Abstract
Neonatal meningitis Escherichia coli (NMEC) is one of the top causes of neonatal meningitis worldwide. Here, 85 NMEC and 204 fecal E. coli isolates from healthy humans (HFEC) were compared for possession of traits related to virulence, antimicrobial resistance, and plasmid content. This comparison was done to identify traits that typify NMEC and distinguish it from commensal strains to refine the definition of the NMEC subpathotype, identify traits that might contribute to NMEC pathogenesis, and facilitate choices of NMEC strains for future study. A large number of E. coli strains from both groups were untypeable, with the most common serogroups occurring among NMEC being O18, followed by O83, O7, O12, and O1. NMEC strains were more likely than HFEC strains to be assigned to the B2 phylogenetic group. Few NMEC or HFEC strains were resistant to antimicrobials. Genes that best discriminated between NMEC and HFEC strains and that were present in more than 50% of NMEC isolates were mainly from extraintestinal pathogenic E. coli genomic and plasmid pathogenicity islands. Several of these defining traits had not previously been associated with NMEC pathogenesis, are of unknown function, and are plasmid located. Several genes that had been previously associated with NMEC virulence did not dominate among the NMEC isolates. These data suggest that there is much about NMEC virulence that is unknown and that there are pitfalls to studying single NMEC isolates to represent the entire subpathotype.
INTRODUCTION
Despite advances in antimicrobial therapy, Gram-negative neonatal bacterial meningitis continues to be a major cause of mortality and morbidity worldwide, with neonatal meningitis Escherichia coli (NMEC) being one of the top two causes of neonatal bacterial meningitis in industrialized countries, along with group B streptococci (22, 23, 26). Most of the survivors of Gram-negative neonatal bacterial meningitis suffer neurologic sequelae or developmental abnormalities (23), and if left untreated, this disease is usually fatal. Better control of NMEC diseases is desirable in order to alleviate human suffering and to decrease the financial burden caused by bacterial meningitis.
Unfortunately, current knowledge of NMEC pathogenesis is incomplete, complicating efforts to control or eliminate this disease. In the review of E. coli pathogenicity by Croxen and Finlay (5), deficits in our understanding of NMEC pathogenesis become apparent. For instance, it is clear that many NMEC strains lack cnf1, a factor thought to be critical in NMEC pathogenesis (19). In addition, we have shown that NMEC strains tend to harbor genes found among plasmid-located pathogenicity islands (PAIs) in avian pathogenic E. coli (APEC) (19). Such plasmids are frequently transferable by conjugation (9, 12, 13, 17, 31) and have been shown to confer on their host E. coli the abilities to survive in the bloodstream, traverse the blood-brain barrier of mammals, and cause bacteremia and meningitis in the rat model of human disease (13, 32). In some cases, such virulence plasmids also harbor resistance genes conferring multidrug resistance on their host bacterium (13). Despite the fact that it has been long known that plasmids occur in NMEC (26, 29, 32) and are linked to E. coli virulence (16), their contributions to NMEC pathogenesis are ill defined and understudied. Thus, there appears to be some deficits, perhaps important ones, in our knowledge of NMEC pathogenesis. Here, we seek to lay the groundwork for addressing these knowledge gaps by determining what traits distinguish NMEC from human fecal commensal E. coli.
MATERIALS AND METHODS
Bacterial strains.
A total of 289 E. coli isolates were used in this study, including 85 from cases of human neonatal meningitis (NMEC) and 204 from the feces of healthy human hosts (HFEC). Seventy of the NMEC isolates came from the cerebrospinal fluid of newborns in The Netherlands and were isolated between 1989 and 1997 (11) (courtesy of L. Spanjaard). The remaining NMEC isolates were isolated over the same time period from patients in the United States and were provided by K. S. Kim and C. DebRoy. The HFEC strains were obtained from two sources: 179 were provided to us by J. Johnson, and the remainder was isolated for this study as follows. Human fecal samples were collected from healthy human volunteers at North Dakota State University using internal review board (IRB)-approved protocols. Swabs were collected from the rectum of healthy volunteers and transferred to Cary Blair transport medium. On receipt at the lab, the swabs were transferred to 5 ml of buffered peptone water (Difco, BD Diagnostics, NJ) and incubated at 37°C for 18 to 24 h. Following incubation, a loopful of the enrichment broth was streaked out on MacConkey agar (MAC; Difco) and eosin methylene blue agar (EMB; Difco) plates and incubated at 37°C for 18 to 24 h. Suspect colonies on MAC (bright pink with a dimple) and EMB (green colonies with a metallic sheen) were selected to tryptone soy agar (TSA; Difco) for purification, and plates were incubated at 37°C for 18 to 24 h. Identification was carried out using the Sensititre GNID panels (Trek Diagnostics, Cleveland, OH), with incubation of the panels at 37°C for 18 h. All organisms were stored at −80°C in brain heart infusion broth (Difco Laboratories, Detroit, MI) with 20% (vol/vol) glycerol until use (28). All NMEC strains were provided to us with the serogrouping already completed, whereas all HFEC strains were serogrouped through the Escherichia coli Reference Center (Pennsylvania State University, University Park, PA).
Hemolytic reaction.
Test and control organisms were plated on 5% sheep blood agar plates (BD Diagnostics) and incubated overnight at 37°C. The plates were then examined for “greening” or clearing of the blood agar around areas of bacterial growth as an indication of hemolytic activity (8).
Fermentation of lactose.
Test and control organisms were plated on MacConkey agar (Difco) and incubated overnight at 37°C. Isolates were considered positive for lactose utilization if pink colonies were observed (8).
Phylogenetic typing.
Isolates were assigned to phylogenetic groups according to the PCR amplification method described by Clermont et al. (4).
Multiplex PCR genotyping and plasmid replicon typing.
Test and control organisms were examined for the presence of a number of ExPEC virulence genes, including different allelic variants, genes associated with the pathogenicity islands (PAIs) of large ExPEC plasmids, plasmid replicons, and genes of unknown function found in genomic islands of an ExPEC isolate known as APEC O1 (14) using a number of multiplex PCR assays (see Table S1 in the supplemental material). Some of these multiplex panels have been previously described (12, 17, 18, 20, 27). All primers were obtained from Integrated DNA Technologies (Coralville, IA). In all, multiplex panels targeting 182 gene products were used, making this study the most extensive comparative genotypic analysis of NMEC and HFEC populations to date. PCR was performed as previously described (27). Strains known to possess or lack the genes of interest were examined with each amplification procedure. Reactions were performed twice. An isolate was considered to contain a gene of interest if it produced an amplicon of the expected size.
Antimicrobial susceptibility profiles.
All isolates were subjected to antimicrobial susceptibility testing using the broth microdilution assay and the National Antimicrobial Resistance Monitoring Scheme (NARMS) panels (CMV1AGNF, Sensititre; Trek Diagnostics, Cleveland, OH), as described previously (15). Procedures, control strains (including Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, and Staphylococcus aureus ATCC 29213) and interpretative criteria were as specified by the Clinical and Laboratory Standards Institute (CLSI) and NARMS (1). The NARMS panel tests antimicrobial susceptibility to the following antimicrobials: amikacin (0.5 to 64 μg/ml), ampicillin (1 to 32 μg/ml), amoxicillin-clavulanic acid (1/0.5 to 32/16 μg/ml), ceftriaxone (0.25 to 64 μg/ml), chloramphenicol (2 to 32 μg/ml), ciprofloxacin (0.015 to 4 μg/ml), trimethoprim-sulfamethoxazole (0.12/2.38 to 4/76 μg/ml), cefoxitin (0.5 to 32 μg/ml), gentamicin (0.25 to 16 μg/ml), kanamycin (8 to 64 μg/ml), nalidixic acid (0.5 to 32 μg/ml), sulfisoxazole (15 to 256 μg/ml), streptomycin (32 to 64 μg/ml), tetracycline (4 to 32 μg/ml), and ceftiofur (0.12 to 8 μg/ml). Resistance or susceptibility of the strain to an antimicrobial was determined according to the observed MIC and breakpoints recommended by CLSI and NARMS (1).
Breakpoints used in this study were amikacin, ≥64 μg/ml; ampicillin, ≥32 μg/ml; amoxicillin-clavulanic acid, ≥32/16 μg/ml; ceftriaxone, ≥4 μg/ml; chloramphenicol, ≥32 μg/ml; ciprofloxacin, ≥4 μg/ml; trimethoprim-sulfamethoxazole, ≥4/76 μg/ml; cefoxitin, ≥32 μg/ml; gentamicin, ≥16 μg/ml; kanamycin, ≥64 μg/ml; nalidixic acid, ≥32 μg/ml; sulfisoxazole, ≥512 μg/ml; streptomycin, ≥64 μg/ml; tetracycline, ≥16 μg/ml; and ceftiofur, ≥8 μg/ml.
Biostatistics.
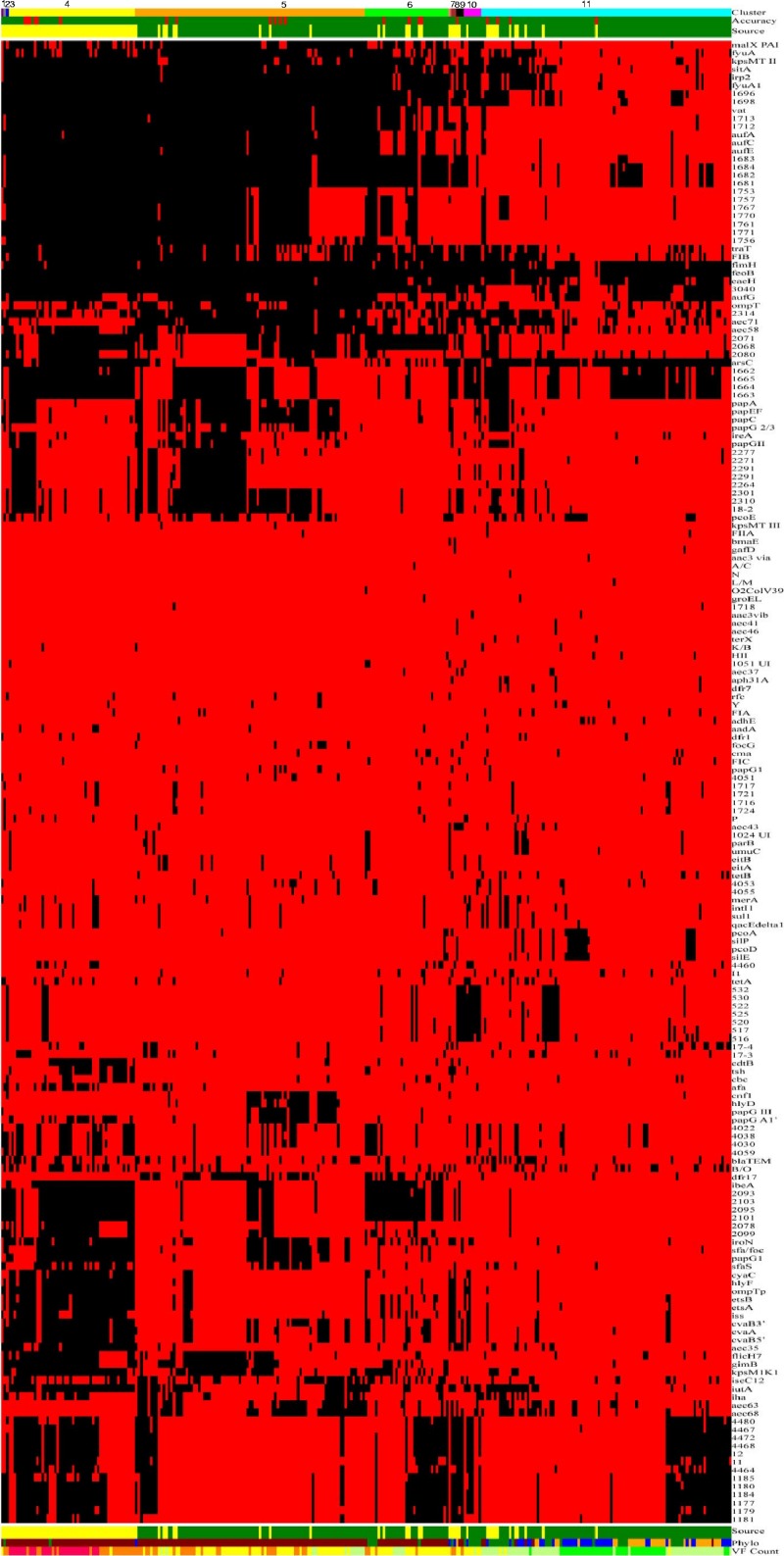
A chi-square test of homogeneity was used for comparison between groups, and Fisher's exact test was used where the assumptions of the chi-square test did not hold (30). In a further attempt to discern patterns among all isolates based on their content of virulence genes, a linear discriminant analysis (LDA) was used to determine if an isolate type (NMEC or HFEC) could be predicted based on the virulence genes present (10). Although the use of data from binary variables in an LDA, as done here, violates the assumption of multivariate normality, LDA was used since parametric LDA can be very robust in spite of such violations (24). Additionally, a cluster analysis of the isolates was performed using the average linkage method based upon Jaccard's dissimilarity coefficient calculated from the presence of virulence genes (SAS 9.22) (2). In order to better discern patterns among the isolates, results of the cluster and discriminant analyses and the isolates' virulence genotypes and phylogenetic groups were used to construct a single figure based on the principles of Eisen et al. (6) (Fig. 1).
Fig 1.
Gel diagram merging the cluster analysis and LDA of the genotyping results of NMEC and HFEC strains. The first row (the uppermost row at the top of the figure) identifies the clusters to which the isolates were assigned based on their genotypes. There are 11 identified clusters: 1, magenta; 2, gray; 3, navy blue; 4, yellow; 5, orange; 6, lime green; 7, salmon; 8, brick red; 9, black; 10, bright pink; and 11, cyan. The second row shows the accuracy of prediction from the linear discriminant analysis (LDA) of an isolate type based on possession of genes/traits. Green indicates a correct prediction as to whether an isolate is an NMEC or an HFEC strain, whereas red indicates a misprediction, with 21 instances observed. The third row indicates the source of the isolates, where HFEC strains are denoted as green, and NMEC strains are identified in yellow. The next set of rows of red and black bars (rows 4 through 185; not numbered) shows the results for a single gene or trait. The identity of each gene tested is shown in the column at the far right of the diagram (below cluster, accuracy, and source). In the body of the figure, a black line means that the gene of interest is present in a particular isolate, whereas a red line means the gene is absent. The row following the black and red pattern (row 186) shows the source of the isolates (same as the third row from the top). Row 187 (made up of brown, green, blue, and orange colors) indicates the phylogenetic groups of each isolate: blue, phylogenetic group A; orange, B1; brown, B2; and green, D. Row 188 (the last row of the figure) indicates the gene/trait content for each strain, where isolates in category 1 contain 1 to 20 of the tested genes/traits and are identified as dark lime; category 2 isolates are identified in light lime and possess 20 to 40 genes; category 3 isolates are identified in yellow and possess 40 to 60 genes; category 4 isolates are identified in orange and possess 60 to 80 genes; and category 5 isolates are identified in bright pink and possess 80 to 100 genes. This figure is also available online at http://ecoli.cvm.iastate.edu:81/blast/images/Figure1nmec.jpg.
RESULTS
Serogroups.
Of the 85 NMEC isolates, only 65.9% (56 isolates) could be classified to a single O serogroup using standard antisera, while 77.5% of the HFEC isolates classified to a single O serogroup (Table 1). A large number of NMEC and HFEC isolates could not be serogrouped because they were rough, autoagglutinated, grouped to multiple serogroups, or untypeable. Among the NMEC isolates that were serogrouped, 18 serogroups were represented, with seven of these being unique to NMEC. Among the HFEC isolates, 54 different serogroups were found, with 43 of these unique to HFEC. Around 54% (46) of the typeable NMEC isolates and 30.9% (63) of the typeable HFEC isolates shared serogroups. The most common serogroups found among the NMEC isolates were O18, followed by O83, O7, O12, and O1; among the HFEC, the most commonly occurring serogroups were O6, followed by O2, O25, O1, O75, O18, O8, O4, and O15.
Table 1.
Serogroups among NMEC and HFEC isolates
| Serogroup | No. (%) of isolates |
|
|---|---|---|
| NMEC (n = 85) | HFEC (n = 204) | |
| Shared | ||
| O1 | 4 | 9 |
| O4 | 1 | 5 |
| O6 | 2 | 21 |
| O7 | 6 | 1 |
| O12 | 4 | 1 |
| O16 | 1 | 3 |
| O18 | 17 | 7 |
| O21 | 2 | 2 |
| O25 | 1 | 11 |
| O78 | 1 | 1 |
| O83 | 7 | 2 |
| Subtotal | 46 | 63 |
| % total shared | (54.1) | (30.9) |
| Unshared | ||
| O2 | 0 | 21 |
| O8 | 0 | 6 |
| O9 | 1 | 0 |
| O11 | 0 | 3 |
| O14 | 2 | 0 |
| O15 | 0 | 4 |
| O19 | 0 | 3 |
| O23 | 2 | 0 |
| O29 | 0 | 1 |
| O37 | 0 | 2 |
| O38 | 0 | 1 |
| O44 | 0 | 1 |
| O45 | 2 | 0 |
| O46 | 0 | 1 |
| O48 | 0 | 1 |
| O54 | 0 | 2 |
| O55 | 0 | 1 |
| O68 | 0 | 1 |
| O73 | 0 | 3 |
| O74 | 0 | 2 |
| O75 | 0 | 8 |
| O77 | 0 | 1 |
| O81 | 0 | 1 |
| O84 | 1 | 0 |
| O86 | 0 | 3 |
| O88 | 0 | 1 |
| O90 | 0 | 1 |
| O97 | 0 | 1 |
| O101 | 0 | 2 |
| O102 | 0 | 1 |
| O104 | 0 | 1 |
| O105 | 0 | 1 |
| O106 | 0 | 2 |
| O109 | 0 | 1 |
| O113 | 0 | 3 |
| O117 | 0 | 2 |
| O118 | 0 | 1 |
| O120 | 0 | 2 |
| O131 | 0 | 1 |
| O135 | 1 | 0 |
| O138 | 0 | 1 |
| O147 | 0 | 1 |
| O148 | 0 | 1 |
| O153 | 0 | 1 |
| O154 | 0 | 1 |
| O156 | 0 | 1 |
| O166 | 1 | 0 |
| O167 | 0 | 1 |
| O168 | 0 | 1 |
| O171 | 0 | 1 |
| Subtotal | 10 | 95 |
| % total unshared | (11.8) | (46.6) |
| Total no. (%) typed | 56 (65.9) | 158 (77.5) |
| None (not serogrouped) | ||
| Nontypeable | 29 | 45 |
| Multiple serogroups | 0 | 1 |
| Subtotal | 29 | 46 |
| % total untyped | (34.1) | (22.5) |
Lactose utilization and hemolytic activity.
NMEC and HFEC isolates did not differ significantly (P = 0.43) in their abilities to hemolyze blood, with a large majority of both being nonhemolytic. Although over 90% of both NMEC and HFEC isolates fermented lactose, significantly (P = 0.01) more NMEC isolates than HFEC isolates were nonfermenters.
Phylogenetic typing of NMEC and HFEC isolates.
NMEC and HFEC isolates differed significantly in their assignments to phylogenetic groups (P = 0.0002), despite the fact that the majority of both (79% of NMEC and 54% of HFEC isolates) were assigned to group B2 (Table 2).
Table 2.
Assignment of isolates to phylogenetic groupsa
| Isolate | No. (%) of isolates by group |
Total no. of isolates | |||
|---|---|---|---|---|---|
| A | B1 | B2 | D | ||
| NMEC | 9 (10.6) | 1 (1.2) | 67 (78.8) | 8 (9.4) | 85 |
| HFEC | 33 (16.2) | 32 (15.7) | 110 (53.9) | 29 (14.2) | 204 |
| Total | 42 | 33 | 174 | 37 | 289 |
Chi-square test for homogeneous assignment to phylogenetic group: χ2 = 19.51, df = 3, P = 0.0002.
Genotypes and plasmid replicon content.
Based on a cluster analysis of their genotypes, NMEC and HFEC isolates were assigned to 11 statistically distinct clusters (Fig. 1), with one or more NMEC strain assigned to each cluster. The majority of NMEC isolates were assigned to cluster 4, a cluster whose members were characterized by their possession of genes of plasmid PAIs, unknown function, the yersiniabactin operon, known PAIs, and putative PAIs.
Linear discriminant analysis of genotyping data found that 21 of 289 isolate assignments were mispredictions. That is, 18 NMEC isolates were incorrectly predicted to be HFEC, while three HFEC isolates were inaccurately predicted to be NMEC.
Virulence gene/trait count analysis of the isolates examined in the study found that the numbers of genes/traits were greatest among the NMEC isolates (pink group; 80 to 100 genes possessed), while the HFEC isolates were likely to possess considerably fewer of the genes/traits (green and yellow groups; 40 to 60 genes possessed).
Several genes were identified in the genotype analysis that occurred in the majority of NMEC isolates (≥50%) but were found infrequently in HFEC isolates (Table 3 and Fig. 1; see also Table S2 in the supplemental material), making them good discriminators of NMEC from HFEC. Identified genes included those associated with ExPEC plasmid PAIs, i.e., ompTp, hlyF, cvaC, etsA, cvaA, etsB, cvaB, iss, iutA, and tsh (Table 3). Also, certain genes of unknown function found in the genomic islands of ExPEC strain APEC O1 (i.e., 2093, 2095, 2102, and 2103), aec35 (an lacI family transcriptional regulator of APEC strain BEN2908's PAI), and certain known chromosomal virulence genes (i.e., sfaS, cdtB, and papGI) were also found to be good discriminators, though not all occurred in over half of the NMEC isolates studied (Table 3). In addition, NMEC isolates were more likely to harbor the FIB replicon, a replicon type that typifies ExPEC virulence plasmids but is also common in many wild-type E. coli isolates (16, 20).
Table 3.
The 20 traits with the greatest differential in percent positives between NMEC and HFEC isolates
| Gene/traita | No. (%) of isolates containing trait |
Chi-square value | P | |
|---|---|---|---|---|
| NMEC (n = 85) | HFEC (n = 204) | |||
| aec35 | 64 (75.3) | 4(2) | 179.33 | <0.0001 |
| ompTp | 57 (67.1) | 8 (3.9) | 137.20 | <0.0001 |
| hlyF | 53 (62.4) | 8(3.9) | 123.02 | <0.0001 |
| cvaC | 49 (57.6) | 8 (3.9) | 109.38 | <0.0001 |
| etsA | 54 (63.5) | 14 (6.9) | 107.08 | <0.0001 |
| cvaA | 61 (71.8) | 29 (14.2) | 92.67 | <0.0001 |
| etsB | 52 (61.2) | 17 (8.3) | 92.18 | <0.0001 |
| cvaB5′ | 58 (68.2) | 27 (13.2) | 87.42 | <0.0001 |
| iss | 48 (56.5) | 15 (7.4) | 84.91 | <0.0001 |
| sfaS | 42 (49.4) | 14 (6.9) | 69.53 | <0.0001 |
| iutA | 68 (80) | 55 (27) | 69.04 | <0.0001 |
| 2093 | 55 (64.7) | 33 (16.2) | 66.72 | <0.0001 |
| 2101 | 55 (64.7) | 33 (16.2) | 66.72 | <0.0001 |
| cdtB | 32 (37.7) | 5 (2.5) | 66.58 | <0.0001 |
| tsh | 28 (32.9) | 2 (0.9) | 65.88 | <0.0001 |
| 2103 | 55 (64.7) | 34 (16.7) | 64.71 | <0.0001 |
| 2095 | 55 (64.7) | 34 (16.7) | 64.701 | <0.0001 |
| cvaB3′ | 55 (64.7) | 35 (17.2) | 64.71 | <0.0001 |
| 2078 | 33 (38.8) | 7 (3.4) | 63.02 | <0.0001 |
| papG1* | 48 (56.5) | 28 (13.7) | 56.56 | <0.0001 |
The traits are listed in descending order based on the magnitude of the chi-square value, with the most powerful trait being aec35, which contributes to the virulence of APEC BEN2908. The next 8 traits have been found among the PAIs of large APEC virulence plasmids. The tenth trait listed, sfaS, occurs in just under half of the NMEC isolates examined. The next two traits, 2093 and 2101, were originally found in a genomic island of APEC O1 and have not previously been ascribed a function or associated with ExPEC virulence. The remaining traits had smaller differences in prevalence rates and thus lower chi-square values, either because they occurred relatively more frequently in HFEC isolates or relatively less frequently in NMEC isolates than the more discriminating traits. Interestingly, many of the better-studied NMEC virulence traits, such as gimB, ibeA, or cnf1, did not make this list, suggesting that there is much yet unknown about NMEC pathogenesis.
It was especially interesting that NMEC and HFEC were significantly different in their possession of genes from APEC genomic islands. In fact, other than the plasmid PAI genes, several of these genomic island genes were among the best discriminators of NMEC and HFEC. For instance, gene target aec35 occurred in ∼75% of NMEC isolates but only in 2% of HFEC isolates. This difference was very highly significant (χ2 = 179.3282, P < 0.0001), suggesting that this gene or the island in which it is contained may be involved in NMEC pathogenesis or fitness.
Several genes known for their contributions to NMEC pathogenesis were not as widespread among NMEC isolates as other traits implicated in pathogenesis, including cnf1, which occurred in less than 5% of NMEC isolates; ibeA and gimB, which occurred in 60% of NMEC isolates; and sfa-foc, which occurred in only 55.3% of NMEC isolates. Several other traits occurred in significantly more NMEC than HFEC isolates but did not occur in the majority of NMEC isolates tested (bmaE, papGI′, cdtB, B/O, and P replicon types and certain genomic island genes of unknown function) (see Table S2 in the supplemental material and Table 3). In addition, some genes that occurred in a high number of NMEC isolates showed a significant association with NMEC but were also found in a majority of HFEC isolates, rendering them less useful in accurately predicting NMEC or HFEC status but still typical of the NMEC subpathotype (e.g., sitA, traT, eaeH, fyuA, irp2, kpsMTII, malX PAI, vat, FIB replicon type, and other genes of unknown function) (Table 3).
Antimicrobial susceptibility profiles.
NMEC and HFEC isolates were also compared by their susceptibility to several antimicrobial agents (Table 4). Almost one-third of the NMEC isolates were resistant to streptomycin and sulfisoxazole, while about one-fifth were resistant to ampicillin, 15% were resistant to tetracycline, and a few were resistant to chloramphenicol, kanamycin, and trimethoprim-sulfamethoxazole. Antimicrobial resistance was also observed in some of the HFEC isolates examined, with resistance to tetracycline (13.7%), sulfisoxazole (12.3%), and trimethoprim-sulfamethoxazole (7.4%) being the most common.
Table 4.
Antimicrobial susceptibility profiles of NMEC and HFEC
| Antimicrobial agent | No (%) of isolates resistant |
|
|---|---|---|
| NMEC | HFEC | |
| Amikacin | 0 (0) | 0 (0) |
| Amoxicillin-clavulanic acid | 3 (3.5) | 4 (2) |
| Ampicillin | 20 (23.5) | 37 (18.2) |
| Cefoxitin | 0 (0) | 3 (1.5) |
| Ceftiofur | 0 (0) | 0 (0) |
| Ceftriaxone | 0 (0) | 0 (0) |
| Chloramphenicol | 2 (2.4) | 6 (3) |
| Ciprofloxacin | 0 (0) | 1 (0.5) |
| Gentamicin | 0 (0) | 2 (1) |
| Kanamycin | 2 (2.4) | 0 (0) |
| Nalidixic acid | 0 (0) | 1 (0.5) |
| Streptomycin | 26 (30.6) | 13 (6.4) |
| Sulfisoxazole | 25 (29.4) | 25 (12.3) |
| Tetracycline | 13 (15.3) | 28 (13.7) |
| Trimethoprim-sulfamethoxazole | 6 (7.1) | 15 (7.4) |
DISCUSSION
If the E. coli strains causing meningitis in human neonates use a common set of virulence factors to do so, they would constitute an ExPEC subpathotype (21). In an effort to identify common traits that unite the NMEC subpathotype, 85 E. coli isolates from cases of human neonatal bacterial meningitis were compared to 204 isolates from the feces of healthy human hosts for possession of various traits, including those associated with ExPEC virulence, resistance, and genomic and plasmid islands. As it turns out, typical NMEC isolates ferment lactose but do not hemolyze blood. This pattern is also true of HFEC, meaning that lactose fermentation and hemolytic ability cannot be used to discriminate HFEC and NMEC. Although NMEC and HFEC strains differ significantly in their distributions among the phylogenetic groups, their patterns of distribution are similar to the majority of NMEC and HFEC strains being assigned to the B2 phylogenetic group. Thus, while assignment to the B2 phylogenetic group is typical of NMEC, phylogenetic grouping also fails to distinguish NMEC from HFEC. This finding is interesting since ExPEC strains are thought to belong predominately to group B2 and to a lesser extent to group D (4), suggesting that several of the HFEC isolates studied here might harbor pathogenic potential or that the association between phylogenetic group and pathogenicity is not distinct in the case of HFEC. For further discussion of this issue, see the work of Bailey et al. (3).
Although there were a few NMEC isolates that belonged to serogroups not found among the HFEC isolates tested, none of these serogroups were common. Indeed, the more common serogroups occurring in NMEC, such as O18, O83, and O7, were also found in HFEC. Antimicrobial susceptibility patterns also failed to provide a clear distinction between NMEC and HFEC, despite the fact that most of the NMEC isolates were harvested from infants in Europe from the late 1980s through the 1990s (11), while the HFEC isolates were harvested from people in North America more recently.
A greater percentage of NMEC isolates than HFEC isolates showed resistance to the antimicrobials streptomycin, sulfisoxazole, and tetracycline, with the biggest differential occurring for streptomycin and sulfisoxazole; but still, these differences were not dramatic, and neither group had 31% or more of its members resistant to any antimicrobial tested. Also, about a fifth of the NMEC isolates were predicted to be HFEC based on their possession of the assessed traits, while less than 2% of HFEC isolates were predicted to be NMEC. Such a high percentage of mispredictions of NMEC as HFEC complicates making a clear definition of the NMEC subpathotype, but it also presents hypotheses for future testing, i.e., that the mispredicted NMEC isolates are opportunistic pathogens, whereas the correctly assigned NMEC isolates are frank pathogens or that NMEC harbor as-of-yet-unknown virulence traits lacking in HFEC that enable NMEC's pathogenic lifestyle.
There were some distinct differences in the occurrence of different plasmid replicons in NMEC and HFEC isolates, with Inc types B/O, P, and FIB significantly more likely (P < 0.01) to occur in NMEC than in HFEC isolates. However, Inc P replicons occurred in few strains overall, and Inc B/O replicons occurred in less than 50% of NMEC isolates and <15% of HFEC isolates. The difference in distribution of the Inc FIB replicon is intriguing since it occurs in over 90% of NMEC isolates and in only 55% of HFEC isolates, a difference that is very highly significant. It is also interesting from a biological standpoint since it is this replicon type that is typical of large ExPEC virulence plasmids, which harbor PAIs (16). The validity of this observation was also reinforced by the findings that most of the plasmid PAI genes assessed in this study were significantly more likely to occur in NMEC than in HFEC isolates and occurred in most of the NMEC and few HFEC isolates, making them among the most discriminatory traits studied here. Thus, it would appear that virulence plasmids typify the NMEC subpathotype.
This finding is significant in that the role of virulence plasmids in the pathogenesis of E. coli-caused neonatal meningitis has been given only limited scrutiny, with much of what we know of them found in literature published over 3 decades ago (29) or based on APEC plasmids and their contributions to meningitis (13). These findings may have important implications for the future study of NMEC pathogenesis, since at least two of the most frequently studied NMEC strains, whose genomes have been completely sequenced (NMEC IHE3034 and RS218) (25), lack these plasmid PAIs. Thus, it would appear that in order to gain a more complete and/or accurate understanding of NMEC pathogenesis, it will be necessary to include plasmid PAI-containing strains in future studies.
Indeed, until it is routinely possible to perform full genome-wide functional analyses of multiple NMEC strains simultaneously, in-depth analysis of a few NMEC strains, selected for their NMEC typicality, might be a useful strategy to maximize the generalizability of the results obtained. Based on the findings of this study, a typical NMEC strain would belong to the B2 phylogenetic group and O18:K1:H7 serotype; be nonhemolytic; ferment lactose; contain a large PAI-containing plasmid, likely of the Inc FIB replicon type; and lack cnf1 and hlyD, while harboring vat and ibeA genes, part of the pap, sfa, auf, and yersiniabactin operons, and certain genes of previously unknown genomic islands. For such a strain to prove a useful reagent in future studies of NMEC pathogenesis, it would also need to cause bacteremia and meningitis in the rat model of human neonatal meningitis and traverse cultured human brain microvascular endothelial cells. Cases could also be made for using other strains to represent the NMEC subpathotype based on these data, but regardless of which strain or strains we use to represent this group, recognition that there are limitations on the generalizability of data generated from their study is in order.
In addition to guiding the selection of NMEC for future study, identification of traits that occur commonly among NMEC strains and that discriminate them from HFEC strains may have value in tracking NMEC strains back to their point of origin, enabling new interventions to prevent transmission of NMEC to neonates. It might also provide useful targets that could be exploited in vaccine design. Thus, determining what traits define the NMEC subpathotype and distinguish them from human commensals would seem to be a useful effort. Assuming this is so, a community effort to collate and expand our NMEC and HFEC collections and assess them for distinguishing traits is highly desirable.
Finally, the results of this study reinforce previous findings (7, 14, 19, 32) that NMEC and APEC subpathotypes share significant similarities. For instance, both contain plasmid PAIs as a dominant and defining trait, and several NMEC strains contain genes of ill-defined function that were originally identified in the genomic islands of APEC O1, the only APEC strain whose full genomic sequence is publicly available (14). Whether these similarities indicate that NMEC and APEC share a lineage or harbor zoonotic potential is not clear, but future comparative studies of representative NMEC and APEC strains will be useful in deciphering the significance of these observed similarities.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the kind gifts of strains from James R. Johnson (University of Minnesota and the VA Medical Center, Minneapolis, MN), Kwang S. Kim (Division of Pediatric Infectious Diseases, School of Medicine, Johns Hopkins University, Baltimore, MD), Chitrita DebRoy (E. coli Reference Center, Pennsylvania State University), and Lodewijk Spanjaard (Netherlands Reference Laboratory for Bacterial Meningitis, Center of Infection and Immunity, Amsterdam, The Netherlands).
This project was funded by Iowa State University College of Veterinary Medicine incentive funds.
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Anonymous 2009. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS) 2009 annual report. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 2. Anonymous 2010. SAS/STAT(R) 9.22 user's guide. SAS Institute Inc., Cary, NC [Google Scholar]
- 3. Bailey JK, Pinyon JL, Anantham S, Hall RM. 2010. Distribution of human commensal Escherichia coli phylogenetic groups. J. Clin. Microbiol. 48:3455–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8:26–38 [DOI] [PubMed] [Google Scholar]
- 6. Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ewers C, et al. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163–176 [DOI] [PubMed] [Google Scholar]
- 8. Forbes BA, Sahm DF, Weissfeld AS, Baron EJ. 1998. Bailey & Scott's diagnostic microbiology, 10th ed Mosby, St. Louis, MO [Google Scholar]
- 9. Ginns CA, et al. 2000. Colonization of the respiratory tract by a virulent strain of avian Escherichia coli requires carriage of a conjugative plasmid. Infect. Immun. 68:1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huberty CJ. 1994. Applied discriminant analysis. Wiley, New York, NY [Google Scholar]
- 11. Johnson JR, Oswald E, O'Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185:774–784 [DOI] [PubMed] [Google Scholar]
- 12. Johnson TJ, Johnson SJ, Nolan LK. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 188:5975–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson TJ, et al. 2010. Sequence analysis and characterization of a transferrable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli. Infect. Immun. 78:1931–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson TJ, et al. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 189:3228–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson TJ, et al. 2012. Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia coli from humans and poultry. Foodborne Pathog. Dis. 9:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73:750–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson TJ, Siek KE, Johnson SJ, Nolan LK. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J. Bacteriol. 188:745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnson TJ, Wannemeuhler YM, Scaccianoce JA, Johnson SJ, Nolan LK. 2006. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3929–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson TJ, et al. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74:7043–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson TJ, et al. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73:1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 22. Kim KS. 2006. Meningitis-associated Escherichia coli. ASM Press, Washington, DC [Google Scholar]
- 23. Korhonen TK, et al. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48:486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McLachlan GJ. 1992. Discriminant analysis and statistical pattern recognition. Wiley, New York, NY [Google Scholar]
- 25. Moriel DG, et al. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peigne C, et al. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect. Immun. 77:2272–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodriguez-Siek KE, et al. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097–2110 [DOI] [PubMed] [Google Scholar]
- 28. Sanderson KE, Zeigler DR. 1991. Storing, shipping, and maintaining records on bacterial strains. Methods Enzymol. 204:248–264 [DOI] [PubMed] [Google Scholar]
- 29. Silver RP, Aaronson W, Sutton A, Schneerson R. 1980. Comparative analysis of plasmids and some metabolic characteristics of Escherichia coli K1 from diseased and healthy individuals. Infect. Immun. 29:200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Snedecor GW, Cochran WG. 1989. Statistical methods, 8th ed Iowa State University Press, Ames, IA [Google Scholar]
- 31. Tivendale KA, Allen JL, Browning GF. 2009. Plasmid-borne virulence-associated genes have a conserved organization in virulent strains of avian pathogenic Escherichia coli. J. Clin. Microbiol. 47:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tivendale KA, et al. 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78:3412–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.