutions, PDMs, and biocontaminants on extraction efficiency were evaluated using traditional culture-based methods. An additional aim of the study was to identify the most effective extraction method, which was robust over the three different organisms. A fluorescence-based viability assay was developed to assess whether differences in recovery between extraction methods could be attributed to loss of cell viability.
MATERIALS AND METHODS
Test organisms and culture methods.
Burkholderia thailandensis (ATCC 700388), a surrogate for the Gram-negative, category B biothreat agent Burkholderia pseudomallei (24), and Bacillus cereus (ATCC 10987), a common surrogate for the Gram-positive category A biothreat agent B. anthracis, were purchased from ATCC (Manassas, VA). An attenuated Escherichia coli O157:H7 (ATCC 700728) strain was also obtained from ATCC. All three organisms were maintained on Luria-Bertani (LB) agar (Acros Organics, Fair Lawn, NJ) plates with incubation at 30°C.
Inoculum preparation.
To prepare inocula for use in the extraction experiment, 10 ml of LB broth (Acros Organics, Fair Lawn, NJ) was inoculated with a single isolated colony and cultured overnight (∼15 h) with shaking at 30°C (B. thailandensis and B. cereus) or 37°C (E. coli). The stationary-phase cultures were harvested by centrifugation (10,000 × g for 3 min for B. cereus and E. coli and for 5 min for B. thailandensis due to difficulties in pelleting cells), washed once in phosphate-buffered saline (PBS) at pH 7.4 containing 0.04% (vol/vol) Tween 80 (PBST), and concentrated 3-fold by resuspension in PBST at one-third the original culture volume. Inoculum concentrations were quantified using a drop plate method (25). Briefly, following 10-fold serial dilution of inoculums in PBST, for each inoculum dilution, five 10-μl drops of the cell suspension were deposited within a quadrant on LB agar plates. Plates were incubated at 30°C for ∼18 h (E. coli and B. cereus) or ∼42 h (B. thailandensis). CFU were enumerated for each replicate drop in the countable dilutions (i.e., colony counts between 3 and 30), and the average value from the five replicate drops was used to calculate the CFU/ml. The drop plate method was chosen over the more commonly applied spread plate method as it allowed all extraction conditions for a particular organism to be tested in a single day by reducing the time required for plating and the number of agar plates needed to plate multiple dilutions for each sample. This reduces the potential that day effects will confound observed differences in recovery efficiency results among extraction methods. Additionally, using this method, average colony counts for a single sample could be calculated from five replicate drops, as opposed to the two or three replicate plates commonly used with spread plating. Previous studies have shown the equivalence of drop and spread plate methods for obtaining cell recovery estimates (26). An electronic repeat pipettor was used to dispense the 10-μl drops in order to minimize variation in counts among replicate drops caused by slight differences in drop volumes.
Experimental design.
This study was designed to evaluate the impact of different experimental parameters on the efficiency with which bacterial cells are transferred from a sampling material to an extraction solution, defined here as the extraction efficiency. Three factors were evaluated for their impact on extraction efficiency: (i) organism, including one Gram-positive (B. cereus) and two Gram-negative cell types (B. thailandensis and E. coli), (ii) extraction solution, and (iii) physical dissociation method (PDM), including sonication, vortexing, or a combination thereof, for multiple time periods (Table 1). Extraction solutions and PDMs were chosen from those commonly found in the literature regarding extraction of biological agents from sampling materials (4, 13, 42, 48). Inclusion of phosphate-buffered saline (PBS; pH 7.4) and Butterfield's buffer (BB; pH 7.2) allowed comparison of high- and low-salt solutions. Each of these buffers was evaluated in the presence and absence of 0.04% of the polysorbate Tween 80. Maximum recovery diluent (MRD), which contains peptone and has been shown to help maintain cell viability during sample processing (53), was also evaluated for use as an extraction solution. Three experimental replicates were performed for each extraction condition.
Table 1.
Experimental factors evaluated for impact on extraction efficiency
| Organism (factor 1) | Extraction solution (factor 2)a | PDMb (factor 3) |
|---|---|---|
| E. coli | PBS | Touch vortexing for 10 s (TV) |
| B. cereus | PBST | Vortexing for 2 min (V2) |
| B. thailandensis | BB | Vortexing for 10 min (V10) |
| BBT | Sonication for 1 min (S1) | |
| MRD | Sonication for 2 min (S2) | |
| Sonication for 5 min (S5) | ||
| Sonication for 1 min + vortexing for 1 min (S1V1) | ||
| Sonication for 2 min + vortexing for 2 min (S2V2) |
PBS, phosphate-buffered saline; PBST, PBS plus 0.04% Tween 80; BB, Butterfield's buffer; BBT, BB plus 0.04% Tween 80; MRD, maximum recovery diluent.
PDM, physical dissociation method.
Wipe extraction procedure.
Sterile nonwoven polyester-rayon blend wipes (2 by 2 in.) (Kendal Versalon, catalog no. 8042; Tyco Healthgroup LP, Mansfield, MA), previously shown to efficiently release Bacillus anthracis Sterne spores into extraction solutions (13), were used in the present study. Wipes were premoistened with 1 ml of neutralizing buffer (Hardy Diagnostics, Santa Maria, CA) and subsequently inoculated with 100 μl of inoculum (∼109 CFU of a single organism). Neutralizing buffer, which inactivates chlorine and quaternary ammonium compounds commonly found in disinfectants, is currently recommended by the Centers for Disease Control and Prevention (CDC) as a wetting agent for sampling of B. anthracis spores from smooth, nonporous surfaces (10) and has been shown to be an effective wetting agent for recovery of Yersinia pestis from inoculated swabs (46). Following inoculation, wipes were left in partially covered sterile petri dishes in a biological safety cabinet for 1 h to promote adherence of cells to the wipes. Incorporation of wipe holding times into sampling material inoculation procedures has been described previously (46, 60). Following the 1-h holding period, wipes were transferred into sterile 50-ml screw-top conical polypropylene tubes (catalog no. 23-2262; Crystalgen, Plainview, NY) containing 30 ml of the extraction solution. After subjection of the extraction tube to the PDM for the specified time period (Table 1), 100-μl aliquots were removed and serially diluted in PBST to 10−4. Due to high inoculum cell concentrations, an additional 1:10 dilution was performed on the extracted B. thailandensis cell suspensions prior to the 4-log serial dilutions by transferring the 100-μl aliquot from the extraction tube into 900 μl of PBST. Using the drop plate method (25), 10−1 through 10−4 dilutions (10−2 through 10−5 for B. thailandensis) were plated onto LB agar, applying five 10-μl drops per dilution. Plates were incubated at 30°C for all three bacteria for ∼18 h (E. coli and B. cereus) or ∼42 h (B. thailandensis). CFU were enumerated for each replicate drop, and the average value from the five replicates was used to calculate the CFU/ml as a function of the suspension volume plated and the dilution factor.
Direct tube inoculation controls.
A reference control was run for each experimental condition (organism-extraction solution-PDM combination). For each reference, the inoculum was prepared as described above, and 100 μl was transferred directly into sterile 50-ml screw-top conical tubes containing 30 ml of the extraction solution. Reference control tubes were subjected to the wipe extraction procedure described above (i.e., application of PDM followed by serial dilution and plating to determine cell concentration in the extraction solution), and the result theoretically represents the maximum expected recovery for a given extraction method, accounting for potential cell losses due to adherence to tube walls, aggregation, or losses in viability. Reference control experiments were run on separate days from the wipe extractions and consequently, inocula for reference control and wipe experiments evaluating the same experimental conditions (organism-extraction solution-PDM combination) may have varied in concentration slightly. However, cell recoveries from both wipes and reference control samples were normalized to the starting inoculum concentrations to allow comparison.
Calculation of extraction efficiency.
Extraction efficiency, represented as the percentage of recovery from the inoculated wipes, was calculated as the CFU/ml in the extraction solution after wipe processing relative to the concentration of the initial inoculum. A secondary response measure, the recovery ratio (RR), provided information on the efficiency with which bacteria were released from the wipe surface. RR is defined by the equation RR = mean % recovery from wipes/mean % recovery from reference controls.
Viability assay.
A fluorescence-based assay utilizing a commercial LIVE/DEAD BacLight bacterial viability kit (catalog no. L7012; Invitrogen, Eugene, OR) was developed to measure viability loss associated with the extraction conditions and the 1-h wipe holding time. Fluorescence measurements were collected using a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA). A calibration curve was generated for each experimental replicate in order to calculate the percentage of viable cells remaining after exposure of cells to the extraction method. To generate the calibration curve, live and dead cells were mixed in the following ratios: 0:100, 10:90, 25:75, 50:50, 75:25, 90:10, and 100:0. Dead cells were prepared by resuspending pelleted cells in 70% ethanol and incubating them for 1 h at room temperature with constant shaking, as per the manufacturer's instructions. Prior to mixing, live and dead cell pellets were washed one time in 0.85% NaCl and resuspended in 0.85% NaCl containing 0.02% Tween 80 (NaCl-T). For cell staining using the LIVE/DEAD BacLight bacterial viability kit components, a 2× dye mixture solution was prepared in sterile deionized (DI) water by adding SYTO 9 and propidium iodide (1:2 ratio of the two dyes, respectively), as per the manufacturer's instructions. The dye mixture was added to the live-dead cell mixtures at a 1:1 ratio and incubated for 15 min in the dark. Subsequently, 200 μl of each sample was pipetted into a 96-well black Costar microplate and analyzed in triplicate. The fluorescence emission was measured at 530 nm with a cutoff at 515 nm for SYTO 9 (green emission) and 620 nm with a cutoff at 590 nm for propidium iodide (red emission). Both dyes were excited at 485 nm. The percentage of live cells in the suspension was plotted as a function of the ratio between the green and red fluorescence intensities.
Viability losses associated with the 1-h holding period following wipe inoculation were assessed for E. coli and B. cereus. Wipes were premoistened and inoculated as described above for the wipe extraction procedure. Immediately (time [t] = 0 h) and 1 h (t = 1 h) after inoculation, wipes were placed into 50-ml screw-top conical polypropylene tubes containing 30 ml of PBST. As controls for viability losses associated with the processing method, tubes containing 30 ml of PBST were directly inoculated using the same inoculum as that used for the wipes. All tubes were vortexed for 2 min, and triplicate 100-μl aliquots were transferred into 1.5-ml centrifuge tubes. The tubes were centrifuged (10,000 × g, 3 min), and the pellets were washed once in 300 μl of 0.85% NaCl and resuspended in 100 μl of NaCl-T. An equal volume of dye mixture (prepared as described above) was added to the tubes, and samples were incubated for 15 min in the dark. Fluorescence emission was measured as described above, and the ratio of green to red fluorescence was compared to the calibration curve in order to determine losses in viability associated with the extraction method. Due to assay sensitivity problems for B. cereus, the above protocol was modified as follows to increase cell yields prior to cell staining. Wipes were cut into 2.5-cm2 pieces and premoistened with 250 μl of neutralizing buffer prior to inoculation as described above. The inoculated wipe pieces were processed in 15-ml screw-top polystyrene tubes (catalog no. UP2018; United Laboratory Plastics, St. Louis, MO) containing 5 ml PBST. Following processing by vortexing as described above, the 5 ml containing extracted cells was split among five 1-ml microcentrifuge tubes. Tubes were centrifuged (10,000 × g, 2 min), and cells were washed in 500 μl 0.85% NaCl and then combined in 300 μl of NaCl-T. Thirty microliters of the combined cell suspension was diluted into 150 μl of NaCl-T, and an equal volume of the 2× dye solution was added to stain the cells as described above.
Viability losses for E. coli and B. cereus associated with the extraction method were assessed for all five extraction solutions (only BB and BB with 0.04% Tween 80 [BBT] for B. cereus), and three of the PDMs (touch vortexing, vortexing for 2 min, and sonication for 5 min). To ensure roughly the same starting cell concentration for each of the experimental replicates, overnight cultures were adjusted to a set optical density at 670 nm (OD670) value (∼1.3 and 3.4 for E. coli and B. cereus, respectively) at the start of the experiment. Thirty microliters of the adjusted overnight culture was added to 1.5-ml microcentrifuge tubes containing 500 μl of 0.85% NaCl. Tubes were centrifuged (10,000 × g, 3 min), and pellets were resuspended in 300 μl of the appropriate extraction solution. After subjecting cell suspensions to one of the three PDMs, cells were pelleted and resuspended in 150 μl of NaCl-T. An equal volume of dye mixture (prepared as described above) was added to tubes, and samples were incubated for 15 min in the dark. Fluorescence emission was measured, and the percentage of viable cells was calculated from the calibration curve as described above.
Statistical analysis.
Statistical analyses were performed using the free, public domain software system Dataplot (Statistical Engineering Division, National Institute of Standards and Technology [http://www.itl.nist.gov/div898/software/dataplot]) and R (R Development Core Team [http://www.R-project.org/]). Differences in mean recovery efficiency between experimental groups were assessed by analysis of variance (ANOVA), with a significance level set at 0.05. Ninety-five percent confidence intervals were calculated to delineate equivalent groups. Coefficients of variation (CV) were calculated as a measure of the variation within experimental groups (i.e., precision). A Levene's test for homogeneity of variance was used to identify differences in precision between experimental groups. Reproducibility, sometimes defined as variability in results from different laboratories, was not evaluated in the present study.
RESULTS
Extraction efficiency and recovery ratio.
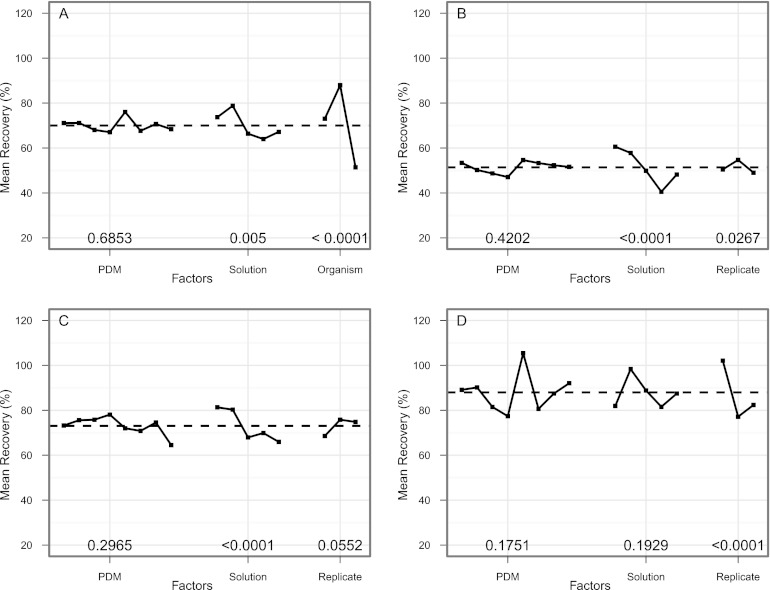
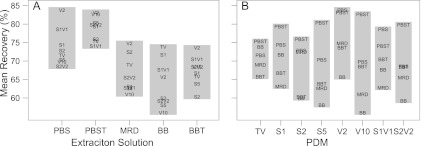
Mean extraction efficiency for directly inoculated wipes, expressed as a percentage of the initial inoculum, varied dramatically across experimental conditions, ranging from 35.1 to 127%, with an overall average of 70%. Efficiencies slightly greater than 100% likely reflect an underestimation of the initial inoculum concentration and have been reported in previous similar studies (42, 48). A sensitivity analysis (Fig. 1A) indicated that the primary factor driving extraction efficiency was the target organism. Analysis of variance (ANOVA) results indicated statistically significant differences in mean percentages of recovery among the three microorganisms (P < 0.0001), when averaging across all extraction methods for a given organism. Mean recovery was highest for B. thailandensis (88%), lowest for E. coli (51%), and 73% for B. cereus. The extraction solution had the second largest impact on extraction efficiency, whereas PDM had the smallest effect (Fig. 1A). A rank analysis of PDMs showed that when averaging across all three organisms, the mean percentage of recovery from wipes was highest when vortexing for 2 min for four out of the five extraction solutions (Fig. 2A). The binomial probability of this occurring by chance alone is less than 1 in 1,000. However, one-way analysis of variance failed to identify statistically significant differences between PDMs for any individual organism, and consequently, these groups were collapsed for subsequent analysis to provide larger sample sizes.
Fig 1.
Main effects plots showing the mean value of the percentages of recovery from extracted wipes as a function of the experimental factors evaluated in the study for all three organisms (A), E. coli (B), Bacillus cereus (C), and Burkholderia thailandensis (D). PDM, physical dissociation method; solution, extraction solution. Dashed lines represent grand mean. Numbers above experimental factors are P values from one-way analysis of variance. P < 0.05 was considered statistically significant.
Fig 2.
Rank analysis for physical dissociation method (PDM) (A) and extraction solution (B). Block plots show mean percentage of recovery averaged across all three organisms for each extraction method. TV, touch vortexing; S1, sonication for 1 min; S2, sonication for 2 min; S5, sonication for 5 min; V2, vortexing for 2 min; V10, vortexing for 10 min; S1V1, sonication and then vortexing for 1 min each; S2V2, sonication and then vortexing for 2 min each; BB, Butterfield's buffer; BBT, Butterfield's buffer with 0.04% Tween 80; PBS, phosphate-buffered saline; PBST, phosphate-buffered saline with 0.04% Tween 80; MRD, maximum recovery diluent.
Statistically significant differences (P < 0.05 for one-way ANOVA) in mean percentage of recovery among extraction solutions were observed for E. coli (Fig. 1B) and B. cereus (Fig. 1C). For both organisms, extraction efficiency was significantly higher in PBS and PBST compared to those of the other three buffers. Extraction efficiency for each organism as a function of the extraction solution is presented in Table 2. For B. cereus, the percentages of recovery in BB, BBT, and MRD were equivalent, whereas for E. coli, extraction of wipes in BB resulted in significantly lower cell recovery. Although differences in mean percentages of recovery across the five extraction solutions were not statistically significant for B. thailandensis (Fig. 1D), likely due to the substantial variability in recovery results, extraction efficiency for this organism was highest in PBST. Averaging across all three organisms, rank analysis showed that the mean percentage of recovery was highest for wipes processed in PBST for six out of the eight PDMs (Fig. 2B). The binomial probability of this occurring by chance alone is 1 in 10,000. The percentages of recovery data for each experimental condition are provided in Table S1 and Fig. S1 in the supplemental material.
Table 2.
Extraction efficiency for E. coli, B. cereus, and B. thailandensis inoculated onto polyester-rayon wipes using data pooled from all PDMs
| Organism | Extraction solution | % recovery (95% CI)a | CV (%)b | Recovery ratioc | Sample sized |
|---|---|---|---|---|---|
| E. colie | PBS | 60.6 (57.2–64.0) | 13.3 | 0.89 | 24 |
| PBST | 57.8 (54.9–60.7) | 11.8 | 0.76 | 24 | |
| BB | 40.5 (37.7–43.3) | 16.5 | 0.68 | 24 | |
| BBT | 48.2 (45.4–50.9) | 13.5 | 0.73 | 24 | |
| MRD | 49.9 (47.3–52.5) | 12.4 | 0.62 | 24 | |
| Allf | 51.4 (49.6–53.2) | 19.2 | 0.73 | 120 | |
| B. cereuse | PBS | 81.3 (75.9–86.7) | 15.7 | 0.71 | 24 |
| PBST | 80.3 (74.2–86.4) | 18.0 | 0.68 | 24 | |
| BB | 69.9 (64.9–74.9) | 16.9 | 0.70 | 24 | |
| BBT | 65.9 (60.4–71.4) | 19.8 | 0.63 | 24 | |
| MRD | 67.9 (61.8–74.1) | 21.4 | 0.60 | 24 | |
| Allf | 73.1 (70.4–75.7) | 20.1 | 0.67 | 120 | |
| B. thailandensis | PBS | 82.0 (69.7–94.2) | 28.1 | 0.95 | 16 |
| PBST | 98.3 (88.4–108) | 24.0 | 1.11 | 24 | |
| BB | 81.6 (69.6–93.5) | 34.7 | 0.95 | 24 | |
| BBT | 87.4 (79.1–95.8) | 22.6 | 0.98 | 24 | |
| MRD | 88.8 (70.2–107) | 39.2 | 1.01 | 16 | |
| Allf | 88.0 (82.9–93.1) | 29.8 | 1.00 | 104 |
95% CI, 95% confidence interval for the mean.
CV, coefficient of variation.
The recovery ratio was calculated as the ratio of the mean percentage of recovery from inoculated wipes to the mean percentage of recovery from directly inoculated extraction solution reference controls.
Sample size was based on pooled PDM data (n = 8) from two (n = 16) or three (n = 24) independent experimental replicates. Although three independent experimental replicates were performed for each extraction condition, in some cases, data from only two replicates were used in the analysis due to deviations from the extraction protocol.
There were significant differences in mean percentages of recovery among extraction solutions for that organism by ANOVA (P < 0.0001).
Average from combined data from all extraction methods.
Measurements of efficiency of bacterial cell release from wipes may be confounded by cell losses during processing caused by aggregation, adhesion to other surfaces (e.g., tube walls, pipette tips, etc.), and/or viability loss. To account for these potential losses not associated with entrapment in the wipe, the ratio of mean percentage of recovery from the wipe to mean percentage of recovery from the reference control was calculated (Table 2). The overall mean recovery ratio for B. thailandensis was 1.0, indicating that this organism was released from the wipe with close to 100% efficiency. Although the mean percentage of recovery from the reference control tube was highest for B. cereus, the overall recovery ratio (0.67) was lowest, indicating that losses to the wipe were highest for this organism.
Precision.
Variability in recovery data between replicates was highest for B. thailandensis and smallest for E. coli (see coefficients of variation in Table 2 and residual standard deviations in Fig. S1 in the supplemental material). A Levene's test for homogeneity of variance confirmed that recovery precision was driven by the target organism and not the extraction method. For E. coli and B. cereus, no difference in variance across extraction solutions was observed for any of the PDMs, while for B. thailandensis, the Levene's test failed for only one of the eight PDMs. Levene's test results and variation between replicates for all extraction conditions can be seen in the supplemental material (see Fig. S1).
Viability assay.
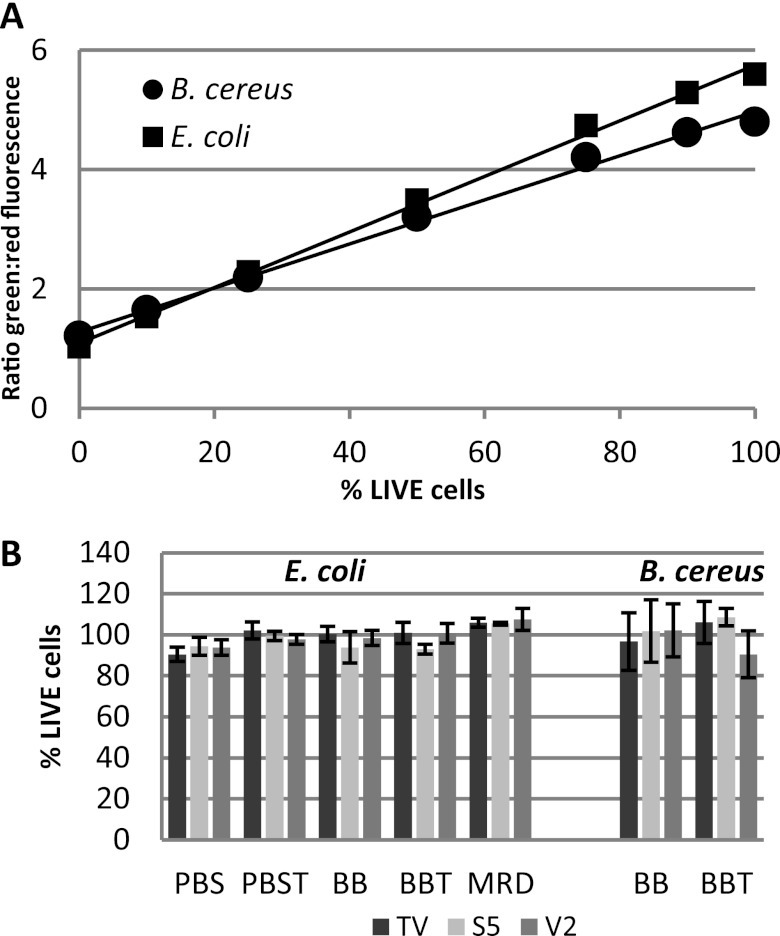
To better understand the reduced recovery of E. coli and B. cereus from wipes relative to recovery from the reference control tubes, viability loss associated with the 1-h wipe holding time was assessed using a fluorescence-based viability assay. As the recovery ratio for B. thailandensis was approximately 1.0, there was no indication of losses associated with the wipe, and, therefore, viability loss was not assessed for this organism. No reduction in the percentage of live cells recovered from wipes after the 1-h holding time was observed for either organism when compared to control wipes processed by the same extraction method (i.e., vortexing for 2 min in PBST) immediately after inoculation. Approximately 100% of cells recovered from the wipe at both time points were viable based on comparison of the green/red fluorescence ratio to the calibration curve (data not shown). Additionally, no loss in viability associated with the extraction method was observed for either organism (Fig. 3).
Fig 3.
Viability as a function of extraction method. A calibration curve (A) was constructed and used to calculate the percentage of recovered cells that were viable after sample processing (B). Error bars represent the standard deviation of the mean (n = 3). BB, Butterfield's buffer; BBT, Butterfield's buffer with 0.04% Tween 80; PBS, phosphate-buffered saline; PBST, phosphate-buffered saline with 0.04% Tween 80; MRD, maximum recovery diluent. TV, touch vortexing for 10 s; S5, sonication for 5 min; V2, vortexing for 2 min.
DISCUSSION
Effective surface sampling for microbiological contaminants is critical to the success of environmental characterization and monitoring processes utilized across diverse fields of applications and for myriad purposes. Studies designed to optimize and validate performance of surface sampling methodologies often focus on a single microorganism of interest (e.g., B. anthracis spores and Listeria spp.). Although highly optimized sampling procedures are of use when the presence of a specific organism is expected, many times the identity of microbial surface contaminants is unknown. Additionally, the combination of limited funding and time makes optimization of sampling methods to develop a best practice for every possible target organism infeasible; therefore, the development of methods that are broadly effective for recovery and analysis of numerous organisms is highly desirable. A thorough understanding of how different factors, both controlled and uncontrolled, impact sampling performance for different microorganisms is requisite to the development of best practices. In the present study, the identity of the target microbe had the largest impact on sampling performance, for both recovery efficiency (Fig. 1A) and precision (Table 2). However, the negligible impact of PDM on recovery efficiency and the efficacy of PBST across all three organisms suggest that although recovery performance may vary considerably for different microbes, development of broadly effective extraction procedures is possible, eliminating the need for optimization of organism-specific methods.
The organisms utilized in this study were chosen based on relevance to several different fields, including food safety, clean room applications and biodefense, as well as diversity of phenotypes (i.e., cell wall structure, spore-forming ability, and size). As such, variability in sampling performance was expected and desirable in order to evaluate sampling method robustness. The wide range in mean extraction efficiencies observed in the present work (35.1 to 127%) is consistent with extraction efficiencies previously reported for swabs directly inoculated with E. coli (24.3 to 80.3%) or Staphylococcus aureus (46.3 to 105.3%) (42). Of interest, the organism for which mean recovery efficiency from inoculated wipes was greatest, B. thailandensis, also produced the most variable results, with CV over 20% (Table 2). Conversely, extraction efficiency was lowest for E. coli but precision (i.e., variation among experimental replicates) was greatest for this organism. These results emphasize the need to evaluate recovery efficiency and precision as separate performance metrics. Like recovery efficiency, sampling method precision is difficult to compare between studies due to differences in experimental conditions. In a recent swab validation study for B. anthracis spores, coefficients of variation (CV) for spores recovered from directly inoculated macrofoam swabs ranged from 9.4 to 32.3% (27), depending on the inoculum concentration, with greater precision at higher concentrations. In the present study, sampling materials were inoculated at high cell densities (∼109 CFU/wipe) to allow viability assessment in parallel using a fluorescence-based microplate assay (Fig. 3). Although lower inoculum loads (102 to 104 CFU) more similar to expected natural contamination levels are often used in studies relevant to the food industry or clinical and clean room settings (42, 43), large inoculums are appropriate for studies on biothreats or BT surrogates (7, 18, 29) given the potential for highly concentrated contaminants in bioterrorism situations. However, the effect of inoculum load on sampling performance should be considered when comparing results between studies.
Teasing out a biological explanation for the significant differences in sampling performance among organisms is challenging due to the complexity of the system; however, known phenotypic characteristics of the different microbes can offer some insights. When comparing the mean percentage of recovery from the inoculated wipe to that of the reference control, the high recovery ratio observed for B. thailandensis (∼1.0) indicates that (i) there was no viability loss associated with the 1-h wipe holding time and (ii) the bacteria were efficiently released from the wipe surface. One possible explanation for the high extraction efficiency is that production of biosurfactants by B. thailandensis (16) or another secretion product may have reduced adherence of the cells to the wipe. For example, B. thailandensis lacks the polysaccharide capsule produced by pathogenic Burkholderia species, including B. pseudomallei (65); however, the presence of an exopolysaccharide (EPS) gene cluster at the chromosomal location of the B. pseudomallei capsular polysaccharide (CPS) gene cluster indicates the potential for production and secretion of polysaccharide (33, 51). The role of these polysaccharides in pathogen recovery has not yet been investigated; however, the possibility of an organism-specific secretion product influencing recovery makes protocol optimization more challenging.
The recovery from reference control tubes provides a means of estimating the ability of a solution to disperse and extract a given organism from sampling materials. Notably, the mean percentage of recovery from the reference control of B. thailandensis was less than 100% (i.e., 88%) (see Table S1 in the supplemental material), indicating that there were losses to the system during processing (e.g., losses due to adhesion to tube walls, pipette tips, viability loss associated with processing, etc.). In contrast, the mean percentage of recovery of B. cereus from reference control tubes was approximately 100%, but the mean recovery ratio for this organism was only 0.67 (Table 2), indicating that the lower observed extraction efficiency was due to losses associated with the wipe. Since no loss in viability was observed for cells recovered from wipes following the 1-h holding time, these losses are likely related to a strong interaction of B. cereus with the wipe surface. For E. coli, mean percentage recoveries of ∼70% and ∼50% (Table 2; see Table S1 in the supplemental material) for reference control tubes and inoculated wipes, respectively, indicate losses both to the wipe and during processing in the extraction tubes. As was observed for B. cereus, no reduction in viability of recovered cells was noted following the 1-h wipe holding period, indicating that the low extraction efficiency is related to a tendency for the bacterium to adhere to surfaces. In a recent study evaluating extraction efficiency for B. anthracis Sterne spores directly inoculated onto wipes, surface thermodynamics was used to explain extraction performance (13). The effect of surface thermodynamics on aggregation and adhesion to substrata has been widely studied for a variety of microorganisms, including B. cereus and E. coli (21). Given the evidence for losses to the wipe and/or extraction tube surfaces, microbial surface chemistry, including hydrophobicity and surface charge measurements, may in part explain the organism-specific differences in extraction performance observed in the present study and is under investigation.
After the factor organism, extraction solution had the next greatest impact on extraction efficiency (Fig. 1A). Although the impact of the extraction method on recovery of vegetative cells from sampling materials has not been well studied, this finding is consistent with a previous study utilizing bacterial spores that showed the importance of extraction solution and the relatively small contribution of PDM to extraction method efficacy (13). Processing of wipes in BB resulted in the lowest extraction efficiency for E. coli and B. thailandensis (Table 2). Unlike PBS, BB is not a physiological solution, and osmotic stress resulting from exposure to the extremely low salt concentration may have induced some cells into a viable but nonculturable (VBNC) state, as indicated by reduced recovery using culture-based methods in the absence of observed viability losses from the fluorescence-based viability assay (Fig. 3). Although Butterfield's buffer is used extensively in food (15) and water microbiology (11) and is considered a superior diluent compared to water, viability losses for bacteria diluted in BB have been reported (53). However, such assumed viability losses associated with exposure to BB were determined using culture-based methods and may instead represent induction into a VBNC state. Induction of a VBNC state under conditions of osmotic stress, as measured by fluorescence-based cell viability assays and colony counts, has been reported previously (63). Interestingly, recovery of E. coli and B. cereus from wipes in MRD, which contains peptone and saline at physiological levels, was significantly lower than recovery in PBS and PBST and equivalent to recovery in BBT (Table 2). Given the inferior performance compared to PBS-containing solutions and the potential for growth of some microorganisms in the presence of peptone (42), MRD is not an ideal extraction solution, especially if delays in sample processing could result in long contact times.
One surprising observation was the excellent extraction performance for all three organisms in PBS. In a previous study examining extraction efficiency of B. anthracis spores (13), sample processing by vortexing in PBS resulted in poor sampling performance, with recoveries less than 10%, even from directly inoculated control tubes. The authors attributed the poor performance to spore adhesion to the tube walls during processing and found that this effect could be ameliorated through the addition of Tween 80 to the extraction solution. They hypothesized that the strong attractive forces between the Tween 80 molecules and the polypropylene tube would result in the formation of a Tween 80 film on the tube walls, which would prevent spore adhesion. Contrastingly, in the present study, extraction efficiencies were highest for both E. coli and B. cereus when wipes were processed in PBS, and the addition of Tween 80 failed to improve performance. Improved performance in the presence of Tween 80 was observed only for B. thailandensis (82.0% and 98.3% recovery for PBS and PBST, respectively), but analysis of variance failed to show any statistical differences in mean recovery by extraction solution for this organism. Improved recovery efficiency in the presence of surfactant for spores but not vegetative cells has been observed previously. Kim et al. (34) found that while a Tween 80-containing solution improved recovery of Bacillus globigii spores from artificially loaded HVAC filters, recovery of the vegetative cells Mannheimia haemolytica and Yersinia ruckeri was higher when filters were eluted in PBS. In that study, recovery efficiencies for M. haemolytica and Y. ruckeri in PBS were 87% and 80%, respectively, which are similar to the extraction efficiencies for B. cereus and B. thailandensis observed in the present study.
Polysorbates like Tween 80 and Tween 20 have been used extensively in biotherapeutic formulations to prevent surface adsorption and aggregation (32). The same properties make Tween and other surfactants attractive as additives to solutions used in surface sampling, and in recent years, use of polysorbates has become increasingly common in surface sampling and sample processing procedures. Surfactants are often used in wetting agents to improve release of adherent microorganisms from surfaces (although this function requires further experimental verification) and in extraction solutions to reduce losses caused by aggregation and adhesion to tube walls during sample processing (13). For studies utilizing high inoculum concentrations, sample dilution prior to plating in solutions lacking surfactant may dramatically bias measurements of the inoculum concentration and subsequently, recovery efficiency estimates. In the present study, results from pilot experiments where dilutions were conducted in extraction solutions lacking Tween 80 showed recovery efficiency estimates as high as 1,000% (data not shown), likely due to inaccurate estimation of the initial inoculum concentration. It is probable that a substantial number of cells were lost to tube walls or pipette tips during the serial dilution of the inoculum. These losses likely resulted in inoculum concentration estimates that were as much as 10-fold lower than actual concentrations, resulting in percentage of recovery estimates that were off by up to factor of 10. This effect was corrected by the use of PBST as the dilution buffer for the inoculum in all subsequent experiments.
Despite the many advantages of surfactants, caution should be taken regarding potential organism-specific effects, particularly when there is a potential for long contact times, as is the case for transport media. Polysorbates such as Tween 80 and Tween 20 are highly susceptible to autooxidation and hydrolysis of the fatty acid ester bond, producing hydroperoxide and short-chain acids, which can substantially lower a solution's pH (32). Based on observations made during the pilot study, we found it necessary to prepare BBT just prior to the start of each experiment due to the pH instability of the solution over time. In one extreme case, the pH of a BBT solution that was greater than 1 month old had dropped from 7.2 to 3.8. The pH of BB was also unstable, likely due to BB's lack of buffering capacity, but to a lesser extent than that observed for BBT, with no observed drop in pH below 6.0 during a similar time period. The use of the older BBT resulted in substantial viability loss for B. cereus, but not E. coli or B. thailandensis, with the magnitude of the loss related to the age of the solution, as determined by plate counts (data not shown). The observed growth inhibition for B. cereus could be related to low pH, sensitivity to Tween 80 degradation products, or a combination of these two factors. Sensitivity of bacteria to oleic acid (degradation product of Tween 80) and linoleic acid (degradation product of Tween 20) has been reported and may be enhanced at low pH (52, 54, 64, 66). Impacts of low-nutrient solutions containing Tween 80 on viability of Gram-positive bacteria have also been noted previously (42, 55). Although the sensitivity of B. cereus to older BBT was not observed for PBST, for which no drop in pH was observed, the innocuous nature of this solution should be verified using a broader range of microorganisms, given its potential use as a transport medium (30, 46). Proper storage of Tween and Tween-containing solutions, away from light and oxygen (i.e., sealed) and at lower temperatures (≤25°C), may help prevent degradation from occurring (32). The addition of Tween 80 and other polysorbates to solutions just prior to use is also recommended to mitigate potential toxic effects of degradation products that accumulate over time.
This work has shown that organism-specific characteristics (e.g., structural and physiological properties) of a microbe will likely have a greater impact on recovery efficiency than the extraction method; therefore, a thorough understanding of these effects is needed to properly interpret and apply study results and to ensure the selection of appropriate surrogates for use in laboratory studies. Although in this study, organisms were selected for diversity of phenotype, even differences between strains of the same species (e.g., pathogenic and nonpathogenic strains [29] or laboratory versus environmental isolates [41]) can substantially influence sampling performance. When the nature of the surface-associated microorganism(s) is unknown, it may be useful to employ methods that have demonstrated robustness across multiple species. In this study, processing of wipes by vortexing for 2 min resulted in the highest mean percentage of recovery when averaging across all three organisms, but PDM had comparatively little effect on extraction efficiency, so logistical considerations such as availability of equipment or throughput capability may drive the choice of processing methods. Of the five extraction solutions benchmarked in this study, PBST appears to be optimal for vegetative cells, with robust recovery across all three organisms. Losses to the system as measured by recovery from reference control tubes were highest for Butterfield's buffer, which has traditionally been used in microbiological analysis of food (15) and dairy products (40), as well as water and wastewater (11). For this reason and the potential for growth inhibition for some bacteria with the addition of Tween 80, caution should be taken when using BB and BBT as extraction solutions.
Sampling data are inherently noisy, such that substantial variability among replicates results in large uncertainties in expected recovery efficiencies. Thoughtful study design and incorporation of proper controls are critical to partitioning sources of variability between those that can and cannot be controlled. This comprehensive study delineated the relative impact of different parameters on extraction efficiency, including commonly used extraction solutions and physical dissociation methods, which have rarely been subjected to side-by-side comparisons, particularly for vegetative cells. While most previous studies have evaluated method performance for a single microorganism, the testing of multiple bacterial species in this study demonstrated the robustness of some extraction methods across microbes with varied phenotypes. The use of reference controls helped inform where losses of cells were occurring in the system, which can inform optimization of sample processing methods. However, despite these strengths, the results from this study are subject to several limitations. The small numbers of replicates performed for each combination of experimental factors impacted our ability to detect statistically significant differences among groups, necessitating the pooling of recovery data from the different PDMs to evaluate the impact of extraction solution for each organism. Extraction efficiency estimates derived for the laboratory bacterial strains used in this work may not be representative of method performance for environmental isolates. Additionally, recovery efficiency and method precision for low inoculum concentrations may differ from those observed for the high inoculum levels applied in this study. Finally, samples collected from contaminated surfaces may behave differently than directly inoculated sampling materials, so future work must look at surface sampling performance for both single organisms and mixed communities. Despite these limitations, the results of this work inform the development of standardized sampling and sample processing methods, which will improve comparison across studies and the development of best sampling practices. Validated methodologies can then increase confidence in real world sampling results, facilitating risk characterization and environmental monitoring.
Supplementary Material
ACKNOWLEDGMENTS
The Department of Homeland Security (DHS) Science and Technology Directorate sponsored the production of this material under Interagency Agreement HSHQDC-09-X-00457 with the National Institute of Standards and Technology (NIST).
We thank Laura Rose from CDC, Atlanta, GA, for advice regarding wipe inoculation procedures. We thank Kenneth Cole at NIST for use of his laboratory equipment.
All opinions expressed in this paper are the authors' and do not necessarily reflect the policies and views of DHS or NIST or affiliated venues. Certain pieces of commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Anderson RL. 1969. Biological evaluation of carpeting. Appl. Microbiol. 18:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angelotti R, Foter MJ, Busch KA, Lewis KH. 1958. A comparative evaluation of methods for determining the bacterial contamination of surfaces. Food Res. 23:175–185 [Google Scholar]
- 3. Bright KR, Boone SA, Gerba CP. 2010. Occurrence of bacteria and viruses on elementary classroom surfaces and the potential role of classroom hygiene in the spread of infectious diseases. J. Sch. Nurs. 26:33–41 [DOI] [PubMed] [Google Scholar]
- 4. Brown GS, et al. 2007. Evaluation of a wipe surface sample method for collection of Bacillus spores from nonporous surfaces. Appl. Environ. Microbiol. 73:706–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown GS, et al. 2007. Evaluation of rayon swab surface sample collection method for Bacillus spores from nonporous surfaces. J. Appl. Microbiol. 103:1074–1080 [DOI] [PubMed] [Google Scholar]
- 6. Buggy BP, Wilson KH, Fekety R. 1983. Comparison of methods for recovery of Clostridium difficile from an environmental surface. J. Clin. Microbiol. 18:348–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buttner MP, Cruz P, Stetzenbach LD, Cronin T. 2007. Evaluation of two surface sampling methods for detection of Erwinia herbicola on a variety of materials by culture and quantitative PCR. Appl. Environ. Microbiol. 73:3505–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buttner MP, et al. 2004. Determination of the efficacy of two building decontamination strategies by surface sampling with culture and quantitative PCR analysis. Appl. Environ. Microbiol. 70:4740–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buttner MP, et al. 2004. Evaluation of the biological sampling kit (BiSKit) for large-area surface sampling. Appl. Environ. Microbiol. 70:7040–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention September 2010, posting date Surface sampling procedures for Bacillus anthracis spores from smooth, non-porous surfaces. http://www.cdc.gov/niosh/topics/emres/surface-sampling-bacillus-anthracis.html
- 11. Clesceri LS, Eaton AD, Greenberg AE, Franson MA. (ed). 1998. Standard methods for the examination of water and wastewater, 20th ed American Public Health Association, Washington, DC [Google Scholar]
- 12. Dancer SJ. 2004. How do we assess hospital cleaning? A proposal for microbiological standards for surface hygiene in hospitals. J. Hosp. Infect. 56:10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Da Silva SM, Filliben JJ, Morrow JB. 2011. Parameters affecting spore recovery from wipes used in biological surface sampling. Appl. Environ. Microbiol. 77:2374–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desai R, et al. 2011. Survival and transmission of community-associated methicillin-resistant Staphylococcus aureus from fomites. Am. J. Infect. Control 39:219–225 [DOI] [PubMed] [Google Scholar]
- 15. Downes FP, Ito K. (ed). 2001. Compendium of methods for the microbiological examination of foods. American Public Health Asssociation, Washington, DC [Google Scholar]
- 16. Dubeau D, Deziel E, Woods DE, Lepine F. 2009. Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol. 9:263 doi:10.1186/1471-2180-9-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edmonds JM. 2009. Efficient methods for large-area surface sampling of sites contaminated with pathogenic microorganisms and other hazardous agents: current state, needs, and perspectives. Appl. Microbiol. Biotechnol. 84:811–816 [DOI] [PubMed] [Google Scholar]
- 18. Edmonds JM, et al. 2009. Surface sampling of spores in dry-deposition aerosols. Appl. Environ. Microbiol. 75:39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eickhoff TC. 1974. Microbiologic sampling of the hospital environment. Health Lab. Sci. 11:73–75 [PubMed] [Google Scholar]
- 20. Estill CF, et al. 2009. Recovery efficiency and limit of detection of aerosolized Bacillus anthracis Sterne from environmental surface samples. Appl. Environ. Microbiol. 75:4297–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Faille C, et al. 2002. Adhesion of Bacillus spores and Escherichia coli cells to inert surfaces: role of surface hydrophobicity. Can. J. Microbiol. 48:728–738 [DOI] [PubMed] [Google Scholar]
- 22. Finch JE, Prince J, Hawksworth M. 1978. A bacteriological survey of the domestic environment. J. Appl. Bacteriol. 45:357–364 [DOI] [PubMed] [Google Scholar]
- 23. Frawley DA, et al. 2008. Recovery efficiencies of anthrax spores and ricin from nonporous or nonabsorbent and porous or absorbent surfaces by a variety of sampling methods. J. Forensic Sci. 53:1102–1107 [DOI] [PubMed] [Google Scholar]
- 24. Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. 2008. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect. Immun. 76:5402–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herigstad B, Hamilton M, Heersink J. 2001. How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods 44:121–129 [DOI] [PubMed] [Google Scholar]
- 26. Hoben HJ, Somasegaran P. 1982. Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. Appl. Environ. Microbiol. 44:1246–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hodges LR, Rose LJ, O'Connell H, Arduino MJ. 2010. National validation study of a swab protocol for the recovery of Bacillus anthracis spores from surfaces. J. Microbiol. Methods 81:141–146 [DOI] [PubMed] [Google Scholar]
- 28. Hodges LR, Rose LJ, Peterson A, Noble-Wang J, Arduino MJ. 2006. Evaluation of a macrofoam swab protocol for the recovery of Bacillus anthracis spores from a steel surface. Appl. Environ. Microbiol. 72:4429–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong-Geller E, et al. 2010. Evaluation of Bacillus anthracis and Yersinia pestis sample collection from nonporous surfaces by quantitative real-time PCR. Lett. Appl. Microbiol. 50:431–437 [DOI] [PubMed] [Google Scholar]
- 30. Hubbard K, Emanuel P, Pellar G. 30 October 2011. Suitability of commercial transport media for biological pathogens under nonideal conditions. Int. J. Microbiol. [Epub ahead of print.] doi:10.1155/2011/463096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Julian TR, Tamayo FJ, Leckie JO, Boehm AB. 2011. Comparison of surface sampling methods for virus recovery from fomites. Appl. Environ. Microbiol. 77:6918–6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kerwin BA. 2008. Polysorbates 20 and 80 used in the formulation of protein biotherapeutics: structure and degradation pathways. J. Pharm. Sci. 97:2924–2935 [DOI] [PubMed] [Google Scholar]
- 33. Kim HS, et al. 2005. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics 6:174 doi:10.1186/1471-2164-6-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim SW, et al. 2008. Optimizing the recovery of surrogates for bacterial bioterrorism agents from ventilation filters. Clean 36:601–608 [Google Scholar]
- 35. Kirschner LE, Puleo JR. 1979. Wipe-rinse technique for quantitating microbial contamination on large surfaces. Appl. Environ. Microbiol. 38:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kovacevic J, et al. 2009. Evaluation of environmental sampling methods and rapid detection assays for recovery and identification of Listeria spp. from meat processing facilities. J. Food Prot. 72:696–701 [DOI] [PubMed] [Google Scholar]
- 37. Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 6:130 doi:10.1186/1471-2334-6-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Letant SE, et al. 2011. Rapid-viability PCR method for detection of live, virulent Bacillus anthracis in environmental samples. Appl. Environ. Microbiol. 77:6570–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lewandowski R, Kozlowska K, Szpakowska M, Stepinska M, Trafny EA. 2010. Use of a foam spatula for sampling surfaces after bioaerosol deposition. Appl. Environ. Microbiol. 76:688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marshall RT. (ed). 2001. Standard methods for the examination of dairy products, 17th ed American Public Health Association, Washington, DC [Google Scholar]
- 41. Moore G, Griffith C. 2002. A comparison of surface sampling methods for detecting coliforms on food contact surfaces. Food Microbiol. 19:65–73 [Google Scholar]
- 42. Moore G, Griffith C. 2007. Problems associated with traditional hygiene swabbing: the need for in-house standardization. J. Appl. Microbiol. 103:1090–1103 [DOI] [PubMed] [Google Scholar]
- 43. Nellen J, Rettberg P, Horneck G, Streit WR. 2006. Planetary protection—approaching uncultivable microorganisms. Adv. Space Res. 38:1266–1270 [Google Scholar]
- 44. Nyachuba DG, Donnelly CW. 2007. Comparison of 3M Petrifilm environmental Listeria plates against standard enrichment methods for the detection of Listeria monocytogenes of epidemiological significance from environmental surfaces. J. Food Sci. 72:M346–M354 [DOI] [PubMed] [Google Scholar]
- 45. Obee P, Griffith CJ, Cooper RA, Bennion NE. 2007. An evaluation of different methods for the recovery of meticillin-resistant Staphylococcus aureus from environmental surfaces. J. Hosp. Infect. 65:35–41 [DOI] [PubMed] [Google Scholar]
- 46. Perry A, et al. 2010. Evaluation of swab processing methods for recovery of Yersinia pestis, p 9 Abstr. 4th Natl. Bio-Threat Conf., New Orleans, LA [Google Scholar]
- 47. Rhodes KA, Government Accountability Office US 2005. Anthrax detection: agencies need to validate sampling activities in order to increase confidence in negative results. GAO-05-251 Government Accountability Office, Washington, DC: http://www.gao.gov/new.items/d05251.pdf [Google Scholar]
- 48. Rose L, Jensen B, Peterson A, Banerjee SN, Arduino MJ. 2004. Swab materials and Bacillus anthracis spore recovery from nonporous surfaces. Emerg. Infect. Dis. 10:1023–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rose LJ, Hodges L, O'Connell H, Noble-Wang J. 2011. National validation study of a cellulose sponge wipe-processing method for use after sampling Bacillus anthracis spores from surfaces. Appl. Environ. Microbiol. 77:8355–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scott E, Bloomfield SF, Barlow CG. 1982. An investigation of microbial contamination in the home. J. Hyg. (Lond.) 89:279–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sim BM, et al. 2010. Genomic acquisition of a capsular polysaccharide virulence cluster by non-pathogenic Burkholderia isolates. Genome Biol. 11:R89 doi:10.1186/gb-2010-11-8-r89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skrivanona E, Marounek M, Benda V, Brezina P. 2006. Susceptibility of Escherichia coli, Salmonella sp. and Clostridium perfringens to organic acids and monolaurin. Vet. Med. (Prague) 51:81–88 [Google Scholar]
- 53. Straka RP, Stokes JL. 1957. Rapid destruction of bacteria in commonly used diluents and its elimination. Appl. Microbiol. 5:21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tomioka H, Saito H. 1984. Mechanism of mycobacteriocin and Tween 80 mediated antimycobacterial activity. J. Gen. Microbiol. 130:3085–3089 [DOI] [PubMed] [Google Scholar]
- 55. Tsukamura M. 1991. Some observations on the mechanism of bactericidal activity of Tween 80 on mycobacteria. Kekkaku 66:75–79 [PubMed] [Google Scholar]
- 56. Uhlemann AC, et al. 2011. The environment as an unrecognized reservoir for community-associated methicillin resistant Staphylococcus aureus USA300: a case-control study. PLoS One 6:e22407 doi:10.1371/journal.pone.0022407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. US Department of Agriculture, Food Safety and Inspection Service 2003. Control of Listeria monocytogenes in ready-to-eat meat and poultry products; final rule. 9 CFR, part 430 US Department of Agriculture, Washington, DC [Google Scholar]
- 58. Valentine NB, et al. 2008. Evaluation of sampling tools for environmental sampling of bacterial endospores from porous and nonporous surfaces. J. Appl. Microbiol. 105:1107–1113 [DOI] [PubMed] [Google Scholar]
- 59. Vorst KL, Todd EC, Rysert ET. 2004. Improved quantitative recovery of Listeria monocytogenes from stainless steel surfaces using a one-ply composite tissue. J. Food Prot. 67:2212–2217 [DOI] [PubMed] [Google Scholar]
- 60. Walker RE, Petersen JM, Stephens KW, Dauphin LA. 2010. Optimal swab processing recovery method for detection of bioterrorism-related Francisella tularensis by real-time PCR. J. Microbiol. Methods 83:42–47 [DOI] [PubMed] [Google Scholar]
- 61. Walsh RL, Camilli A. 2011. Streptococcus pneumoniae is desiccation tolerant and infectious upon rehydration. mBio 2(3):e00092–11 doi:10.1128/mBio.00092-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wiener-Well Y, et al. 2011. Nursing and physician attire as possible source of nosocomial infections. Am. J. Infect. Control 39:555–559 [DOI] [PubMed] [Google Scholar]
- 63. Wong HC, Wang P. 2004. Induction of viable but nonculturable state in Vibrio parahaemolyticus and its susceptibility to environmental stresses. J. Appl. Microbiol. 96:359–366 [DOI] [PubMed] [Google Scholar]
- 64. Yang J, Hou X, Mir PS, McAllister TA. 2010. Anti-Escherichia coli O157:H7 activity of free fatty acids under varying pH. Can. J. Microbiol. 56:263–267 [DOI] [PubMed] [Google Scholar]
- 65. Yu Y, et al. 2006. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 6:46 doi:10.1186/1471-2180-6-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y, Zhang H, Sun Z. 2003. Susceptibility of Mycobacterium tuberculosis to weak acids. J. Antimicrob. Chemother. 52:56–60 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.