Abstract
Here, high-throughput sequencing was employed to reveal the highly diverse bacterial populations present in 62 Irish artisanal cheeses and, in some cases, associated cheese rinds. Using this approach, we revealed the presence of several genera not previously associated with cheese, including Faecalibacterium, Prevotella, and Helcococcus and, for the first time, detected the presence of Arthrobacter and Brachybacterium in goats' milk cheese. Our analysis confirmed many previously observed patterns, such as the dominance of typical cheese bacteria, the fact that the microbiota of raw and pasteurized milk cheeses differ, and that the level of cheese maturation has a significant influence on Lactobacillus populations. It was also noted that cheeses containing adjunct ingredients had lower proportions of Lactococcus species. It is thus apparent that high-throughput sequencing-based investigations can provide valuable insights into the microbial populations of artisanal foods.
INTRODUCTION
High-throughput sequencing has revolutionized the field of microbial ecology, allowing for a more accurate identification of microbial taxa, including those which are difficult to culture and/or are present in low abundance (43). These technologies have provided detailed insights into the microbial compositions of a wide variety of different ecosystems, including sea (43), soil (38), and gut environments (2, 9), as well as that of a relatively small selection of food-associated niches (17, 33, 39). One group of complex microbial environments not assessed, to date, in this way are artisanal cheeses. The complex, fermentation-based nature of cheese means that the microbiota of different cheeses vary considerably. Many of these microbes are also hugely influential with respect to the textural and organoleptic properties of a cheese (31). Thus, unsurprisingly, there have been considerable efforts made to characterize the microbial populations of cheeses. Traditional culture-independent molecular methods, most frequently the analysis of 16S rRNA genes through denaturing or temperature gradient gel electrophoresis (DGGE/TGGE) (21, 35), single-stranded conformation polymorphisms (SSCP) (7), and/or Sanger sequencing (20), have improved our understanding of cheese microbial populations (36). However, we anticipated that the application of high-throughput sequencing could provide an even more detailed understanding of the microbial composition of cheese. Thus, we have applied this technology to investigate the microbiota of 62 soft, semihard, and hard artisanal cheeses, manufactured from unpasteurized or pasteurized cow, goat, and sheep milk, and of 11 associated naturally developed or smear-ripened rinds.
MATERIALS AND METHODS
Cheese collection and nucleic acid extraction.
A total of 62 handmade cheeses, including 18 soft cheeses, 31 semihard cheeses, and 13 hard cheeses, manufactured from unpasteurized or pasteurized cow, goat, or sheep milk were obtained from artisanal cheese producers and farmer's markets throughout Ireland (see Table S1 in the supplemental material). To facilitate the culture-independent analysis of the bacterial compositions of these cheeses, their associated rinds, naturally developed or smear-ripened cheese rinds, were also analyzed. One gram of cheese or 1g of cheese rind (6, 13, 16, 20, 22) was combined with 9 ml 2% trisodium citrate and homogenized before DNA was extracted using the PowerFood microbial DNA isolation kit (MoBio Laboratories Inc.).
PCR amplification of the microbial community 16S rRNA genes.
The DNA extracts were used as a template for PCR amplification according to the methods described by Quigley et al. (36). Here, universal 16S primers targeting the V4 region (239 nucleotides long) predicted to bind to 94.6% of all 16S genes were incoporated, i.e., the forward primer F1 (5′-AYTGGGYDTAAAGNG) and a combination of four reverse primers, R1 (5′-TACCRGGGTHTCTAATCC), R2 (5′-TACCAGAGTATCTAATTC), R3 (5′-CTACDSRGGTMTCTAATC), and R4 (5′-TACNVGGGTATCTAATC) (RDP pyrosequencing pipeline; http://pyro.cme.msu.edu/pyro/help.jsp). The primers incorporated a proprietary 19-mer sequence (GCCTGCCAGCCCGCTCAG) at the 5′ end to allow emulsion-based clonal amplification for the 454 pyrosequencing system. Unique molecular identifier (MID) tags were incorporated between the adaptamer and the target-specific primer sequence, to allow identification of individual sequences from pooled amplicons. The PCR mixture contained 25 μl GoTaq Green master mix (Promega), 1 μl of each primer (200 nmol liter−1), 5 μl DNA template, and nuclease-free H2O to give a final reaction volume of 50 μl. PCR amplification was performed using a G-Storm thermal cycler (Gene Technologies, United Kingdom). The amplification program consisted of an initial denaturation step at 94°C for 2 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1 min. A final elongation step at 72°C for 2 min was also included. Amplicons were cleaned using the AMPure XP purification system (Beckman Coulter, Takeley, United Kingdom). The quantity of DNA extracted was assessed using the Quant-It Picogreen dsDNA reagent (Invitrogen) in accordance with the manufacturer's instructions and a Nanodrop 3300 fluorospectrometer (Thermo Fisher Scientific Inc.).
High-throughput sequencing and bioinformatics analysis.
The 16S rRNA V4 amplicons were sequenced on a 454 genome sequencer FLX platform (Roche Diagnostics Ltd., Burgess Hill, West Sussex, United Kingdom) according to Roche 454 protocols. Read processing was performed using techniques implemented in the RDP pyrosequencing pipeline (11). Sequences not passing the FLX quality controls were discarded, the 454-specific portions of the primers were trimmed, the raw sequences were sorted according to tag sequences, and reads with low quality scores (quality scores below 40) and short lengths (less than 150 bp for the 16S rRNA V4 region) were removed, as were reads that did not have exact matches with respect to primer sequence. Statistical analysis to measure the sequencing diversity, included Choa1 richness, Shannon diversity, and Good's coverage results, as well as monitoring results for sequencing abundance using rarefaction, were performed using the MOTHUR package (42). Principal coordinate analysis (PCoA), measuring dissimilarities at phylogenetic distances based on weighted Unifrac analysis, was performed using the QIIME suite of programs (8). Trimmed Fasta sequences were assessed by BLAST analysis (1) against information in a previously published 16S rRNA-specific database (45) using default parameters. The resulting BLAST output was parsed using MEGAN (27). MEGAN assigns reads to NCBI taxonomies by employing the lowest common ancestor algorithm, which assigns each RNA tag to the lowest common ancestor in the taxonomy from a subset of the best-scoring matches in the BLAST result. Bit scores were used from within MEGAN for filtering the results prior to tree construction and summarization (absolute cutoff, BLAST bit score of 86; relative cutoff, 10% of the top hit) (45). Statistical significance was determined by using the nonparametric Kruskal-Wallis test (29) in the Minitab statistical package.
RESULTS
Sequencing and bioinformatic analysis.
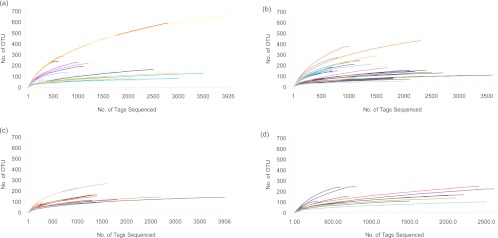
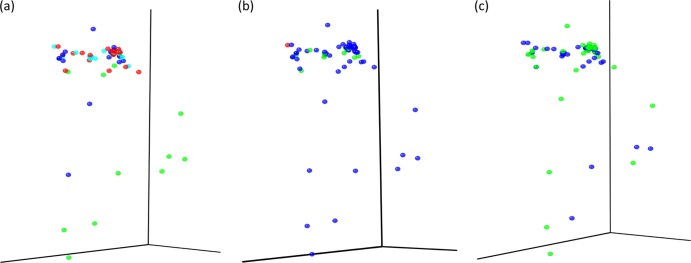
DNA was extracted from a 1-g samples from 62 cheeses and from the rinds of 11 of the cheeses (see Table S1 in the supplemental material). Following total genomic DNA extraction, amplicons of the V4 16S rRNA gene were generated, and a total of 116,238 pyrosequencing reads were obtained through 454 sequencing, corresponding to 32,322, 48,388, and 18,340 reads from soft, semihard, and hard cheeses, respectively, and 17,188 reads corresponding to cheese rinds. Diversity, richness, and coverage estimations were calculated for each data set (Table 1; individual sample diversity results are presented in Table S2 of the supplemental material). The Chao1 estimator of species richness indicated good sample richness throughout. The Shannon diversity index, a measurement of overall diversity, indicated a diverse microbiota, while the Good's coverage result, an estimator of completeness of sampling, highlighted good overall sampling, with levels of 89 to 95%. Rarefaction curve analysis, which assesses species richness from the results of sampling, showed all samples approached being parallel with the x axis, revealing that the overall bacterial diversity was well represented (Fig. 1). Principal coordinate analysis, which clusters the communities according to different parameters, in this case cheese type, animal source of milk, and whether the milk was pasteurized, was conducted according to weighted UniFrac distances (Fig. 2). Regardless of the community parameters, there was no definitive split in the microbiota of the cheeses. However, the most extreme outliers generally tended to be cheese rinds from cow's milk cheeses. No statistical differences were found in operational taxonomic units at the phylum level; however, a number of statistical differences were determined at the genus level. The gene sequence information has been summarized in a MiXS-MIMARKS metatable (46), which is shown in Table S3 of the supplemental material.
Table 1.
Sequencing richness, diversity, and coverage of artisanal cheesesa
| Data set | Result for cheese type |
||
|---|---|---|---|
| Soft | Semihard | Hard | |
| Similarity (%) | 97 | 97 | 97 |
| Chao1 richness estimation | 295 | 315 | 254 |
| Shannon index for diversity | 3.8 | 4.3 | 3.7 |
| Good's coverage (%) | 92 | 90 | 91 |
Average results of the statistical analysis of artisanal cheese, at two different similarity levels, for sequencing richness, diversity, and coverage as analyzed with MOTHUR software. The analyses were separated on the basis of cheese type.
Fig 1.
Rarefaction curves of microbial populations from artisanal cheeses and cheese rinds. Each line represents a cheese sampled and sequenced. (a) Soft cheeses; (b) semihard cheeses; (c) hard cheeses; (d) cheese rinds. The curvature of a line toward the right (or x) axis shows that a reasonable number of sequences were obtained, and thus sampling was sufficient.
Fig 2.
Principal coordinate analysis graphs for weighted UniFrac results. Samples were assessed for different community parameters. (a) Cheese type. Soft cheeses, light blue; semihard cheeses, red; hard cheeses, dark blue; rinds, green. (b) Animal sources. Cow, dark blue; goat, green; sheep, red. (c) Milk. Unpasteurized, green; pasteurized, blue.
Microbial compositions of artisanal cheeses as revealed by pyrosequencing.
In silico analysis of high-throughput sequence data revealed microorganisms corresponding to five phyla in the soft, semihard and hard cheeses (Table 2). These phyla were representatives of four bacterial phyla, i.e., Firmicutes, Proteobacteria, Bacteroidetes, and Acintobacteria. Surprisingly, a fifth phylum detected was the fungal phylum Ascomycota. The latter was detected occasionally throughout the cheese samples at a subdominant level, i.e., 0.37 to 0.50%, and at the genus level its detection corresponded almost exclusively with Penicillium. Further examination of the Penicillium sequence established that it corresponds to that of the mitochondrial 16S RNA gene of Penicillium. Of the four bacterial phyla, Firmicutes dominated in the three cheese types, corresponding to 96%, 95%, and 91% of the reads from the soft, semihard, and hard cheeses, respectively. Proteobacteria (0.91, 3.31, and 1.79%), Bacteroidetes (0.27, 0.25, and 0.21%), and Actinobacteria (1.22, 0.12, and 4.45%) were detected at various levels throughout (Table 2).
Table 2.
Summary of reads calculated from total phylum reads for each variable assessed
| Phylum or genus | % of reads in the phylum or genus per: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cheese type |
Animal source |
Milk type |
|||||||
| Soft | Semihard | Hard | Rind | Cow | Goat | Sheep | Unpasteurized | Pasteurized | |
| Phyla | |||||||||
| Proteobacteria | 0.91 | 3.31 | 1.79 | 26.99 | 95.5 | 89.6 | 98.6 | 96.1 | 93.1 |
| Firmicutes | 96.03 | 95.49 | 91.41 | 32.14 | 0.69 | 1.4 | 0 | 0.74 | 0.83 |
| Acintobacteria | 1.22 | 0.12 | 4.45 | 26.02 | 1.4 | 6.8 | 0 | 1.9 | 2.5 |
| Bacteroidetes | 0.27 | 0.25 | 0.21 | 5.47 | 0.19 | 1.1 | 0 | 0.22 | 0.48 |
| Ascomycota | 0.49 | 0.37 | 0.50 | 7.13 | 0.50 | 0.89 | 1.38 | 0.61 | 0.60 |
| Genera | |||||||||
| Lactococcus | 89.83 | 84.45 | 49.56 | 25.80 | 77.2 | 76.0 | 98.5 | 72.8 | 84.4 |
| Leuconostoc | 1.79 | 0.51 | 1.80 | 2.57 | 1.0 | 2.2 | 0 | 1.2 | 1.1 |
| Lactobacillus | 0.65 | 7.30 | 17.80 | 0.20 | 8.1 | 1.0 | 0.08 | 11.3 | 0.82 |
| Pseudomonas | 0.11 | 0.03 | 0.49 | 0.14 | 0.07 | 0.64 | 0 | 0.09 | 0.25 |
| Psychrobacter | 0.58 | 0.02 | 0.53 | 9.92 | 0.28 | 0 | 0 | 0.21 | 0.23 |
| Staphylococcus | 0.06 | 0.17 | 0.73 | 1.98 | 0.31 | 0 | 0 | 0.30 | 0.17 |
| Arthrobacter | 0.28 | 0.08 | 1.10 | 4.14 | 0.39 | 0.85 | 0 | 0.49 | 0.39 |
| Pseudoalteromonas | 0.06 | 0.03 | 0 | 3.80 | 0.03 | 0 | 0 | 0.03 | 0.02 |
| Vibrio | 0.02 | 0 | 0 | 2.84 | 0.004 | 0 | 0 | 0 | 0.008 |
| Faecalibacterium | 0.02 | 0.08 | 0.05 | 0 | 0.07 | 0 | 0 | 0.07 | 0.04 |
| Bifidobacterium | 0.02 | 0.03 | 0 | 0 | 0.02 | 0 | 0 | 0.02 | 0.009 |
| Brevibacterium | 0.02 | 0 | 2.10 | 9.22 | 0.57 | 5.4 | 0 | 0.82 | 1.9 |
| Brachybacterium | 0.81 | 0 | 1.45 | 3.56 | 0.39 | 0.50 | 0 | 0.57 | 0.17 |
| Pediococcus | 0.03 | 0.27 | 0 | 0 | 0.12 | 0 | 0 | 0.17 | 0.01 |
| Prevotella | 0 | 0.15 | 0.34 | 0.03 | 0.07 | 0.048 | 0 | 0.10 | 0.16 |
| Halomonas | 0 | 0.25 | 0 | 2.46 | 0.12 | 0 | 0 | 0.1 | 0 |
| Enterococcus | 0 | 0 | 0.10 | 0 | 0.02 | 0 | 0 | 0.03 | 0 |
| Helcococcus | 0 | 0.07 | 0 | 0 | 0.03 | 0 | 0 | 0.05 | 0 |
| Tetragenococcus | 0 | 0 | 0.05 | 0.18 | 0.01 | 0 | 0 | 0.02 | 0 |
| Corynebacterium | 0 | 0 | 0 | 1.20 | 0 | 0 | 0 | 0 | 0 |
| Streptococcus | 0 | 0.04 | 0 | 0.24 | 0.01 | 0 | 0 | 0.02 | 0 |
| Clostridium | 0 | 0 | 0.06 | 0 | 0.01 | 0 | 0 | 0 | 0.02 |
| Facklamia | 0 | 0 | 0 | 0.60 | 0 | 0 | 0 | 0 | 0 |
| Flavobacterium | 0 | 0 | 0 | 0.19 | 0 | 0 | 0 | 0 | 0 |
| Cronobacter | 0 | 0 | 0 | 0.05 | 0 | 0 | 0 | 0 | 0 |
| Penicillium | 0.49 | 0.37 | 0.51 | 12.96 | 0.50 | 0.89 | 1.38 | 0.61 | 0.60 |
| Other genera | 5.16 | 6.14 | 23.34 | 17.91 | 10.67 | 12.47 | 0.04 | 11.1 | 9.7 |
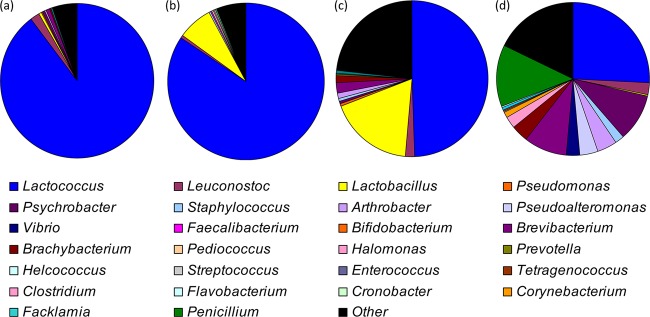
The bacteria detected corresponded to 21 different genera (Fig. 3; Table 2), with Lactococcus dominating. At the depth of analysis carried out, a total of eight genera were found to be common in all three cheese types (soft, semihard, and hard). In addition to Lactococcus (89.93, 84.45, and 49.56%), Lactobacillus (0.65, 7.30, and 17.8%), Leuconostoc (1.79, 0.51, and 1.8%), Pseudomonas (0.11, 0.03, and 0.49%), Psychrobacter (0.58, 0.02, and 0.53%), Staphylococcus (0.06, 0.17, and 0.73%), Arthrobacter (0.28, 0.08, and 1.1%) and Faecalibacterium (0.02, 0.08, and 0.05%) were also identified. Statistically significant differences was observed for the levels of Lactococcus (P = 0.031) and Lactobacillus (P = 0.010), with the level of lactococci increasing and the level of lactobacilli decreasing between soft, semihard, and hard cheese types (Fig. 3). Vibrio were found in soft cheeses only (0.02%), and Helcococcus (0.07%), Halomonas (0.25%), and Streptococcus (0.04%) were found in the semihard cheeses only, while Enterococcus (0.1%), Tetragenococcus (0.05%), and Clostridium (0.06%) were found in hard cheeses only. Three genera were shared between soft and semihard cheeses. These were Pseudoalteromonas (0.06 and 0.03%), Pediococcus (0.03% and 0.27%), and Bifidobacterium (0.02% and 0.03%). Brevibacterium (0.81% and 2.10%) was the only genus shared between soft and hard cheeses, and Prevotella (0.15% and 0.34%) was the only genus common to semihard and hard cheeses.
Fig 3.
Assignment of cheese microbiota at the genus level, according to MEGAN. (a) soft cheese; (b) semihard cheese; (c) hard cheese; (d) cheese rinds.
Some interesting observations were made regarding the influence of the animal source of milk and pasteurization on the microbial populations present in the resultant cheeses. We noted that cow's milk cheese contained 21 different bacterial genera (Table 2), whereas goat milk contained only 8 different bacterial genera, and only two bacterial genera, Lactococcus and Lactobacillus, were detected in sheep's milk cheese. Also, by comparing the bacterial genera present in artisanal cheeses manufactured from unpasteurized milk relative to those made from pasteurized milk, it was apparent that Halomonas, Helcococcus, Streptococcus, Enterococcus, and Tetragenococcus were detected in raw milk cheeses only and that Clostridium and Vibrio were detected from pasteurized milk cheeses only. A significant difference was noted in the levels of Lactococcus (P = 0.025) and Lactobacillus (P = 0.002) between unpasteurized and pasteurized milk cheeses. Further comparisons provided some interesting findings. Cheeses S2 and S3 were produced with milk from the same herd using similar protocols, but they differed in that S3 was a feta-style cheese and thus had a higher salt content, which may explain the absence of Leuconostoc and Pseudomonas from this cheese. Similarly, S7 and H9 were produced in the same farmhouse but differed with respect to the level of maturation, with associated differences in the proportions of Lactobacillus (0.65% in soft cheese, 17.8% in hard cheese). This is reflective of the aforementioned overall greater number of lactobacilli in hard relative to semihard and, in turn, soft cheeses. This pattern was also apparent when cheeses SH1 and H3 to H6, all from the same producers, were compared. In addition, the results of a specific comparison of H4 and H5 were also interesting, as these cheeses differed solely on the basis of H5 containing an adjunct ingredient, fenugreek seeds, the presence of which coincided with a reduction in the proportion of lactococci (from 61 to 2%) and increase in lactobacilli (from 34 to 95%). The inclusion of herbs, spices, or seaweed was also found to coincide with reduced proportions of lactococci in sample SH8 relative to SH10 and SH11 and in SH28 relative to SH26.
Revealing the microbial compositions of the rinds of artisanal cheeses.
We again used high-throughput sequencing to analyze the microbiota of 11 rinds of the artisanal cheeses (samples R1 to 11) (Fig. 3; see Table S1). These included smear/wash-ripened rinds, i.e., R1, R7, R8, and R9; naturally developed rinds, i.e., R2 to R6 and R11; and one mold-ripened rind, R10. In silico analysis of sequence data revealed the presence of 19 different genera (Fig. 3; Table 2). While some of these genera, including Lactococcus, Leuconostoc, and Lactobacillus, corresponded to those also detected in the cheese core, a selection was identified in the cheese rinds only. These included Corynebacterium (1.2%), Facklamia (0.60%), Flavobacterium (0.19%), and Cronobacter (0.05%). While Lactococcus remained the most common genus in cheese rinds, the relative proportions of this genus were significantly lower in the rind than in the core. Generally, smear/wash-ripened rinds had particularly low levels of lactococci (1.9 to 4.8%), while naturally developed rinds had levels of lactococci of up to 98%. It was also apparent that Psychrobacter and Brevibacterium represented a considerable proportion, i.e., 0.29 to 57% and 0.67 to 54.6%, respectively, or ∼10% on average, of the total population. The other genera detected, i.e., Leuconostoc, Lactobacillus, Pseudomonas, Psychrobacter, Pseudoalteromonas, Brachybacterium, Prevotella, Arthrobacter, Streptococcus, Tetragenococcus, and Facklamia, corresponded to between 0.03 and 4.14% of reads (Table 2). Brevibacterium and Brachybacterium were present at significantly different levels in the rinds of soft, semihard, and hard cheeses compared to cheese core (P = 0.040 and P = 0.014, respectively). Penicillium was also detected in the cheese rind and in higher proportions than were detected in the cheese core. Finally, we also detected the presence of Prevotella, a genus that has previously not been detected in cheese or cheese rinds, and we noted that Vibrio was only identified in rinds of the smear/wash-developed variety (P = 0.009).
DISCUSSION
In this study, pyrosequencing-based 16S rRNA profiling provided detailed insights into the complex microbiota of artisanal cheeses. Its use effectively revealed the presence of a number of taxa not previously associated with specific cheese types or, indeed, of any cheeses. Among those identified for the first time were the genera Prevotella and Faecalibacterium. Prevotella spp. are Gram-negative bacteria from the phylum Bacteroidetes that thrive in anaerobic environments. They are commensals of the rumen and hind gut in cattle and sheep, but they can also be the cause of periodontal disease as well as other human infections. Members of the genus Faecalibacterium are strict anaerobes and have been shown to produce butyrate, d-lactate, and formate, as well as utilize acetate (18). While butyrate can contribute positively to cheese development, at high levels this product can induce the late-blowing defect in cheese (10). d-Lactate and acetate are also produced during the development of cheese (25). Further investigations will be required to determine if, at the levels present in cheese, these microbes contribute flavor in a significant way. A third genus which is typically associated with anaerobic gastrointestinal environments, Helcococcus, was also detected, but only in one cheese, a semihard cheese made from unpasteurized cow milk. Helcococcus spp. have been associated with clinical problems in humans (12), cows (30), sheep (47), and horses (41) and thus, in this instance, may reflect the sourcing of contaminated milk from an infected animal. Given that in this study these insights were gained through the analysis of merely a 1-g sample per cheese, it may be that further investigations of even larger sample sizes and at a greater depth of sequencing will uncover additional genera not previously associated with cheese. Nevertheless, the detection of these anaerobes reveals that the microbiota of cheeses are more diverse than previously appreciated, thus further highlighting the benefits of high-throughput sequencing investigations.
We also detected a number of genera not previously associated with specific cheese types. Arthrobacter and Brachybacterium were detected for the first time in goat cheese; these are commonly detected on the cheese surface but not, to our knowledge, in the core of goat cheese. The presence of Pseudoalteromonas in soft and semihard cow milk cheeses, as well as cheese rinds, was also unexpected. Pseudoalteromonas species are usually regarded as marine bacteria (5) and have been detected on the surface of a smear-ripened cheese on only one previous occasion (23). This is the first instance in which this genus has been detected in a cheese core.
In addition to identifying taxa not previously associated with cheeses or specific cheese types, there were a number of other interesting observations. We noted that the milk source impacted the number of genera detected, i.e., 21 genera from cow milk cheeses, 8 from goat milk cheeses, and 2 from sheep milk cheeses, although this observation may have been influenced by differences in the number of samples within each group. Notably, a number of studies have previously established that milk source can influence the type and number of microbes present in a cheese (13, 37). Previous studies have also highlighted the dramatic impact of milk pasteurization on the microbiota of resultant cheeses (4, 13, 19). Here, through the use of high-throughput sequencing, we also observed differences in cheeses produced from raw and pasteurized milks (Table 2) in that, for example, significant differences in levels of Lactococcus and Lactobacillus organisms were apparent when these cheese types were compared. For further studies to investigate these differences, it would be interesting to focus on RNA (and thus cDNA), or to employ stains that inactivate DNA from cells that have been killed by the temperature treatment, to determine if such approaches provide different results.
Several previous studies have employed other technologies to investigate the impact of salt (26), ripening (32), and additional ingredients (3, 14, 44) on the microbiota of cheese. Here we noted that neither Leuconostoc nor Pseudomonas was detected in cheeses with a high salt content, that significantly increased Lactobacillus populations were detected in cheeses from the same farmhouse but which had been ripened to various degrees, and that the inclusion of adjunct ingredients, such as herbs, spices, or seaweed, did impact microbial composition.
As a consequence of its exposure to the external environment and, in some cases, steps taken during the manufacturing process, the microbiota of the rind of a cheese will frequently differ dramatically from that of the rest of the cheese (24). This presumably reflects the exposure of the cheese rind to the environment. Many of the bacterial genera detected are commonly identified in cheese rinds and, indeed, Corynebacterium, Arthrobacter, Brevibacterium, and Halomonas have previously been identified on the surface of Irish artisanal cheeses (34). The high proportions of Psychrobacter and Brevibacterium in the cheese rinds studied here were particularly notable. Brevibacterium is known to be involved in the development of cheese rind flavors and smear rind color (28). Although Psychrobacter is frequently detected on cheese surfaces, its specific role is unclear. It may contribute to flavor, given the ability of strains from this genus to produce branched-chain aldehydes, alcohols, and esters (15). The impact of the presence of such high proportions of these bacteria on the cheese surface will require further investigation. The previously unreported or rare phenomenon of Prevotella, Facklamia (40), or Vibrio being detected on cheese rinds further highlights the benefits of employing high-throughput sequencing to investigate these populations.
Thus, in conclusion, we have employed high-throughput sequencing to investigate the microbiota of 62 Irish artisanal cheeses in a greater depth than ever before. We have highlighted for the first time the presence of a number of genera previously undetected in cheeses, as well as detecting genera not typically associated with specific cheese types. These analyses also provide insights into the influence of different factors on the composition of the artisanal cheese microbiota, which can now be investigated in greater depth through the study of cheeses prepared in the laboratory.
Supplementary Material
ACKNOWLEDGMENT
Lisa Quigley is funded by the Teagasc Walsh Fellowship Scheme.
Footnotes
Published ahead of print 8 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Andersson AF, et al. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836 doi:10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ankri S, Mirelman D. 1999. Antimicrobial properties of allicin from garlic. Microbes Infect. 1:125–129 [DOI] [PubMed] [Google Scholar]
- 4. Bonetta S, Bonetta S, Carraro E, Rantsiou K, Cocolin L. 2008. Microbiological characterisation of Robiola di Roccaverano cheese using PCR-DGGE. Food Microbiol. 25:786–792 [DOI] [PubMed] [Google Scholar]
- 5. Bozal N, Montes MJ, Tudela E, Guinea J. 2003. Characterization of several Psychrobacter strains isolated from Antarctic environments and description of Psychrobacter luti sp nov and Psychrobacter fozii sp nov. Int. J. Syst. Evol. Microbiol. 53:1093–1100 [DOI] [PubMed] [Google Scholar]
- 6. Callon C, Delbes C, Duthoit F, Montel MC. 2006. Application of SSCP-PCR fingerprinting to profile the yeast community in raw milk Salers cheeses. Syst. Appl. Microbiol. 29:172–180 [DOI] [PubMed] [Google Scholar]
- 7. Callon C, et al. 2007. Stability of microbial communities in goat milk during a lactation year: molecular approaches. Syst. Appl. Microbiol. 30:547–560 [DOI] [PubMed] [Google Scholar]
- 8. Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Claesson MJ, et al. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669 doi:10.1371/journal.pone.0006669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cocolin L, Innocente N, Biasutti M, Comi G. 2004. The late blowing in cheese: a new molecular approach based on PCR and DGGE to study the microbia1 ecology of the alteration process. Int. J. Food Microbiol. 90:83–91 [DOI] [PubMed] [Google Scholar]
- 11. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins MD, Falsen E, Brownlee K, Lawson PA. 2004. Helcococcus sueciensis sp. nov., isolated from a human wound. Int. J. Syst. Evol. Microbiol. 54:1557–1560 [DOI] [PubMed] [Google Scholar]
- 13. Coppola S, Blaiotta G, Ercolini D, Moschetti G. 2001. Molecular evaluation of microbial diversity occurring in different types of mozzarella cheese. J. Appl. Microbiol. 90:414–420 [DOI] [PubMed] [Google Scholar]
- 14. Dash BK, Sultana S, Sultana N. 2011. Antibacterial activities of methanol and acetone extracts of fenugreek (Trigonella foenum) and coriander (Coriandrum sativum). Life Sci. Med. Res. 2011:LSMR-27 http://astonjournals.com/manuscripts/Vol2011/LSMR-27_Vol2011
- 15. Deetae P, Bonnarme P, Spinnler HE, Helinck S. 2007. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl. Microbiol. Biotechnol. 76:1161–1171 [DOI] [PubMed] [Google Scholar]
- 16. Delbes C, Ali-Mandjee L, Montel MC. 2007. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 73:1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dobson A, O'Sullivan O, Cotter PD, Ross P, Hill C. 2011. High-throughput sequence-based analysis of the bacterial composition of kefir and an associated kefir grain. FEMS Microbiol. Lett. 320:56–62 [DOI] [PubMed] [Google Scholar]
- 18. Duncan SH, Hold GL, Harmsen HJM, Stewart CS, Flint HJ. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:2141–2146 [DOI] [PubMed] [Google Scholar]
- 19. Duthoit F, Callon C, Tessier L, Montel MC. 2005. Relationships between sensorial characteristics and microbial dynamics in “registered designation of Origie” Salers cheese. Int. J. Food Microbiol. 103:259–270 [DOI] [PubMed] [Google Scholar]
- 20. Duthoit F, Godon JJ, Montel MC. 2003. Bacterial community dynamics during production of registered designation of origin Salers cheese as evaluated by 16S rRNA gene single-strand conformation polymorphism analysis. Appl. Environ. Microbiol. 69:3840–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ercolini D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297–314 [DOI] [PubMed] [Google Scholar]
- 22. Ercolini D, Hill PJ, Dodd CER. 2003. Bacterial community structure and location in Stilton cheese. Appl. Environ. Microbiol. 69:3540–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feurer C, Irlinger F, Spinnler HE, Glaser P, Vallaeys T. 2004. Assessment of the rind microbial diversity in a farm house-produced vs a pasteurized industrially produced soft red-smear cheese using both cultivation and rDNA-based methods. J. Appl. Microbiol. 97:546–556 [DOI] [PubMed] [Google Scholar]
- 24. Feurer C, Vallaeys T, Corrieu G, Irlinger F. 2004. Does smearing inoculum reflect the bacterial composition of the smear at the end of the ripening of a French soft, red-smear cheese? J. Dairy Sci. 87:3189–3197 [DOI] [PubMed] [Google Scholar]
- 25. Fox PF. 1999. Cheese: chemistry, physics and microbiology, vol 1 Chapman and Hall, London, United Kingdom [Google Scholar]
- 26. Fox PF, McSweeney PLH, Cogan TM, Guinee TP. 2004. Cheese: chemistry, physics and microbiology (3rd ed), p 1–17 Elsevier, Amsterdam, Netherlands [Google Scholar]
- 27. Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krubasik P, Sandmann G. 2000. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol. Gen. Genet. 263:423–432 [DOI] [PubMed] [Google Scholar]
- 29. Kruskal WH, Wallis AW. 1952. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47:583–621 [Google Scholar]
- 30. Kutzer P, Schulze C, Engelhardt A, Wieler LH, Nordhoff M. 2008. Helcococcus ovis, an emerging pathogen in bovine valvular endocarditis. J. Clin. Microbiol. 46:3291–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marilley L, Casey MG. 2004. Flavours of cheese products: metabolic pathways, analytical tools and identification of producing strains. Int. J. Food Microbiol. 90:139–159 [DOI] [PubMed] [Google Scholar]
- 32. Martin-Platero AM, Maqueda M, Valdivia E, Purswani J, Martinez-Bueno M. 2009. Polyphasic study of microbial communities of two Spanish farmhouse goats' milk cheeses from Sierra de Aracena. Food Microbiol. 26:294–304 [DOI] [PubMed] [Google Scholar]
- 33. Masoud W, et al. 2011. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. Int. Dairy J. 21:142–148 [Google Scholar]
- 34. Mounier J, et al. 2005. Surface microflora of four smear-ripened cheeses. Appl. Environ. Microbiol. 71:6489–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogier JC, et al. 2004. Molecular fingerprinting of dairy microbial ecosystems by use of temporal temperature and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:5628–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quigley L, et al. 2011. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 150:81–94 [DOI] [PubMed] [Google Scholar]
- 37. Randazzo CL, Vaughan EE, Caggia C. 2006. Artisanal and experimental Pecorino Siciliano cheese: microbial dynamics during manufacture assessed by culturing and PCR-DGGE analyses. Int. J. Food Microbiol. 109:1–8 [DOI] [PubMed] [Google Scholar]
- 38. Roesch LF, et al. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roh SW, et al. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 4:1–16 [DOI] [PubMed] [Google Scholar]
- 40. Roth E, Schwenninger SM, Eugster-Meier E, Lacroix C. 2011. Facultative anaerobic halophilic and alkaliphilic bacteria isolated from a natural smear ecosystem inhibit Listeria growth in early ripening stages. Int. J. Food Microbiol. 147:26–32 [DOI] [PubMed] [Google Scholar]
- 41. Rothschild CM, et al. 2004. Helcococcus ovis isolated from a pulmonary abscess in a horse. J. Clin. Microbiol. 42:2224–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schloss PD, et al. 2009. Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tajkarimi MM, Ibrahim SA, Cliver DO. 2010. Antimicrobial herb and spice compounds in food. Food Control 21:1199–1218 [Google Scholar]
- 45. Urich T, et al. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527 doi:10.1371/journal.pone.0002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yilmaz P, et al. 2011. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29:415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y, et al. 2009. Isolation of Helcococcus ovis from sheep with pleuritis and bronchopneumonia. J. Vet. Diagn. Invest. 21:164–166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.