Abstract
The phytopathogenic prokaryote Xanthomonas oryzae pv. oryzae is the causal agent of bacterial leaf blight (BB) of rice and utilizes a type III secretion system (T3SS) to deliver T3SS effectors into rice cells. In this report, we show that the ketoglutarate transport protein (KgtP) is secreted in an HpaB-independent manner through the T3SS of X. oryzae pv. oryzae PXO99A and localizes to the host cell membrane for α-ketoglutaric acid export. kgtP contained an imperfect PIP box (plant-inducible promoter) in the promoter region and was positively regulated by HrpX and HrpG. A kgtP deletion mutant was impaired in bacterial virulence and growth in planta; furthermore, the mutant showed reduced growth in minimal media containing α-ketoglutaric acid or sodium succinate as the sole carbon source. The reduced virulence and the deficiency in α-ketoglutaric acid utilization by the kgtP mutant were restored to wild-type levels by the presence of kgtP in trans. The expression of OsIDH, which is responsible for the synthesis of α-ketoglutaric acid in rice, was enhanced when KgtP was present in the pathogen. To our knowledge, this is the first report demonstrating that KgtP, which is regulated by HrpG and HrpX and secreted by the T3SS in Xanthomonas oryzae pv. oryzae, transports α-ketoglutaric acid when the pathogen infects rice.
INTRODUCTION
The ability to acquire carbohydrates from plants is essential for pathogens to establish successful infections (7, 54). To reach the cell densities required to overcome the host immune system, the pathogen has to adapt to the intercellular environment and utilize available nutrients, especially carbohydrates, from the host plant (51, 54). The plant apoplast lacks hexose sugars but is rich in gluconeogenic substrates, including intermediates of the tricarboxylic acid (TCA) cycle (40, 54). C4-dicarboxylates (DCAs), such as oxaloacetate, malate, fumarate, and succinate, are the principal carbon and energy sources for Xanthomonas spp. during infection (54, 61). This is also true for Rhizobium spp., in which C4-dicarboxylic acids are the principal carbon sources for the bacteroid within nodules (3, 8, 9). TCA intermediates were also preferred carbon sources in various Pseudomonas spp. (30).
The dicarboxylate transport (DctA) system mediates the uptake of C4-dicarboxylates in Escherichia coli under aerobic conditions (14). However, in Corynebacterium glutamicum, malate transport by DctA is inhibited by α-ketoglutarate, indicating that substrates may also regulate nutrient uptake (68). A mutation in the DctA system in Pseudomonas syringae pv. tomato had a partial effect on bacterial growth and virulence in planta (30), indicating that other uptake systems also function in nutrient acquisition.
Sodium-dependent dicarboxylate transporters, including ketoglutarate permease KgtP, are responsible for transferring succinate, α-ketoglutaric acid, citrate, and other TCA cycle intermediates into the bacterial cell and play an important role in maintaining dicarboxylic acid concentrations (69). KgtP is a hydrophobic membrane protein that transports α-ketoglutarate into E. coli cells and consists of membrane-spanning domains connected with hydrophobic loops (45, 46). This structure resulted in the development of a membrane topology model in which KgtP functions in transport across the cell membrane (47). Site-directed mutagenesis of KgtP domains abolished α-ketoglutarate transport (46). In support of this model, the expression of heterogeneous E. coli kgtP in Synechococcus conferred the ability to incorporate 2-oxoglutarate through active transport (57). However, it is unclear whether phytopathogenic bacteria can use KgtP to transport DCAs for gluconeogenesis.
Xanthomonas spp. secrete effector proteins via a type III secretion system (T3SS) directly into plant cells, where they interfere with host cellular processes to suppress defense (16, 32, 60). Previous reports showed that the expression of T3SS effectors was induced in planta and in the hrp-inducing medium XOM3 (65) and was regulated by HrpG and HrpX (38, 63). HrpG belongs to the OmpR family of two-component signal transduction response regulators, and HrpX is an AraC-type transcriptional regulator (63, 64). HrpG and HrpX form a regulatory cascade in which HrpG regulates the expression of hrpX and HrpX regulates the expression of downstream hrp operons and some T3SS effectors (62). HrpX-regulated genes usually possess a PIP box (plant-inducible promoter), TTCGB-N15-TTCGB (B refers to the base C, G, or T but not A), which is bound by HrpX (35, 37, 49, 62). An exception is hrpF, which contains an imperfect PIP box (TTCGC-N8-TTCGT) that is also bound by HrpX (22, 55). In Xanthomonas oryzae pv. oryzae, an imperfect PIP box is present in the promoter region of kgtP; however, it is not clear whether kgtP is regulated by HrpX.
The Gram-negative bacterium X. oryzae pv. oryzae is the causal agent of bacterial leaf blight (BB), one of the most destructive diseases of rice (33, 39). The pathogen enters rice leaves through hydathodes or wounds, propagates in the intercellular spaces of the underlying epidermis, and then spreads throughout the plant in the xylem, where it presumably interacts with xylem parenchyma cells (17, 33, 34, 39). In this study, we demonstrate that kgtP in X. oryzae pv. oryzae is involved in the assimilation of dicarboxylates in minimal medium, bacterial growth, and virulence in rice. The expression of kgtP was strongly induced in planta and by exogenous dicarboxylates and positively regulated by HrpX. KgtP was secreted in an HpaB-independent manner through the T3SS and localizes to the host cell membrane to facilitate α-ketoglutaric acid import.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli strains were routinely grown in Luria-Bertani (LB) medium at 37°C (31). X. oryzae pv. oryzae PXO99A strains were grown at 28°C in NB (0.1% yeast extract, 0.3% beef extract, 0.5% polypeptone, and 1% sucrose), NA (NB with 15 g/liter agar), NAN (NA without sucrose), NAS (NA with 10% sucrose), and NY (NB without beef extract and sucrose). In some experiments, strains were grown in MMX minimal medium [0.5% glucose, 0.2% (NH4)2SO4, 0.1% trisodium citrate dihydrate, 0.4% K2HPO4, 0.6% KH2PO4, 0.02% MgSO4 · 7H2O], NCM (noncarbohydrate) medium (15, 54), or XOM3 medium (65) or with rice suspension cells (65). Antibiotics were added at the following concentrations (μg/ml) when required: kanamycin (Km), 25; rifampin (Rif), 50; ampicillin (Ap), 100; spectinomycin (Sp), 50; and streptomycin (Sm), 50.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5α | F− ϕ80dlacZ ΔM15 Δ(lacZYA-argF)U169 endA1 deoR recA1 hsdR17(rK− mK+) phoA supE44 λ− thi-l gyrA96 relA1 | Clontech |
| Agrobacterium tumefaciens | ||
| EHA105 | Rifr | This lab |
| X. oryzae pv. oryzae | ||
| PXO99A | Wild type, Philippine race 6 | This lab |
| PΔhrcU | hrcU deletion mutant of PXO99A | This lab |
| PΔhrpX | hrpX deletion mutant of PXO99A | This lab |
| PΔhrpG | hrpG deletion mutant of PXO99A | This lab |
| PΔhrpF | hrpF deletion mutant of PXO99A | This lab |
| PΔhpaB | hpaB deletion mutant of PXO99A | This lab |
| PΔhpaP | hpaP deletion mutant of PXO99A | This lab |
| PΔhrpE | hrpE deletion mutant of PXO99A | This lab |
| PΔ kgtP | kgtP deletion mutant of PXO99A | This study |
| CPΔkgtP | PΔkgtP containing pCkgtP; Kmr | This study |
| Plasmids | ||
| pMD18-T | pUC ori, cloning vector; Apr | TaKaRa |
| pUFR034 | incW mob(p) mob+ lacZa+; PK2 replicon, cosmid; Kmr | This lab |
| pHM1 | Broad-host-range vector, cos parA incW derivative of pRI40; Spr Smr | This lab |
| pKMS1 | Suicide vector derived from pK18mobGII, sacB+; Kmr | 71 |
| pEGAD | lacZa+, contains GFP; Kmr | This lab |
| pKΔkgtP | 1,052-bp fusion of the left and right regions flanking kgtP locus in pKMS1; Kmr | This study |
| pCkgtP | 1,630-bp fusion of the kgtP promoter and kgtP gene in pUFR034; Kmr | This study |
| pKgtP-c-Myc | pHM1 expressing KgtP with c-Myc tag under the control of the kgtP promoter; Spr Smr | This study |
| pkgtPPGFP | 1,017-bp fusion of the kgtP promoter and the coding region for gfp in pUFR034; Kmr | This study |
| pZWavrXa10 | Coding region for avrXa10 fused to the lacZ promoter of pBluescript II KS(+); Apr | This lab |
| pAvrXa10 | Coding region for avrXa10 with native promoter in pUFR034; Kmr | This lab |
| pΔ28AvrXa10 | Truncated avrXa10 missing the region corresponding to the N-terminal 28 amino acids; under the control of avrXa10 native promoter in pUFR034; Kmr | 27 |
| pNKgtPΔ28AvrXa10 | pUFR034 expressing the N-terminal 50 amino acids of KgtP and Δ28AvrXa10 under the control of the kgtp promoter; Kmr | This study |
| pKgtP-GFP | pEGAD expressing KgtP; Kmr | This study |
| pKgtPΔ6-GFP | pEGAD expressing 780 bp corresponding to the N terminus of KgtP; Kmr | This study |
| pKgtPΔ12-GFP | pEGAD expressing 66 and 72 bp corresponding to regions from the N and C termini of KgtP; Kmr | This study |
| pXopR-GFP | pEGAD expressing XopR; Kmr | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Rifr, rifampin resistance; Spr, spectinomycin resistance.
Recombinant DNA techniques.
DNA manipulations were performed as described previously (42). Mobilization of plasmids into X. oryzae pv. oryzae was performed as described by Turner (56). Kits for isolating DNA and for purifying DNA from agarose gels were purchased from Axygen (Beijing, China). Restriction enzymes and DNA ligases were used according to the manufacturer's instructions (TaKaRa, Dalian, China). Oligonucleotide primers for sequencing or PCR kits were purchased from Jinsite Biotechnology Co. and are listed in Table S1 in the supplemental material. PCR was performed with Ex Taq (TaKaRa). DNA sequences were analyzed with VECTOR NTI software (Invitrogen, Shanghai, China).
Construction of a nonpolar deletion mutation in kgtP.
An in-frame deletion mutation of kgtP was constructed in X. oryzae pv. oryzae PXO99A using homologous recombination and pKMS1 as a suicide vector (71). The kgtP loci referenced under GenBank accession no. PXO_03531 and the genome sequence of X. oryzae pv. oryzae PXO99A (http://www.ncbi.nlm.nih.gov/nuccore/CP000967) were used to design primers for an in-frame deletion. Two fragments flanking the left and right of kgtP (see Fig. S1 in the supplemental material) were amplified using genomic DNA of strain PXO99A as the template and the primer pairs kgtPI-F/kgtPI-R and kgtPII-F/kgtPII-R (Table S1). The amplified fragments were cloned into pMD18-T (TaKaRa), confirmed by sequence analysis, and then digested and subcloned into vector pKMS1 at BamHI and SalI sites, resulting in pKΔkgtP (Table 1). Plasmid pKΔkgtP was introduced into PXO99A by electroporation, and transformants were plated on NAN plates supplemented with kanamycin. Colonies resulting from a single homologous crossover (integration of deletion construct at either the left or right border of kgtP) were then transferred to NBN broth, grown for 12 h at 28°C, and then plated on NAS plates. Single colonies were visible within 3 to 4 days and were transferred to NA and NA plus kanamycin. Since kanamycin-sensitive colonies could be mutants containing a second homologous crossover, these were further examined by PCR amplification with the primer pair kgtPI-F/kgtPII-R (Table S1 and Fig. S1). Southern hybridization with a digoxigenin-labeled probe (Roche) mapping to the left of kgtP was conducted to confirm the deletion of kgtP (Fig. S1). One of the mutants, PΔkgtP (Table 1 and Fig. S1), contained a deletion in kgtP and was used for further study.
Complementation of PΔkgtP.
A 1,630-bp DNA fragment containing kgtP and upstream DNA (from 337 bp upstream of the kgtP start codon and ending with the stop codon) was amplified for complementation studies. Genomic DNA from X. oryzae pv. oryzae PXO99A was used as the template, and full-length kgtP was amplified using primer pair kgtP-F/kgtP-R (Table S1). After confirmation by sequence analysis, the amplified DNA fragment was cloned into pUFR034 at the BamHI and KpnI sites to create the recombinant plasmid pCkgtP (Table 1). Plasmid pCkgtP was then transferred into mutant PΔkgtP by electroporation, and transformants containing pCkgtP were screened on NA plates with kanamycin. A putative transformant was shown to contain pCkgtP by PCR analysis and designated CPΔkgtP (Table 1).
HR and pathogenicity assays.
Assays for the hypersensitive response (HR) and pathogenicity were performed as described previously (15, 70). Briefly, Xanthomonas cells were grown in NB broth with appropriate antibiotics at 28°C and 200 rpm for 16 h, when cells approached the exponential phase of growth. Bacterial cells were then harvested by centrifugation, washed twice, and resuspended in sterile water to an optical density at 600 nm (OD600) of 0.3 (approximately 1 × 108 CFU/ml). Bacteria were then infiltrated into tobacco leaves (Nicotiana tabacum cv. Xanthi) with a needleless syringe to evaluate the HR. Bacteria were inoculated into leaves of adult rice plants (Oryza sativa cv. IR24, susceptible to BB, 2 months old) using leaf clipping for lesion length measurement or to rice seedlings (2 weeks old) for evaluating water-soaked symptoms. All plants were maintained in a greenhouse as described previously (58). Plant phenotypes were scored 24 h postinoculation for the HR in tobacco, 3 days postinoculation (dpi) for water-soaked symptoms, and 14 dpi for lesion length. Five leaves were inoculated for each independent experiment, and each treatment was repeated at least three times.
Bacterial growth in planta and in NCM minimal medium.
Xanthomonas suspensions (1 × 108 CFU/ml) were infiltrated into the intercellular spaces of fully expanded rice leaves (cultivar IR24, 2 weeks old) with needleless syringes at three locations per leaf. Three 0.8-cm-diameter leaf discs were harvested with a cork borer from each inoculated area. After sterilization in 70% ethanol and 30% hypochlorite, leaf discs were homogenized in 1 ml of distilled water. Diluted homogenates were plated on NA plates supplemented with the appropriate antibiotics. The bacterial colonies were counted after incubation at 28°C for 3 to 4 days. The CFU/cm2 of leaf area was then estimated, and the standard deviation was calculated using colony counts from three triplicate spots from each of three samples per time point per inoculation. Experiments were repeated at least three times.
Growth of wild-type PXO99A and that of the kgtP mutant were equivalent in NY and MMX minimal medium (data not shown), indicating that the PΔkgtP mutant was not auxotrophic. To investigate the effect of the mutant on the assimilation of dicarboxylates, Xanthomonas cells were preincubated in 5 ml of NB medium for 16 to 20 h at 28°C at 200 rpm until the OD600 reached 0.6. Next, 2% of this culture was subcultured into 20 ml of fresh NB medium and incubated for 16 to 18 h at 28°C. Bacterial cells were then collected, washed twice in sterile distilled water, and resuspended to an OD600 of 0.1 in 20 ml of NCM minimal medium supplemented with different carbon sources at 0.5%. At each time point, 200 μl of bacterial culture was removed and the OD600 was determined. The data presented are from one representative experiment, and the experiment was repeated independently three times.
Rice suspension cells.
Oryza sativa subsp. indica cv. Shanyou63, which is susceptible to X. oryzae pv. oryzae PXO99A, was used for callus induction. Seeds were hulled and sterilized in 70% ethanol for 10 min, transferred to a solution containing 50% commercial bleach with a few drops of Tween 20 for 30 min, and then soaked in 1% HgCl2 for 15 min. Sterilized seeds were washed five times with sterile distilled water and incubated on N6 medium (6) containing 2,4-dichlorophenoxyacetic acid (2,4-D) (5 mg/liter) at 28°C in darkness. Actively growing calli were then selected and transferred to liquid N6 medium containing 2,4-D and kinetin at 5 mg/liter and 1 mg/liter, respectively. Rice calli were incubated in the dark and transferred by subculturing at 7-day intervals at a dilution of 1:5 (inoculum/fresh medium). Generally, large amounts of rice suspension cells were obtained after 4 to 5 weeks of subculturing; at this time, single, round rice cells could be detected by microscopy.
Determination of kgtP expression using GFP.
The green fluorescent protein (GFP) reporter plasmid pkgtPPGFP was generated by amplifying the native promoter (−1 to −300 bp upstream) of kgtP from genomic DNA of X. oryzae pv. oryzae PXO99A with the primer pair kgtPP-F/kgtPP-R (see Table S1 in the supplemental material). The product was then fused with the 717-bp gfp gene. This fusion was then cloned into the EcoRI and PstI sites of pUFR034, generating recombinant plasmid pkgtPPGFP (Table 1). pkgtPPGFP was then transformed into the wild-type PXO99A strain and the hrcU mutant, PΔhrcU, for expression studies.
To visualize kgtP expression using fluorescence microscopy, X. oryzae pv. oryzae strains were preincubated in NB medium for 16 h at 28°C, harvested at 5,000 rpm for 10 min, and washed twice in sterile distilled water. The pellets were then suspended at an OD600 of 1.0 in sterilized water. Bacteria were cocultivated with rice cell suspensions at 25°C for 16 h, and fluorescence microscopy was then used to monitor the expression of gfp. Microscopic examination of the cocultivated cells of X. oryzae pv. oryzae with rice was performed after staining with Evans blue at a final concentration of 0.001% (wt/vol). When dyed with Evans blue, plant cells emit a red fluorescence when observed in the GFP excitation wavelength range (480 ± 30 nm) with a 520-nm long-pass glass emission filter. Microscopic observations were performed using 10 μl of the cocultivated cells mounted on a glass slide previously coated with poly-l-lysine (Sigma, St. Louis, MO). All images were acquired with the same parameters to allow comparisons between different samples.
Semiquantitative and real-time quantitative RT-PCR.
Gene expression was assayed by semiquantitative or real-time quantitative reverse transcription-PCR (RT-PCR) using primer pairs listed in Table S1 in the supplemental material. Xanthomonas cells were preincubated in 20 ml of NB medium for 16 to 20 h, until the OD600 reached 0.6. Next, 2% of this culture was subcultured into 20 ml of fresh NB medium for 16 to 18 h. The bacterial cells were collected, washed twice, and resuspended to an OD600 of 2.0 in sterile water. Then, 40 μl of the bacterial suspension was inoculated for 16 h of incubation at 25°C in 1.5 ml of one of the following: NB, MMX, or NCM medium supplemented with α-ketoglutaric acid or rice suspension cells. The suspension was then infiltrated into rice leaves for 8 h. As a template, total RNA was extracted using TRIzol reagent (TaKaRa) as instructed by the manufacturer. cDNA synthesis was conducted with avian myeloblastosis virus (AMV) random primers purchased from TaKaRa. Before synthesis of the first-strand cDNA, total RNAs were digested with RNase-free DNase I (TaKaRa) to remove potential traces of genomic DNAs. Semiquantitative RT-PCR was performed using a thermocycler, which was programmed as follows: step 1, 95°C for 3 min; step 2, 95°C for 20 s; step 3, 55°C for 30 s; step 4, 72°C for 40 s; 35 cycles from steps 2 to 4; and step 5, 72°C for 10 min. Real-time quantitative RT-PCR was performed using an Applied Biosystems 7500 real-time PCR system, SYBR Premix Ex Taq (TaKaRa), and the following conditions: denaturation at 95°C for 30 s, 41 cycles at 95°C for 5 s, and 60°C for 34 s. Expression of the 16S rRNA gene and that of the actin gene were used as internal standards for kgtP and OsIDH, respectively, and fragments were amplified using the corresponding primers (Table S1). All RT-PCRs were performed in triplicate.
Type III secretion and HR assays.
To generate KgtP with a c-Myc tag, kgtP with its native promoter (−1 to −331 bp upstream) was amplified from the genomic DNA of X. oryzae pv. oryzae PXO99A by PCR with the primer pair KgtPMyc-F/KgtPMyc-R (Table S1). The amplified PCR product was then cloned into pHM1 at the HindIII and PstI sites in frame with a c-Myc epitope-encoding sequence, generating the recombinant plasmid pKgtP-c-Myc (Table 1). pKgtP-c-Myc was then transformed into X. oryzae pv. oryzae strains PXO99A, PΔhrcU, PΔhrpF, PΔhpaB, PΔhpaP, and PΔhrpE (Table 1), which are the wild types, and hrcU, hrpF, hpaB, hpaP, and hrpE mutants.
For Western blot assays, X. oryzae pv. oryzae PXO99A strains were preincubated in NB medium, washed twice, and suspended to an OD600 of 2.0 in sterile water. Bacterial suspension (40 μl) was then inoculated into 1 ml of the hrp-inducing medium XOM3 and grown at 28°C for 16 h. Cell and supernatant fractions were separated by centrifugation, and the protein in the supernatant fraction was precipitated with 12.5% trichloroacetic acid (24). Proteins were separated by electrophoresis in 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to membranes for immunoblotting using anti-c-Myc primary antibodies (Genescript, Nanging, China). Primary antibodies were recognized by anti-rabbit secondary antibodies (Genescript) and visualized on autoradiographs with the Western-Light chemiluminescence system (Transgene, Beijing, China).
O. sativa cv. IRBB10, containing the Xa10 gene, is resistant to X. oryzae pv. oryzae PXO99A strains if it contains avrXa10 (21, 66). The native promoter of kgtP and the region corresponding to 50 amino acids at the N terminus of KgtP were amplified from genomic DNA of X. oryzae pv. oryzae PXO99A with the primer pair kgtPIII-F/kgtPIII-R (see Table S1 in the supplemental material). This product was then fused with the Δ28AvrXa10 construct, in which the T3SS secretion signal is deleted (26), and the fusion was subcloned into pUFR034 at the BamHI and XbaI sites, generating recombinant plasmid pNKgtPΔ28AvrXa10 (Table 1). pNKgtPΔ28AvrXa10 was then transformed into the wild-type PXO99A strain for detection of secreted proteins.
For detection of the HR in rice, X. oryzae pv. oryzae PXO99A strains were suspended at 1 × 108 CFU/ml and infiltrated into 2-week-old IRBB10 rice seedlings with needleless syringes (21). Plants were scored for water soaking or the HR 3 days after inoculation. All plants were maintained in growth chambers at 25°C with a 12-h photoperiod. Experiments were repeated at least three times.
Subcellular-localization assay.
For subcellular localization, the full-length kgtP coding region, 780 bp of kgtP corresponding to the N terminus, and a fusion fragment containing 66 and 72 bp from the regions corresponding to the N and C termini of KgtP, respectively, were cloned into the pEGAD vector at the EcoRI and HindIII sites. This resulted in the construction of three reporter plasmids, pKgtP-GFP, pKgtPΔ6-GFP, and pKgtPΔ12-GFP (Table 1), respectively, where expression was driven by the cauliflower mosaic virus (CaMV) 35S promoter. As a positive control, the xopR gene (PXO_03819) encoding a membrane-binding protein (1) was amplified with the primers xopRSCL-F/xopRSCL-R (Table S1) and cloned in pEDAD, generating pXopR-GFP (Table 1). pKgtP-GFP, pKgtPΔ6-GFP, pKgtPΔ12-GFP, pXopR-GFP, and pEGAD (vector control) were transiently expressed in tobacco (Nicotiana benthamiana) epidermal cells by Agrobacterium-mediated transformation as described previously (20, 23, 48). After infiltration for 24 h, subcellular localization of the KgtP-GFP, KgtPΔ6-GFP, KgtPΔ12-GFP, and XopR-GFP fusion proteins was observed using an MRC-1024 confocal laser scanning microscope (Bio-Rad, CA).
RESULTS
Identification of kgtP in X. oryzae pv. oryzae PXO99A.
Annotation of the X. oryzae pv. oryzae PXO99A genome revealed the existence of kgtP (PXO_03531), which potentially encodes the ketoglutarate transport protein (KgtP). This protein is responsible for transporting α-ketoglutaric acid from the external environment into the bacterial cell of Rhizobium tropici (4). Phylogenetic comparisons revealed that kgtP in X. oryzae pv. oryzae PXO99A is almost identical to homologues in X. oryzae pv. oryzae KACC10331 (25), X. oryzae pv. oryzae MAFF311018 (36), and X. oryzae pv. oryzicola BLS256 (http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?org=Xoc). Furthermore, KgtP also shares 90% and 79% identity to homologues in Xanthomonas campestris and Xanthomonas albilineans, respectively (data not shown).
kgtP is required for full virulence and growth of X. oryzae pv. oryzae in planta.
To facilitate the functional study of kgtP, a nonpolar deletion mutant, designated PΔkgtP (Table 1), was constructed by integration of a suicide plasmid (Fig. S1). For complementation analysis, a recombinant plasmid containing the entire coding region of kgtP and 337 bp of upstream DNA was constructed and designated pCkgtP (Table 1); this plasmid was introduced into PΔkgtP, resulting in strain CPΔkgtP.
The virulence of X. oryzae pv. oryzae PΔkgtP, CPΔkgtP, PΔhrcU (negative control), and wild-type PXO99A was determined by assessing lesion lengths in 5 adult rice (cultivar IR24) plants for each strain in each replicate 14 days after inoculation. Symptom development in PΔkgtP-inoculated leaves was substantially reduced compared to that in leaves inoculated with wild-type PXO99A (Fig. 1A). The mean lesion lengths in PΔkgtP-inoculated leaves were significantly shorter (P = 0.01, t test) than those observed in leaves inoculated with X. oryzae pv. oryzae PXO99A (Fig. 1B). The T3SS mutant PΔhrcU as a negative control produced no obvious BB symptoms in rice (Fig. 1A and B). The BB lesion lengths for CPΔkgtP were equivalent to those induced by wild-type PXO99A (Fig. 1B), indicating that complementation of PΔkgtP with the kgtP gene restored the wild-type phenotype.
Fig 1.
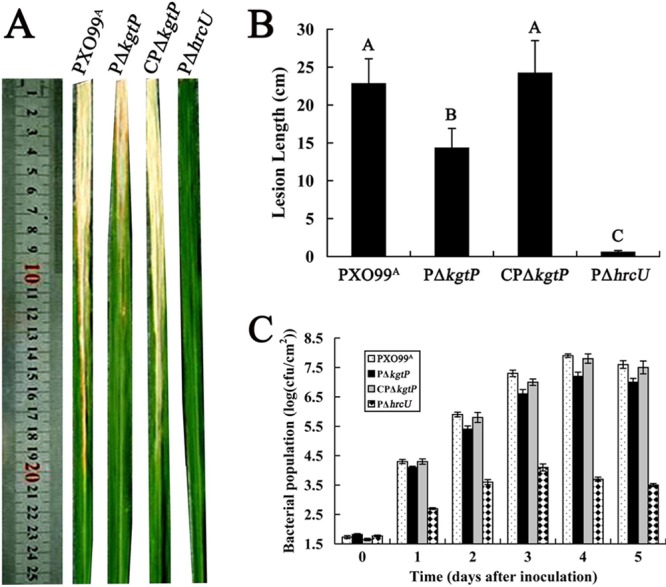
kgtP is required for full virulence and growth of X. oryzae pv. oryzae in planta. (A) Symptoms induced by different X. oryzae pv. oryzae strains inoculated to leaves of 2-month-old rice (IR24, a susceptible cultivar) by the leaf-clipping procedure. Photographs were taken 14 dpi. (B) Bacterial blight lesion lengths in rice. Values represent the means ± standard deviations from three replicates, each containing five leaves. (C) Bacterial growth in inoculated leaves. Inoculated leaves were removed each day for 4 dpi and homogenized in sterile water. The homogenates were diluted and plated on NA plates with appropriate antibiotics. Bacterial CFU were counted after incubation at 28°C for 3 days. Data are the means ± standard deviations from three replicates.
To determine whether kgtP contributes to growth of X. oryzae pv. oryzae in rice, we investigated the population dynamics of PΔkgtP, CPΔkgtP, and wild-type PXO99A in planta. The population of X. oryzae pv. oryzae PΔkgtP was significantly lower than that of wild-type PXO99A at each of the sampling times. Growth of PΔkgtP was restored to wild-type levels when kgtP was present in trans (Fig. 1C). These results indicated that kgtP contributes to the growth of X. oryzae pv. oryzae in planta, although the effect was not as pronounced as seen with the hrcU mutant (PΔhrcU).
To analyze HR induction, tobacco (cultivar Xanthi) leaves were inoculated by infiltrating bacterial suspensions adjusted to a concentration of 1 × 108 CFU/ml. The results showed that PΔkgtP elicited an HR, whereas the T3SS-deficient mutant PΔhrcU did not trigger the HR (data not shown). Thus, kgtP is not involved in HR induction in tobacco, suggesting that the T3SS is not impaired in this mutant.
Functional KgtP is required for growth in NCM medium with α-ketoglutaric acid or sodium succinate.
KgtP is responsible for transferring TCA cycle intermediates and maintaining the concentrations of dicarboxylic acid (14, 30, 68); consequently, we speculated that the kgtP mutant might be impaired for growth on selected carbon sources.
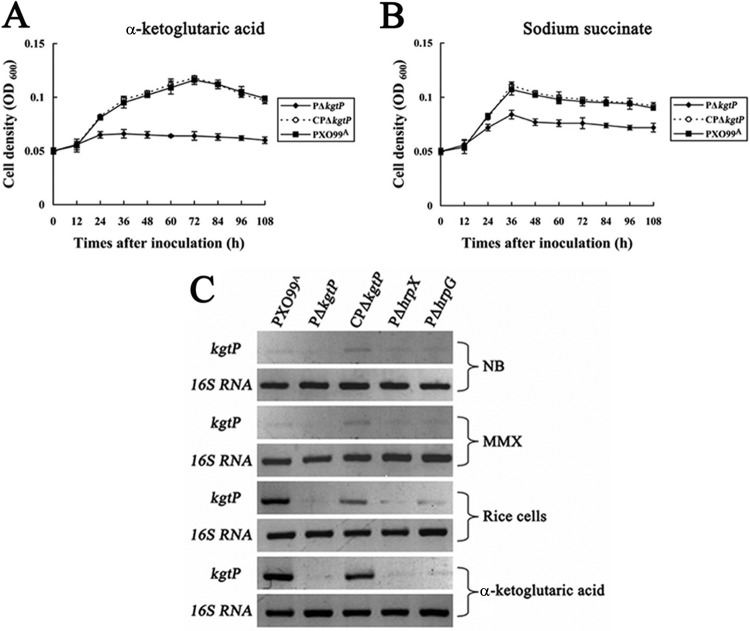
The hrp, hrc, and hpa pathogenicity genes of Xanthomonas are induced in NCM medium, which was developed in an effort to mimic the culture conditions the bacteria encounter in the plant apoplast. To evaluate the contribution of kgtP on carbon source utilization, the growth of PΔkgtP, CPΔkgtP, and wild-type PXO99A was tested in liquid NCM supplemented with 0.5% glucose, pyruvate, malate, citrate, sodium succinate, succinate, α-ketoglutaric acid, or oxaloacetate as the sole carbon source. PΔkgtP was not impaired for growth in NCM medium supplemented with glucose, pyruvate, malate, citrate, succinate, or oxaloacetate (data not shown). However, when PΔkgtP was grown in NCM medium containing sodium succinate or α-ketoglutaric acid as the sole carbon source, its growth was reduced compared to that of wild-type PXO99A (Fig. 2A and B), suggesting that the ability of the mutant to transport and/or utilize these sugars was diminished. The impaired growth was more pronounced when α-ketoglutaric acid was used as a carbon source (Fig. 2A). The growth impairment of the PΔkgtP mutant was restored to wild-type levels by introduction of the pCkgtP complementing clone (Fig. 2A and B).
Fig 2.
Selected carbohydrates impact growth of the kgtP mutant. Growth of X. oryzae pv. oryzae strains in NCM minimal medium containing α-ketoglutaric acid (A) or sodium succinate (B) as the sole carbon source is depicted. The initial concentration of the strains was adjusted to an OD600 of 0.05 in NCM supplemented with α-ketoglutaric acid or sodium succinate. Aliquots were taken in triplicate at 12-h intervals for 108 h. Bacteria were incubated at 28°C, and growth was determined by measuring the OD600. Values given are the means ± standard deviations of triplicate measurements from a representative experiment; similar results were obtained in two other independent experiments. (C) Expression analysis of kgtP in X. oryzae pv. oryzae by RT-PCR. RNAs were isolated from cultures of wild-type PXO99A, PΔkgtP, CPΔkgtP, PΔhrpX, and PΔhrpG strains grown in NB, MMX, rice suspension cells, and NCM supplemented with 0.5% α-ketoglutaric acid for 16 h. The quantity of kgtP mRNA in the tested strains was determined by semiquantitative RT-PCR and the primer kgtP-F1/R1 (see Table S1 in the supplemental material). PCR products were electrophoretically separated on a 1.0% agarose gel. The 16S rRNA PCR product of the pathogen was used as an internal control.
kgtP is positively regulated by HrpX and HrpG.
It is well established that the expression of most T3SS effector genes is repressed in rich media and induced in planta or in minimal media (10–13, 26, 27). Thus, we used RT-PCR to further investigate kgtP expression in X. oryzae pv. oryzae PXO99A, the hrpX mutant (PΔhrpX), and the hrpG mutant (PΔhrpG) in NB, MMX, rice suspension cells, and NCM with α-ketoglutaric acid as the sole carbon source. PΔkgtP and the complemented strain CPΔkgtP were included as negative and positive controls, respectively. As shown in Fig. 2C, the kgtP transcript is highly abundant in wild-type PXO99A and CPΔkgtP when cocultivated with rice cells or grown in NCM supplemented with α-ketoglutaric acid. Regardless of the genetic background, kgtP expression was negligible in rich medium (NB) and MMX (Fig. 2C); thus, the results suggest that kgtP is induced in rice suspension cells and specifically by α-ketoglutaric acid. As predicted, kgtP was not expressed in the mutant PΔkgtP. The lack of kgtP expression in the PΔhrpX and PΔhrpG mutants indicates that kgtP is positively regulated by HrpX and HrpG, respectively.
Analysis of the kgtP promoter region in X. oryzae pv. oryzae PXO99A using promoter prediction software (http://www.fruitfly.org/seq_tools/promoter.html) revealed an imperfect PIP box (TTCGA-N21-TTCGC) upstream of the kgtP start codon (Fig. 3A), suggesting that kgtP may be regulated by HrpX (63, 64). Inspection of the first 50 N-terminal amino acids of KgtP revealed several noteworthy characteristics: a high percentage (>20%) of Pro and Ser residues, the absence of Asp and Glu residues in the first 12 amino acids, and the presence of a Pro residue at the fourth position (Fig. 3A). These features indicated that KgtP may have a targeting signal for the T3SS (10, 43). Collectively, these analyses prompted us to further investigate whether expression of kgtP was modulated by the HrpX regulon and secreted by the T3SS.
Fig 3.
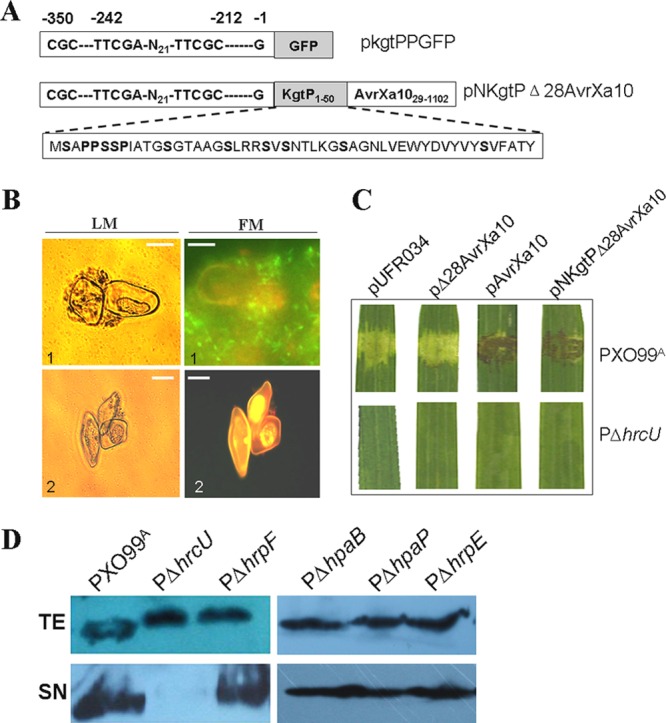
Regulation and secretion of the kgtP gene product. (A) Schematic map showing the kgtP promoter containing the PIP box motifs fused with GFP (top row, in construct pkgtPPGFP), the kgtP promoter, and the N-terminal 50 amino acids of KgtP fused with Δ28AvrXa10 (middle row, in construct pNKgtPΔAvrXa10), and the residue proline (P) and serine (S) consititution of the N-terminal 50 amino acids of KgtP (bottom row). (B) Interaction of X. oryzae pv. oryzae strains with rice suspension cells after a 16-h cocultivation period. Images were acquired by light (LM) or fluorescence (FM) microscopy. X. oryzae pv. oryzae bacterial cells appeared as black dots at rice cell surfaces or gray dots surrounding rice cells under LM and green spindly dots surrounding or near rice cells under FM, while rice cells turned red under FM. The white bar indicates 50 μm. 1, PXO99A(pkgtPPgfp); 2, PΔhrpX(pkgtPPgfp). (C) X. oryzae pv. oryzae strains (3 × 108 CFU/ml) were infiltrated into 2-week-old seedling leaves of IRBB10 rice with needleless syringes. Symptoms were photographed 3 dpi in three independent experiments. The top panels showed symptoms caused by PXO99A(pUFR034), PXO99A(pΔ28AvrX10), PXO99A(pAvrX10), and PXO99A(pNKgtPΔ28AvrXa10), and the bottom panels displayed no symptoms triggered by PΔhrcU(pUFR034), PΔhrcU(pΔ28AvrX10), PΔhrcU(pAvrX10), or PΔhrcU(pNKgtPΔ28AvrXa10). (D) Detection of KgtP secretion by immunoblot analysis. X. oryzae pv. oryzae wild-type PXO99A, PΔhrcU, PΔhrpF, PΔhpaB, PΔhpaP, and PΔhrpE expressing pKgtP-c-Myc were incubated in XOM3 medium. Total protein extracts (TE) and culture supernatants (SN) were analyzed by immunoblotting using anti-c-Myc antibodies.
To determine whether kgtP expression depends on the putative PIP box and HrpX, we constructed a recombinant plasmid, pkgtPPGFP, which contains the kgtP promoter region fused to gfp in pUFR034 (Fig. 3A). Plasmid pkgtPPGFP was introduced into PΔhrpX and wild-type PXO99A by electroporation, and the resulting reporter strains were designated PΔhrpX(pkgtPPGFP) and PXO99A(pkgtPPGFP), respectively. When examined microscopically, strong fluorescence was detected in rice cells cocultivated with the PXO99A(pkgtPPGFP) strain but not in the rice suspension cells inoculated with PΔhrpX(pkgtPPGFP) (Fig. 3B). This result suggests that kgtP expression requires a functional copy of HrpX.
The secretion of KgtP is T3SS dependent.
RT-PCR experiments indicated that the expression of kgtP is positively regulated by HrpX in X. oryzae pv. oryzae PXO99A (Fig. 2C and 3B). Furthermore, the N-terminal sequence of KgtP (Fig. 3A) showed features characteristic of effectors that are secreted via the T3SS. An X. oryzae pv. oryzae effector that is known to be secreted in a T3SS-dependent manner is AvrXa10, a transcription activator-like (TAL) effector that triggers the HR on Oryza sativa cv. IRBB10, which contains the corresponding R gene Xa10 (27, 29, 50). The deletion of the first 28 amino acids at the N terminus of AvrXa10 leads to the failure in secretion to plant cells (27), suggesting that this region contains T3SS secretion signal. To investigate the secretion characteristics of KgtP, the following chimeric constructs were used. pAvrXa10 contains the intact avrXa10 gene and the native avrXa10 promoter in pUFR034, and pΔ28AvrXa10 contains avrXa10 with the region corresponding to the first 28 amino acids deleted and the avrXa10 promoter in pUFR034 (26) (Table 1). pNKgtPΔ28AvrXa10 contains the kgtP promoter (−1 to −359 bp upstream) and the N-terminal 50 amino acids of KgtP fused with Δ28AvrXa10 in pUFR034 (Fig. 3A and Table 1). These three constructs were introduced into PΔhrcU and wild-type PXO99A, and the transformants were inoculated to 2-week-old rice seedlings. Three days postinoculation, PXO99A strains containing pAvrXa10 and pNKgtPΔ28AvrXa10 triggered a typical HR in IRBB10 rice leaves, which contained the Xa10 R gene (Fig. 3C). PXO99A strains containing the empty vector (pUFR034) or the truncated AvrXa10 (pΔ28AvrXa10) did not elicit an HR in IRBB10 rice (Fig. 3C). By comparison, the T3SS mutant PΔhrcU containing pΔ28AvrXa10, pAvrXa10, or pNKgtPΔ28AvrXa10 did not cause any symptoms in rice (Fig. 3C). The results above suggest that the N-terminal 50 amino acids of KgtP enable Δ28AvrXa10 to be secreted into rice cells for HR induction, implying that KgtP may be secreted by the T3SS.
To further study the secretion of KgtP, the gene was expressed in frame with a c-Myc epitope-encoding sequence, and this construct was designated pKgtP-c-Myc (Table 1). This construct was introduced into mutants defective in hrpF (which encodes a translocator that binds to the membranes of host cells) (5), hpaB (which encodes an exit protein in the T3SS) (27, 28), hpaP (which encodes a global chaperone for effector secretion) (10), and hrpE (an hrp pilus protein gene) (59). These mutants were designated PΔhrpF, PΔhpaB, PΔhpaP, and PΔhrpE (Table 1), respectively. Mutant PΔhrcU was included because HrcU is known to encode a two-domain membrane protein that forms an integral part of the T3SS. When the wild-type PXO99A and mutant strains were incubated in XOM3 (hrp-inducing medium) and examined by immunoblotting, the KgtP-c-Myc protein was found to be present in the supernatant (SN) fraction of all mutants except PΔhrcU (Fig. 3D). This result indicates that a fully functional T3SS is needed for secretion of KgtP. The KgtP-c-Myc protein was detected in the SN and total extracts (TE) of PΔhpaF, PΔhpaB, PΔhpaP, and PΔhpaE, indicating that HrpF, HpaB, HpaP, and HrpE are not involved in the secretion of KgtP (Fig. 3D).
KgtP binds plant cell membranes.
Once it was determined that KgtP was secreted via the T3SS of X. oryzae pv. oryzae PXO99A, we then determined where KgtP could localize in plant cells during the Xanthomonas-plant interaction. Homologous to E. coli KgtP protein (45, 57), X. oryzae pv. oryzae KgtP also has 12 transmembrane domains which were evenly interspersed throughout the protein sequence (Fig. 4A), suggesting that it may bind plant cell membrane. To verify this, we generated three GFP-protein fusion constructs: pKgtP-GFP, pKgtPΔ6-GFP, and pKgtPΔ12-GFP (Table 1). The first fusion is the full-length KgtP fused with GFP at the GFP N terminus, the second is the 6 transmembrane domains deleted at the C terminus of KgtP and fused with GFP, and the third the 12 transmembrane domains deleted and fused with GFP (Fig. 4A). XopR-GFP is XopR, which has been verified as a plant membrane-binding protein (1), fused with GFP at the GFP N terminus as a positive control, while pEGAD vector containing GFP was used a negative control. These constructs were transiently expressed in epidermal cells of 5-week-old tobacco plants (N. benthamiana) using Agrobacterium tumefaciens-mediated leaf infiltration (20, 23, 48). The KgtP-GFP and KgtPΔ6-GFP fusion proteins were clearly localized to the plasma membrane of tobacco cells as Xop-GFP (Fig. 4B). However, fluorescence of GFP from the vector pEGAD and the KgtPΔ12-GFP fusion protein could be visualized in the cytosol of the cells (Fig. 4B). These results imply that X. oryzae pv. oryzae KgtP localizes to plant cell membranes.
Fig 4.
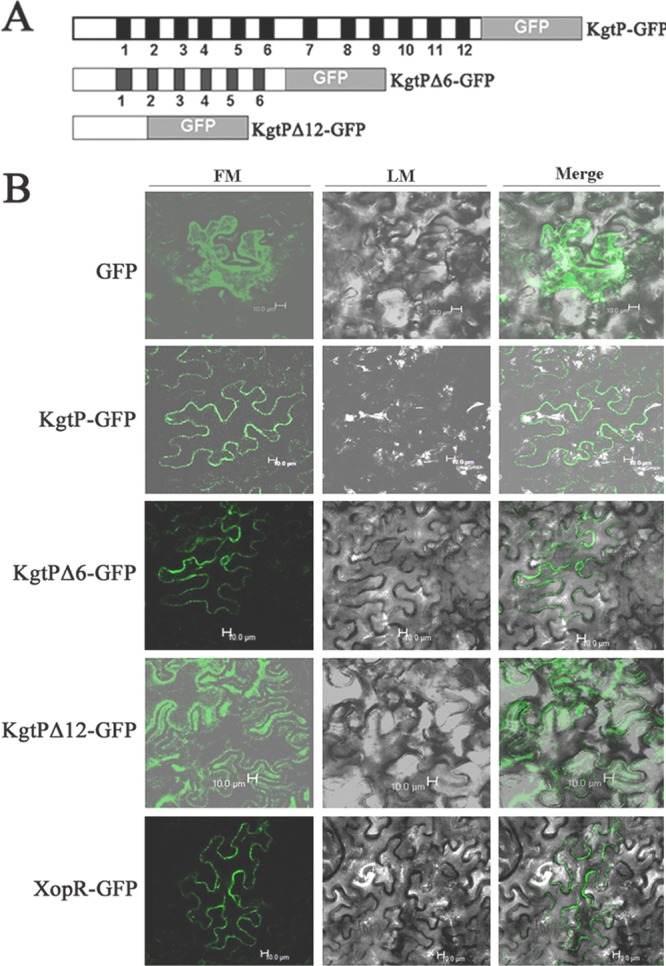
Subcellular localization of KgtP. (A) Transmembrane distribution and schematic map of deletion constructs of KgtP with GFP. The black boxes indicate the membrane-spanning domains in KgtP protein of X. oryzae pv. oryzae based on the homologous KgtP of E. coli (45, 46). KgtPΔ6 and KgtPΔ12 displayed 6 and 12 membrane-spanning domain deletions, respectively, in the KgtP protein of X. oryzae pv. oryzae. (B) Subcellular localization of KgtP-GFP in tobacco epidermal cells. Expression was driven by the CaMV 35S promoter. For confocal laser scanning microscopy, samples were taken 24 h postinoculation. Images were acquired by light microscopy (LM) or fluorescence microscopy (FM).
KgtP is required for wild-type expression of OsIDH in rice.
OsIDH (GenBank accession no. GQ848053.1), which encodes isocitrate dehydrogenase in rice, converts isocitrate to α-ketoglutaric acid. We compared the expression of OsIDH in rice inoculated with X. oryzae pv. oryzae PXO99A, the mutant PΔkgtP, and the T3SS mutant PΔhrcU; the last was used to indicate no secretion of KgtP via the T3SS. In these experiments, we used semiquantitative and real-time quantitative RT-PCR to monitor the expression pattern of OsIDH in rice tissues during X. oryzae pv. oryzae infection. Semiquantitative RT-PCR experiments indicated that the OsIDH transcript level was significantly reduced in rice inoculated with PΔkgtP and PΔhrcU compared to rice plants inoculated with wild-type PXO99A. Real-time quantitative RT-PCR assays for OsIDH expression showed similar results (Fig. 5B). These results indicate that a functional copy of kgtP in X. oryzae pv. oryzae was required for high expression of OsIDH in rice.
Fig 5.
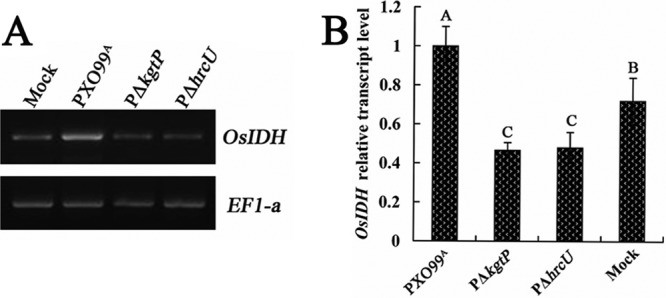
KgtP is required for wild-type levels of OsIDH expression in rice. RNAs were isolated from rice leaves that were infiltrated with cultures of X. oryzae pv. oryzae PXO99A, PΔkgtP, and PΔhrcU strains for 8 h. (A) Semiquantitative RT-PCR analysis. The OsIDH mRNA level of tested strains was determined with the primer OsIDH-F/R (see Table S1 in the supplemental material). PCR products were then electrophoretically separated on a 1% agarose gel. The EF1-a gene of rice was used as an internal standard control. (B) Real-time quantitative RT-PCR analysis. The relative mRNA level of the OsIDH genes in rice inoculated with PΔkgtP was calculated with respect to the level of the corresponding transcripts in rice inoculated with wild-type PXO99A. Values given are the means ± standard deviations of triplicate measurements from a representative experiment. The experiment was repeated at least three times, and similar results were obtained.
DISCUSSION
In this study, we identified a novel X. oryzae pv. oryzae virulence gene, kgtP, which encodes an α-ketoglutarate permease. Annotation of the X. oryzae pv. oryzae PXO99A genome (41) revealed only one copy of kgtP (PXO_03531), which is highly conserved in Xanthomonas spp. KgtP is responsible for importing α-ketoglutaric acid, a TCA cycle intermediate, from the external milieu into the interior of the bacterial cell (Fig. 2 and 6). The dicarboxylate transport (DctA) system for the uptake of C4-dicarboxylates (i.e., oxaloacetate, malate, fumarate, and succinate) has been intensively studied in some bacteria (14, 30, 68). However, little is known about the DctA system in Xanthomonas, despite DCAs being the principal and preferred carbon sources during infection (54, 61). In this study, we demonstrate that mutagenesis of the kgtP gene in X. oryzae pv. oryzae reduced pathogen growth when α-ketoglutaric acid or sodium succinate was used as the sole carbon source (Fig. 2A and B). It is important to note that growth of the kgtP mutant was not impaired on NY, MMX, or NCM containing other carbon sources (e.g., glucose, pyruvate, malate, citrate, succinate, and oxaloacetate), indicating that mutation of this gene does not cause a general metabolic or growth defect. Furthermore, this also suggests that one or more organic acids may be requisite carbon sources during growth in planta. The inactivation of kgtP in X. oryzae pv. oryzae reduced the ability of the pathogen to induce symptoms and grow in rice plants (Fig. 1), possibly because the mutant was impaired in its ability to transport and/or utilize α-ketoglutaric acid. The bacterial growth reduced by the mutation in kgtP on MMX complemented with sodium succinate instead of succinate (Fig. 2C) may support the explanation that X. oryzae pv. oryzae KgtP may also be a sodium-dependent transporter as in E. coli (45, 46, 67). In E. coli, KgtP-mediated α-ketoglutarate transport could be partially inhibited by succinate (46). However, further investigations on whether KgtP is an energy-requiring symporter will facilitate our understanding on how KgtP works as an exporter in host cells for nutrient acquisition.
Fig 6.
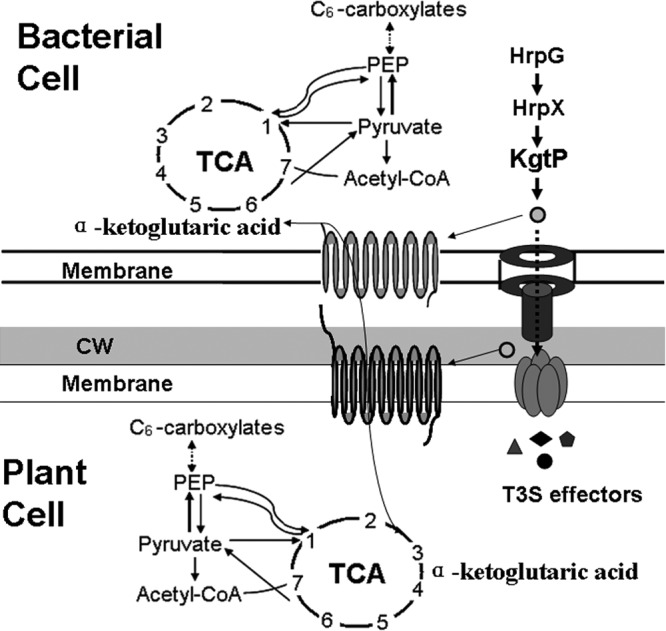
Working model of KgtP in X. oryzae pv. oryzae-rice pathosystem. kgtP, encoding the ketoglutarate transport protein of X. oryzae pv. oryzae PXO99A, is positively regulated by HrpX and HrpG. The protein is presumably secreted in an HpaB-independent manner through the T3SS and localizes to the host cell membrane for α-ketoglutaric acid import. The model speculates that KgtP transports α-ketoglutaric acid to X. oryzae pv. oryzae from external substrates for growth survival and from rice cells for parasitism. PEP, phosphoenolpyruvate; CoA, coenzyme A.
There is an imperfect PIP box in the promoter region of kgtP (Fig. 3A), and the expression of kgtP is positively regulated by HrpX and HrpG (Fig. 2C and 3B). This pattern of regulation follows the established regulatory cascade in which HrpG regulates the expression of hrpX and HrpX activates the expression of the T3SS and some T3SS effectors by binding to the PIP box (13, 19, 22). However, whether HrpX directly activates the expression of kgtP via the imperfect PIP box needs to be validated experimentally. It is well established that the expression of T3SS effectors is suppressed in nutrient-rich media but induced in planta and in apoplast-mimicking media (40, 44, 52). Consistent with this, we found that kgtP expression is induced by rice suspension cells (Fig. 2C). Since many dicarboxylate and tricarboxylate transport genes are induced by their substrates (18, 60), we investigated what conditions stimulate expression of the kgtP promoter. An exciting outcome of these experiments was the discovery that kgtP was induced in the presence of α-ketoglutaric acid but not with other carbon sources. A similar phenomenon exists in the pathogen X. campestris pv. vesicatoria, in which the citH gene functions as a transporter that specifically imports citrate into the cell and is induced by citrate (52).
Although a number of T3SS effectors have been isolated from phytopathogenic bacteria, secretion mechanisms via the T3SS apparatus are not well understood. However, the importance of the N-terminal structures in the secretion of effector gene products has been demonstrated (2, 43, 53), and effector genes modulated by HrpX have been identified (10). The N-terminal region of KgtP showed a motif indicative of secretion by the T3SS (10, 43): Pro and Ser constituted over 20% of the first 50 amino acids, Asp and Glu residues were absent in the first 12 amino acids, and a Pro residue was present at the fourth position. Although universal targeting signals for the T3SS in prokayrotes have not been identified, the above-mentioned features are generally applicable to known T3SS substrates of X. oryzae pv. oryzae, including HpaI, CysP2, and HrpF (11, 12). Thus, we speculated that KgtP is secreted via the T3SS independent of HpaB, HrpE, and HrpF, and our results are consistent with this hypothesis (Fig. 3C and D).
The secretion of KgtP via the T3SS makes it possible that KgtP is involved in α-ketoglutaric acid acquisition from the host cells when X. oryzae pv. oryzae infects rice (Fig. 5A and 6). Sequence analysis revealed that KgtP has 12 transmembrane helices (Fig. 4A) and shows similarity to E. coli KgtP protein, which consists of 12 transmembrane-spanning segments (46). In Sinorhizobium meliloti, previous results suggested that DctA, which belongs to the glutamate transport family, was integrated into the bacterial membrane by three transmembrane domains (69). Therefore, we speculate that KgtP is localized in bacterial membranes and also integrated into the plant cell membrane following secretion through the T3SS (Fig. 6). This hypothesis is based on our subcellular localization analysis with GFP-labeled KgtP, which demonstrated that KgtP localizes to the cell membrane with unique punctate staining as that by XopR, which is a plant membrane-binding protein (Fig. 4B). Our results also suggest that the N-terminal transmembrane helices of KgtP may have a function in binding the membrane of plant cells (Fig. 4B). Since KgtP is secreted via the T3SS and apparently localizes to host plant cell membranes, we speculated that it may play a role in metabolism. Consistent with this hypothesis, the expression of OsIDH, which is responsible for the synthesis of α-ketoglutaric acid, which is an intermediate of the TCA cycle in rice (Fig. 6), was enhanced when KgtP was present in the pathogen (Fig. 5).
On the basis of these findings, we hypothesize that KgtP transports α-ketoglutaric acid to X. oryzae pv. oryzae from external substrates for bacterial growth and from rice cells for parasitism (Fig. 6). Whether KgtP in X. oryzae pv. oryzae PXO99A has other functions in metabolism or pathogenicity is the subject of ongoing studies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Carol Bender, Oklahoma State University, for critically reading the manuscript.
This work was supported by the State Key Basic Research and Development Project of China (2012CB114003), the National Natural Science Foundation of China (31171832) and the Key Basic Research Project of Shanghai Committee of Science and Technology (11JC1406300).
Footnotes
Published ahead of print 8 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akimoto-Tomiyama C, et al. 2012. XopR, a type III effector secreted by Xanthomonas oryzae pv. oryzae, suppresses microbe-associated molecular pattern-triggered immunity in Arabidopsis thaliana. Mol. Plant Microbe Interact. 25:505–514 [DOI] [PubMed] [Google Scholar]
- 2. Alfano JR, Collmer A. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385–414 [DOI] [PubMed] [Google Scholar]
- 3. Arwas R, McKay IA, Rowney FRP, Dilworth MJ, Glenn AR. 1985. Properties of organic acid utilization mutants of Rhizobium leguminosarum strain 300. J. Gen. Microbiol. 131:2059–2066 [Google Scholar]
- 4. Batista S, Patriarca EJ, Tatè R, Martínez-Drets G, Gill PR. 2009. An alternative succinate (2-oxoglutarate) transport system in Rhizobium tropici is induced in nodules of Phaseolus vulgaris. J. Bacteriol. 191:5057–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Büttner D, Nennstiel D, Klüsener B, Bonas U. 2002. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184:2389–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu CC. 1978. The N6 medium and its application to anther culture of cereal crops, p 43–50 In Proceedings of the Symposium on Plant Tissue Culture, May 25–30, 1978 Science Press, Bejing, China [Google Scholar]
- 7. Eisenreich W, Dandekar T, Heesemann J, Goebel W. 2010. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8:401–412 [DOI] [PubMed] [Google Scholar]
- 8. el-Din AK. 1992. A succinate transport mutant of Bradyrhizobium japonicum forms ineffective nodules on soybeans. Can. J. Microbiol. 38:230–234 [DOI] [PubMed] [Google Scholar]
- 9. Finan TM, Wood JM, Jordan DC. 1983. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J. Bacteriol. 154:1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Furutani A, et al. 2009. Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 22:96–106 [DOI] [PubMed] [Google Scholar]
- 11. Furutani A, et al. 2003. HpaI secretion via type III secretion system in Xanthomonas oryzae pv. oryzae. J. Gen. Plant Pathol. 69:271–278 [Google Scholar]
- 12. Furutani A, et al. 2004. Evidence for HrpXo-dependent expression of type II secretory proteins in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 186:1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furutani A, Nakayama T, Ochiai H. 2006. Identification of novel HrpXo regulons preceded by two cis-acting elements, a plant-inducible promoter box and a −10 box-like sequence, from the genome database of Xanthomonas oryzae pv. oryzae. FEMS Microbiol. Lett. 259:133–141 [DOI] [PubMed] [Google Scholar]
- 14. Groeneveld M, Weme RG, Duurkens RH, Slotboom DJ. 2010. Biochemical characterization of the C4-dicarboxylate transporter DctA from Bacillus subtilis. J. Bacteriol. 192:2900–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo W, et al. 2012. Fructose-bisphophate aldolase exhibits functional roles between carbon metabolism and the hrp system in rice pathogen Xanthomonas oryzae pv. oryzicola. PLoS One 7:e31855 doi:10.1371/journal.pone.0031855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He SY, Nomura K, Whittam TS. 2004. Type III protein secretion in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181–206 [DOI] [PubMed] [Google Scholar]
- 17. Hilaire E, et al. 2001. Vascular defense responses in rice: peroxidase accumulation in xylem parenchyma cells and xylem wall thickening. Mol. Plant Microbe Interact. 14:1411–1419 [DOI] [PubMed] [Google Scholar]
- 18. Janausch IG, Zientz E, Tran QH, Kroger A, Unden G. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39–56 [DOI] [PubMed] [Google Scholar]
- 19. Jiang W, et al. 2009. Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant Microbe Interact. 22:1401–1411 [DOI] [PubMed] [Google Scholar]
- 20. Kalantidis K, Tsagris M, Tabler M. 2006. Spontaneous short-range silencing of a GFP transgene in Nicotiana benthamiana is possibly mediated by small quantities of siRNA that do not trigger systemic silencing. Plant J. 45:1006–1016 [DOI] [PubMed] [Google Scholar]
- 21. Kauffman HE, Reddy APK, Hsieh SPV, Merca SD. 1973. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57:537–541 [Google Scholar]
- 22. Koebnik R, Krüger A, Thieme F, Urban A, Bonas U. 2006. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J. Bacteriol. 188:7652–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kościańska E, Kalantidis K, Wypijewski K, Sadowski J, Tabler M. 2005. Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum. Plant Mol. Biol. 59:647–661 [DOI] [PubMed] [Google Scholar]
- 24. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 25. Lee BM, et al. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li YR, et al. 2011. A novel regulatory role of HrpD6 in regulating hrp-hrc-hpa genes in Xanthomonas oryzae pv. oryzicola. Mol. Plant Microbe Interact. 24:1086–1101 [DOI] [PubMed] [Google Scholar]
- 27. Li YR, et al. 2011. HpaII required by HrpF to translocate Xanthomonas oryzae transcriptional activator-like effectors into rice for pathogenicity. Appl. Environ. Microbiol. 77:3809–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo YQ, Zou LF, Li YQ, Zou HS, Chen GY. 2010. hpa1 and hpaB genes contribute pathogenicity to Xanthomonas oryzae pv. oryzicola in rice, but not the hypersensitive response in tobacco. J. Agric. Biotechnol. 18:1038–1045 [Google Scholar]
- 29. Makino S, Sugio A, White FF, Bogdanove AJ. 2006. Inhibition of resistance gene-mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Mol. Plant Microbe Interact. 19:240–249 [DOI] [PubMed] [Google Scholar]
- 30. Mellgren EM, Kloek AP, Kunkel BN. 2009. Mqo, a tricarboxylic acid cycle enzyme, is required for virulence of Pseudomonas syringae pv. tomato strain DC3000 on Arabidopsis thaliana. J. Bacteriol. 191:3132–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Mudgett MB. 2005. New insights in the function of phytopathogenic bacterial type III secretion in plants. Annu. Rev. Plant Biol. 56:509–531 [DOI] [PubMed] [Google Scholar]
- 33. Niño-Liu DO, Ronald PC, Bogdanove AJ. 2006. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7:303–324 [DOI] [PubMed] [Google Scholar]
- 34. Noda T, Kaku H. 1999. Growth of Xanthomonas oryzae pv. oryzae in planta and in guttation fluid of rice. Ann. Phytopathol. Soc. Jpn. 65:9–14 [Google Scholar]
- 35. Noël L, Thieme F, Nennstiel D, Bonas U. 2002. Two novel type III system-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ochiai H, Inoue Y, Takeya M, Sasaki A, Kaku H. 2005. Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39:275–287 [Google Scholar]
- 37. Oku T, et al. 2004. Structural conservation of hrp gene cluster in Xanthomonas oryzae pv. oryzae. J. Gen. Plant Pathol. 70:159–167 [Google Scholar]
- 38. Oku T, et al. 1998. Pathogenicity, non-host hypersensitivity and host defence non-permissibility regulatory gene hrpX is highly conserved in Xanthomonas pathovars. J. Phytopathol. 146:197–200 [Google Scholar]
- 39. Ou SH. 1985. Rice diseases. Commonwealth Agricultural Bureau, Kew, Surrey, United Kingdom [Google Scholar]
- 40. Rahme LG, Mindrinos MN, Panopoulos NJ. 1992. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 174:3499–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salzberg SL, et al. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schulte R, Bonas U. 1992. Expression of the Xanthomonas campestris pv. vesicatoria hrp gene cluster, which determines pathogenicity and hypersensitivity on pepper and tomato, is plant inducible. J. Bacteriol. 174:815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seol W, Shatkin AJ. 1991. Escherichia coli kgtP encodes an alpha-ketoglutarate transporter. Proc. Natl. Acad. Sci. U. S. A. 88:3802–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seol W, Shatkin AJ. 1992. Escherichia coli alpha-ketoglutarate permease is a constitutively expressed proton symporter. J. Biol. Chem. 267:6409–6413 [PubMed] [Google Scholar]
- 47. Seol W, Shatkin AJ. 1993. Membrane topology model of Escherichia coli alpha-ketoglutarate permease by PhoA fusion analysis. J. Bacteriol. 175:565–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1:2019–2025 [DOI] [PubMed] [Google Scholar]
- 49. Staskawicz BJ, Mudgett MB, Dangl JL, Galan JE. 2001. Common and contrasting themes of plant and animal diseases. Science 292:2285–2289 [DOI] [PubMed] [Google Scholar]
- 50. Sugio A, Yang B, White FF. 2005. Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 18:546–554 [DOI] [PubMed] [Google Scholar]
- 51. Tamir-Ariel D, Navon N, Burdman S. 2007. Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J. Bacteriol. 189:6359–6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamir-Ariel D, Rosenberg T, Burdman S. 2011. The Xanthomonas campestris pv. vesicatoria citH gene is expressed early in the infection process of tomato and is positively regulated by the TctDE two-component regulatory system. Mol. Plant Pathol. 12:57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tampakaki AP, Fadouloglou VE, Gazi AD, Panopoulos NJ, Kokkinidis M. 2004. Conserved features of type III secretion. Cell. Microbiol. 6:805–816 [DOI] [PubMed] [Google Scholar]
- 54. Tang DJ, et al. 2005. Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence. J. Bacteriol. 187:6231–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsuge S, et al. 2005. Effects on promoter activity of base substitutions in the cis-acting regulatory element of HrpXo regulons in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 187:2308–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Turner PE. 2004. Phenotypic plasticity in bacterial plasmids. Genetics 167:9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vázquez-Bermúdez MF, Herrero A, Flores E. 2000. Uptake of 2-oxoglutarate in Synechococcus strains transformed with the Escherichia coli kgtP gene. J. Bacteriol. 182:211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang L, Makino S, Subedee A, Bogdanove AJ. 2007. Novel candidate virulence factors in rice pathogen Xanthomonas oryzae pv. oryzicola as revealed by mutational analysis. Appl. Environ. Microbiol. 73:8023–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang YP, Zou LF, Zhou D, Chen GY. 2009. Key roles of hrpE gene of Xanthomonas oryzae pv. oryzicola in formation of Hrp pilus and pathogenicity in rice. Acta Phytopathol. Sinica 39:392–398 [Google Scholar]
- 60. Warner JB, Lolkema JS. 2002. Growth of Bacillus subtilis on citrate and isocitrate is supported by the Mg2+-citrate transporter CitM. Microbiology 148:3405–3412 [DOI] [PubMed] [Google Scholar]
- 61. Watt TF, Vucur M, Baumgarth B, Watt SA, Niehaus K. 2009. Low molecular weight plant extract induces metabolic changes and the secretion of extracellular enzymes, but has a negative effect on the expression of the type-III secretion system in Xanthomonas campestris pv. campestris. J. Biotechnol. 140:59–67 [DOI] [PubMed] [Google Scholar]
- 62. Wengelnik K, Bonas U. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wengelnik K, Van den Ackerveken G, Bonas U. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria, is homologous to two-component response regulators. Mol. Plant Microbe Interact. 9:704–712 [DOI] [PubMed] [Google Scholar]
- 64. Wengelnik K, Rossier O, Bonas U. 1999. Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181:6828–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xiao YL, Li YR, Liu ZY, Xiang Y, Chen GY. 2007. Establishment of the hrp-19 inducing systems for the expression of the hrp genes of Xanthomonas oryzae pv. oryzicola. Acta Microbiol. Sinica 47:396–401 [PubMed] [Google Scholar]
- 66. Yang B, White FF. 2004. Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant Microbe Interact. 17:1192–1200 [DOI] [PubMed] [Google Scholar]
- 67. Yang JR, He YN. 2008. Research progress of sodium-dependent dicarboxylate transporter. Med. Chong Qing 37:427–429 [Google Scholar]
- 68. Youn JW, Jolkver E, Kramer R, Marin K, Wendisch VF. 2009. Characterization of the dicarboxylate transporter DctA in Corynebacterium glutamicum. J. Bacteriol. 191:5480–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yurgel SN, Kahn ML. 2005. Sinorhizobium meliloti dctA mutants with partial ability to transport dicarboxylic acids. J. Bacteriol. 187:1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zou LF, et al. 2006. Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72:6212–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zou LF, Li YR, Chen GY. 2011. A non-marker mutagenesis strategy to generate poly-hrp gene mutants in the rice pathogen Xanthomonas oryzae pv. oryzicola. Agric. Sci. China 10(8):1139–1150 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.