Abstract
Despite major progresses in genetic studies of hyperthermophilic archaea, recombinant protein production in these organisms always suffers from low yields and a robust expression system is still in great demand. Here we report a versatile vector that confers high levels of protein expression in Sulfolobus islandicus, a hyperthermophilic crenarchaeon. Two expression vectors, pSeSD and pEXA, harboring 11 unique restriction sites were constructed. They contain coding sequences of two hexahistidine (6×His) peptide tags and those coding for two protease sites, the latter of which make it possible to remove the peptide tags from expressed recombinant proteins. While pEXA employed an araS promoter for protein expression, pSeSD utilized ParaS-SD, an araS derivative promoter carrying an engineered ribosome-binding site (RBS; a Shine-Dalgarno [SD] sequence). We found that ParaS-SD directed high levels of target gene expression. More strikingly, N-terminal amino acid sequencing of recombinant proteins unraveled that the protein synthesized from pEXA-N-lacS lacked the designed 6×His tag and that translation initiation did not start at the ATG codon of the fusion gene. Instead, it started at multiple sites downstream of the 6×His codons. Intriguingly, inserting an RBS site upstream of the ATG codon regained the expression of the 6×His tag, as shown with pSeSD-N-lacS. These results have yielded novel insight into the archaeal translation mechanism. The crenarchaeon Sulfolobus can utilize N-terminal coding sequences of proteins to specify translation initiation in the absence of an RBS site.
INTRODUCTION
Currently, studying functions of thermophilic archaeal proteins, such as the enzymes involved in DNA replication, gene transcription, and protein translation (10), as well as clustered regularly interspaced palindromic repeat (CRISPR)-associated (Cas) proteins (11, 32), relies almost exclusively on production of recombinant protein from a mesophilic bacterial host such as Escherichia coli and characterization of purified enzymes. However, there is a major drawback in this approach: many thermophilic proteins are insoluble in mesophilic bacterial cells, forming inclusion bodies. Apparently, producing recombinant thermophilic proteins in a homologous host should resolve the problem of protein solubility.
For the past few decades, consistent efforts have been devoted to developing protein expression systems for hyperthermophilic organisms and to establishing genetic tools for studying functions of archaeal genes. As a result, methodologies for gene inactivation and genetic complementation have been established for several model archaeal organisms (reviewed in reference 15), including three hyperthermophilic archaea, the euryarchaea Thermococcus kodakarensis (22) and Pyrococcus furiosus (30), and the crenarchaeon Sulfolobus. While both T. kodakarensis and P. furiosus require an anaerobic condition for growth, Sulfolobus species grow aerobically, which is advantageous for laboratory manipulation. Currently, three Sulfolobus species have been investigated extensively, including Sulfolobus solfataricus (1, 14, 27), S. islandicus (20, 23, 24), and S. acidocaldarius (6, 31).
Despite these exciting developments in genetic studies of hyperthermophilic archaea, the problem of low-level protein expression persists as a bottleneck in producing recombinant protein in hyperthermophilic organisms. Both viral and plasmidic vectors have been employed for expressing recombinant proteins in Sulfolobus, and promoters employed to drive gene expression include those of araS, the heat shock protein gene (tf55α), and a maltose binding protein gene (mbp/mal) (2, 6, 19, 34). Expression systems have been reported for T. kodakarensis (22) and P. furiosus (30), too. Yet, the yield of recombinant protein is always very low in these systems. Consequently, homologous expression still constitutes an unattractive approach to produce a recombinant form of hyperthermophilic enzymes.
In our laboratory, we have focused on developing genetic systems for S. islandicus (reviewed in reference 24), and versatile tools have been established, including routine and novel methods of genetic manipulations (9, 33) and viral and plasmidic shuttle vectors (12, 21, 23). Interestingly, when dissecting the architecture of the Sulfolobus araS promoter using a gene reporter system based on the shuttle plasmid vector pZC1, we found that the araS promoter conferred a high level of expression to a lacS reporter gene in S. islandicus (20, 21). This has prompted us to explore the usefulness of the reporter plasmid to produce recombinant proteins for hyperthermophilic enzymes. Here we report a simple and robust hyperthermophilic protein expression system that facilitates high-yield production of recombinant protein in S. islandicus. The constructed expression vector, pSeSD, is shown to have great versatility in cloning and the ability to remove peptide tags from recombinant proteins. More importantly, the vector carries a synthetic arabinose-inducible promoter that confers high levels of protein expression to several different Sulfolobus genes by a novel translation initiation mechanism.
MATERIALS AND METHODS
Archaeal and bacterial strains used in this work.
The hyperthermophilic archaeal host employed for producing recombinant protein was S. islandicus E233S carrying ΔpyrEF and ΔlacS mutations (9). S. islandicus E233S was derived from S. islandicus REY15A (8). Sulfolobus strains for recombinant protein production (Table 1) were grown at 75 to 78°C in a medium containing basic salts supplemented with 0.2% glucose or sucrose, 0.2% Casamino Acids, and 0.005% yeast extract (GCVy and SCVy, respectively) plus a vitamin mixture. Cells were transferred to ACVy in which d-arabinose was substituted for the other sugar (e.g., sucrose in SCVy) to elevate recombinant protein production.
Table 1.
Sulfolobus strains and vectors used in this study
| Strain or vector | Relevant characteristics | Source or reference |
|---|---|---|
| Sulfolobus strains | ||
| S. islandicus REY15A | Wild type | 7 |
| S. islandicus E233S | REY15A ΔpyrEF ΔlacS | 8 |
| S. solfataricus P2 | Wild type | 20 |
| Vectors | ||
| pZC1 | Sulfolobus-E. coli shuttle vector derived from pHZ2 | 18 |
| pDL1 | Derived from pZC1, lacking PvuII, NdeI, and ZraI sites | 10 |
| pRp | Sulfolobus reporter plasmid constructed with pZC1, containing an arabinose-inducible promoter (−55, +6) and lacS encoding a β-glycosidase as the reporter gene | 18 |
| pEXA | Sulfolobus expression vector derived from pZC1, containing a core araS promoter (ParaS1, −109, +6), an MCS, and a hexahistidine-coding sequence at each terminus, as well as a transcriptional terminator (T6 of SSV1) | 10 |
| pEXA-N-lacS | S. solfataricus lacS was inserted into pEXA to yield a fusion gene coding for N-terminal His tag-LacS fusion protein | This study |
| pEXA-C-lacS | S. solfataricus lacS was inserted into pEXA to yield a fusion gene coding for C-terminal His tag-LacS fusion protein | This study |
| pEXA-N/C-lacS | S. solfataricus lacS was inserted into pEXA to yield a fusion gene coding for a LacS fusion protein fused to a His tag at both ends | This study |
| pSeSD | The synthetic araS-SD promoter was used to substitute for ParaS2 in pEXA | This study |
| pSeSD-estA | An estA gene (SiRe_0290; named pSe-estA in reference 17) was inserted into pSeSD | 17 |
| pSeSD-N-lacS | S. solfataricus lacS was inserted into pSeSD to yield a fusion gene coding for an N-terminal His tag-LacS fusion protein, as for pEXA-N-lacS | This study |
| pSeSD-C-lacS | S. solfataricus lacS was inserted into pSeSD to yield a fusion gene coding for a C-terminal His tag-LacS fusion protein | This study |
| pSeSD-N/C-lacS | S. solfataricus lacS was inserted into pSeSD to yield a fusion gene coding for a LacS fusion protein fused to a His tag at both ends | This study |
| pSeTL1 | ParaS2 of pEXA was replaced with an extended ParaS (−55, +39), the araS promoter plus the coding sequence of the first 11 amino acids of the arabinose-binding protein | This study |
| pSeTL1-estA | S. islandicus estA was inserted into pSeTL1 | This study |
| pSeTL2 | ParaS2 of pEXA was replaced with an extended ParaS (−55, +24), the araS promoter plus the coding sequence of first 6 amino acids of the arabinose-binding protein | This study |
| pSeTL2-estA | S. islandicus estA was inserted into pSeTL2 | This study |
| p10b | The sso-alba gene promoter (−192, +8) was used to substitute for ParaS2 in pEXA | |
| p10b-C-lacS | S. solfataricus lacS was inserted into p10b | This study |
| p7d | The sso7d gene promoter (−231, +7) was used to substitute for ParaS2 in pEXA | This study |
| p7d-C-lacS | S. solfataricus lacS was inserted into p7d | This study |
E. coli DH5α was used for DNA cloning. All E. coli strains were cultured at 37°C in Luria-Bertani (LB) medium, and ampicillin was added to 100 μg/ml if required.
General DNA manipulation.
Restriction and DNA modification enzymes were purchased from New England BioLabs, Fermentas, or Transgen. Plasmid DNA was extracted from E. coli or Sulfolobus cells using either an AxyPrep plasmid minprep kit or a Qiagen plasmid miniprep kit.
The oligonucleotides used in this study (listed in the table in the supplemental material) either were synthesized at Tag Copenhagen or were from Invitrogen, and DNA sequencing of recombinant plasmids was also performed at the same companies.
Construction of Sulfolobus expression vector pSeSD.
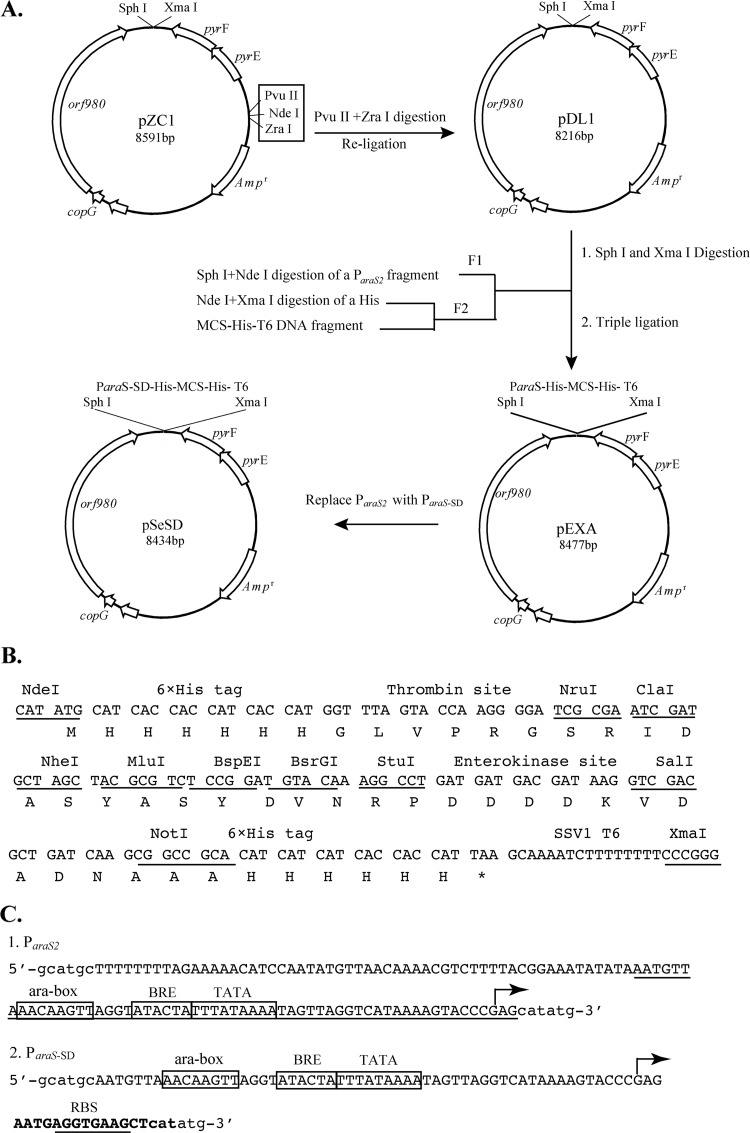
S. islandicus expression vector pEXA was constructed by inserting two DNA fragments, F1 and F2, into the Sulfolobus-E. coli shuttle vector pZC1 (21). DNA fragment F1 was obtained by PCR from S. solfataricus P2 using Pfu DNA polymerase and primers araSF1 and araSR, and it contained a 115-bp promoter fragment of the S. solfataricus arabinose-binding protein gene denoted ParaS2 (−109, +6 [relative to the transcriptional start site]). The F2 fragment was synthesized from annealing of six overlapping oligonucleotides (see the table in the supplemental material) and subsequent gap filling with a DNA polymerase.
A 14-nucleotide sequence containing a ribosome-binding site (RBS; 5′-AGGTGAAG-3′) was incorporated into primer araSSDR by oligonucleotide synthesis and used for PCR with primer araSF2 (see the table in the supplemental material) to yield the synthetic araS-SD (Shine-Dalgarno) promoter (ParaS-SD). Substitution of ParaS-SD for ParaS2 in pEXA gave pSeSD (Fig. 1A).
Fig 1.
Features of the Sulfolobus expression vector pSeSD. (A) Vector construction. Digestion of pZC1 with PvuII and ZraI and subsequent religation eliminated the three restriction sites in the box, giving pDL1. DNA fragment F1 (ParaS2) was obtained by PCR, whereas F2 (MCS) was generated by alignment of 6 oligonucleotides (see the table in the supplemental material) and gap filling with a DNA polymerase (see Materials and Methods). Ligation of F1 and F2 with pDL1 gave pEXA, while replacement of ParaS2 in pEXA with the synthetic inducible promoter ParaS-SD yielded pSeSD. Abbreviations: copG, putative copy-number control protein gene; pyrE, orotate phosphoribosyltransferase gene; pyrF, orotidine 5′-phosphate decarboxylase gene; Ampr, ampicillin resistance gene. (B) Features of the synthetic DNA fragment MCS. The DNA sequence is shown along with the encoded amino acid under each codon. Restriction sites are underlined, with their names indicated above the DNA sequences. Other sequence features, including the 6×His tag, 11 unique restriction sites, thrombin and enterokinase sites, and the SSV1 T6 transcriptional terminator, are also indicated. (C) Sequence features of the ParaS2 and ParaS-SD promoters. Capital letters show the sequence of the promoters, with known promoter elements boxed and indicated above the sequence motifs. Lowercase letters show the restriction sites flanking the promoters. The transcription start site (+1) determined previously (17) is indicated with an arrow. The sequence of ParaS1, which conferred a high level of expression to a reporter gene (21), is underlined within ParaS2. ParaS-SD was constructed by inserting 14 bp between positions +3 and +4 in ParaS1. The 14-bp insertion was generated by oligonucleotide synthesis, and it appears in boldface, with the ribosome-binding site underlined in ParaS-SD. Abbreviations: ara-box, DNA sequence motif conferring arabinose-inducible expression; TATA, AT-rich sequence motif in promoters serving as the binding site for TATA box-binding protein TBP; BRE, transcriptional initiation factor B recognition element; RBS, ribosome-binding site.
Construction of expression plasmids for producing 6×His-tagged β-glycosidase.
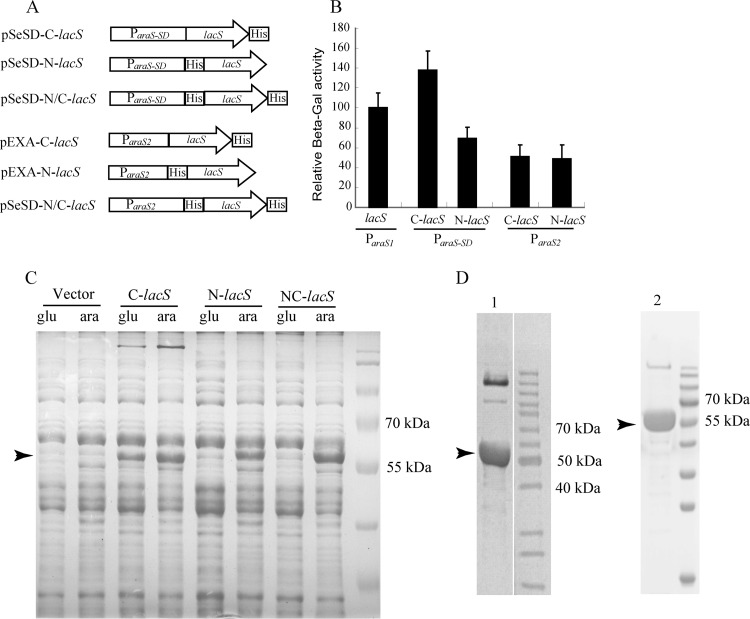
The S. solfataricus lacS gene was amplified by PCR using primers lacSF and lacSR (see the table in the supplemental material). The PCR product was purified, cleaved with NdeI and SalI enzymes, and purified again. The resultant gene fragment was cloned into pEXA (carrying ParaS2) or pSeSD (ParaS-SD), yielding expression plasmids pEXA-C-lacS and pSeSD-C-lacS, respectively. Similarly, expression plasmids pEXA-N-lacS and pSeSD-N-lacS and expression plasmids pEXA-N/C-lacS and pSeSD-N/C-lacS were constructed with the gene fragments generated by PCR with primer sets lacSF/lacSR* (for N-lacS) and lacSF2/lacSR (for N/C-lacS), respectively. Restriction sites used for the cloning were NheI and SalI. Features of these plasmids were summarized in Table 1 and are shown in Fig. 3A.
Fig 3.
Characterization of recombinant β-galactosidase proteins carrying different 6×His tags expressed from ParaS-SD and ParaS2. (A) Schematic diagrams of six plasmids expressing 6×His β-glycosidase fusion proteins from ParaS-SD (pSeSD) or ParaS2 (pEXA). The features of the two vectors and the promoters present in their expression cassettes are shown in Fig. 1. (B) Relative activity of the recombinant β-glycosidase proteins expressed from the two promoters. Strains carrying each plasmid were grown in ACVy to an OD600 of ca. 0.2. Cell mass from which cell extracts were prepared was collected for each culture and used for determining β-galactosidase (β-Gal) activity. The activity of the reporter plasmid pRp (21) was set to 100% and used as a reference to normalize the specific activity of recombinant β-glycosidase enzymes expressed from other plasmids. Assays were conducted in triplicate. (C) SDS-PAGE of recombinant β-glycosidase proteins produced from pSeSD constructs. Strains carrying each plasmid were grown in either GCVy (glu) or ACVy (ara) to an OD600 of ca. 0.2. Cell mass from which cell extracts were prepared was collected for each culture. Equal amounts of cell lysates were loaded onto a 12% SDS-polyacrylamide gel to visualize the recombinant proteins in the context of total protein. The arrowhead indicates the position of the recombinant proteins. (D) Recombinant proteins purified from the overexpression strains carrying pEXA-C-lacS (lane 1) and pEXA-N/C-lacS (lane 2). Cultures were grown in ACVy as described above, and 6×His-tagged recombinant proteins were purified using a nickel-affinity column. Recombinant protein produced from pEXA-N-lacS could not be purified by His tag-affinity chromatography. Arrowheads indicate the protein bands that were transferred onto a membrane and sent for determination of their N-terminal amino acid sequences. The sequencing results are presented in Table 2.
Furthermore, expression vectors carrying two strong constitutive promoters were constructed, namely, p7d (Psso7d amplified with the Sso7dF/R primers) and p10b (Palba amplified with albaF/R primers). This was done by replacement of the promoter module of pSeSD-C-lacS with each of the above-described promoter modules, yielding p7d-C-lacS and p10b-C-lacS, respectively.
Construction of other expression plasmids.
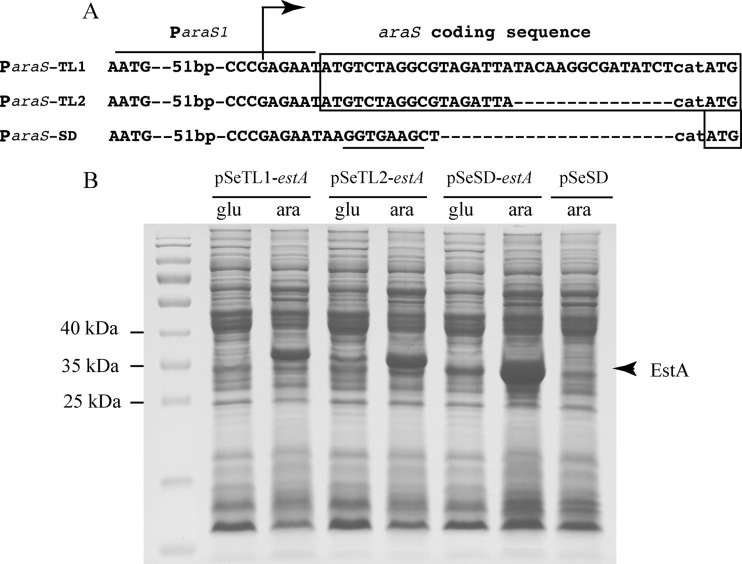
Two fragments containing an extended araS promoter, ParaS-TL1 (−55, +39 [relative to the transcriptional start site]) and ParaS-TL2 (−55, +24), were amplified by PCR using primer sets araSF2/araSR_TL1 and araSF2/araSR_TL2, respectively. The PCR products were digested with SphI and NdeI, and the purified DNA fragments were inserted into pSeSD to replace the original promoter sequence, resulting in expression vectors pSeTL1 and pSeTL2, respectively. A DNA fragment containing S. islandicus estA (SiRe_0290) (13) was amplified from pSeSD-estA (19) by PCR with the corresponding primers and inserted into the above-described vectors, giving expression plasmids pSeTL1-estA and pSeTL2-estA, respectively (Table 1).
Expression and purification of recombinant proteins of His-tagged β-glycosidase in S. islandicus.
Expression plasmids were introduced into S. islandicus E233S via electroporation, and transformants were selected on SCVy plates as described previously (9). A few colonies were grown in liquid medium of the same composition and tested for expression of recombinant proteins. His-tagged recombinant proteins were purified from Sulfolobus cells as described previously (19). One of the transformants was cultured in 0.5 to 1 liter of ACVy until the cell density (optical density at 600 nm [OD600]) reached ca. 0.6. The cell mass was harvested by centrifugation and resuspended in lysis buffer (50 mM phosphate-buffered saline, 500 mM NaCl, 20 mM imidazole). After disruption of the cells with sonication, cell debris was removed by centrifugation (10,000 × g for 30 min at 4°C). Then, the yielded supernatant was applied to an Ni-nitrilotriacetic acid column (Novagen) and His-tagged recombinant proteins were purified by affinity chromatography following the manufacturer's instruction. The quality of the purified proteins was checked by SDS-PAGE.
Determination of N-terminal sequences of recombinant β-glycosidase proteins.
Purified recombinant β-glycosidase proteins were separated according to their sizes in an SDS-polyacrylamide gel. Proteins in the gel were transferred to a polyvinylidene difluoride membrane using a semidry electrophoretic transfer cell (Bio-Rad). After they were stained with Coomassie blue R-250, the membrane pieces containing the recombinant β-glycosidase proteins were excised from the membrane and sent to Alphalyse Inc., Denmark, for determining their N-terminal sequences.
β-Galactosidase assay.
β-Glycosidase activity in the cell extracts of S. islandicus strains was determined with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate as described previously (21). Yields of o-nitrophenol were determined by measurement at 420 nm using Nanodrop1000 spectrophotometers. One specific milliunit (mU) was defined as 1 mmol o-nitrophenol produced per min per mg total protein, and the relative expression activity of each plasmid was normalized with the specific activities of the reporter gene plasmid pRp.
RESULTS AND DISCUSSION
Construction of versatile Sulfolobus protein expression vectors.
Expression vectors have been reported for S. solfataricus (2) and for S. acidocaldarius (6), but none of the systems have been optimized for cloning versatility or for robust protein expression. In a previous work, we found that a reporter plasmid based on the Sulfolobus-E. coli shuttle vector pZC1 and an araS promoter conferred high-level expression of the lacS reporter gene when the β-glycosidase activity in cell extracts was assayed (21). The activity obtained from our reporter gene system is 2- to 6-fold of the activities expressed from Sulfolobus shibatae virus 1 (SSV1)-derived expression plasmids (2). Therefore, we exploited the S. islandicus/pZC1 system for robust protein expression.
First, a DNA fragment containing a multiple-cloning site (MCS) was generated with 6 overlapping oligonucleotides (see Materials and Methods) to facilitate the cloning process. In fact, this fragment contained several sequence modules, including (i) the coding sequences of two 6×His peptide tags, (ii) two sequence modules coding for the cleavage sites of thrombin and enterokinase proteases, and (iii) the transcriptional terminator of transcript 6 (T6) of the Sulfolobus virus SSV1 (27) (Fig. 1B). The sixth module in the resultant vectors is the promoter of the S. solfataricus araS gene (ParaS2), which was inserted in front of the MCS to drive gene expression (Fig. 1C).
Compared to other reported archaeal expression vectors (2, 3, 6, 22), pEXA exhibits a strong cloning versatility since there are 11 unique restriction sites, all of which are suitable for DNA cloning. Furthermore, the promoter and the coding regions of the peptide tags at the N and C termini are all flanked by restriction sites, and this makes it possible to exchange transcriptional promoters, terminators, as well as peptide-coding modules in one-step cloning. At the recombinant protein level, each peptide tag could be removed from the fusion protein by a simple protease digestion (Fig. 1).
Then, we attempted to increase the level of protein expression by genetic engineering of the Sulfolobus araS promoter. The consensus sequence of the RBS (5′-AGGTGA-3′) identified by analyzing the Sulfolobus genomes (28) was inserted into the araS promoter to yield the synthetic promoter denoted ParaS-SD, and this promoter was used to replace the original araS promoter on pEXA, giving the expression vector pSeSD (Fig. 1C).
The RBS sequence strongly elevated the expression of a Sulfolobus esterase gene.
Previously, we expressed a few S. solfataricus recombinant proteins with pEXA, including a β-glycosidase, three DNA polymerase B enzymes, and the DNA double-strand break repair protein Mre11 (unpublished data), but only the S. solfataricus β-glycosidase was expressed to a relatively high level (see below). Since pEXA is based on a transcriptional fusion of the araS promoter and a target gene, we asked if construction of a translational fusion could affect protein expression from the expression system. To test this, two translational fusion expression plasmids, namely, pSeTL1-estA and pSeTL2-estA (Table 1), each containing an extended araS promoter (ParaS-TL1 or ParaS-TL2), were constructed. While ParaS-TL1 retained the coding sequence of the first 11 amino acids (aa) of the arabinose-binding protein (−55, +39), ParaS-TL2 had 6 aa (−55, +24) (Fig. 2A). The third expression plasmid (pSe-estA) was previously constructed with the versatile expression vector pSeSD (19). The target gene was S. islandicus SiRe_0290, encoding an esterase (EstA). Sulfolobus strains harboring these plasmids were obtained, and their recombinant protein synthesis was analyzed by SDS-PAGE under both inducible and noninducible conditions (in ACVy and GCVy, respectively).
Fig 2.
Expression efficiencies of the artificial promoter ParaS-SD and two translational fusion promoters, ParaS-TL1 and ParaS-TL2. (A) Features of the three promoters ParaS-SD, ParaS-ST1, and ParaS-ST2. All three promoters were derived from the 59-bp ParaS1 (−55 to +4) promoter, with its complete sequence shown in Fig. 1C. The coding sequences present in the translational fusion vectors are boxed, whereas dashes indicate gaps in the sequences. The ribosome-binding site (SD) is underlined. The transcriptional start site determined previously for the araS gene (17) is indicated with an arrow. (B) SDS-PAGE analysis of total proteins of strains expressing EstA, an esterase encoded by SiRe_0290 (13). Sulfolobus strains carrying each plasmid (Table 1) were grown in either GCVy (glu) or ACVy (ara) to an OD600 of ca. 0.2. A cell mass with which cell extracts were prepared was collected from each culture. Equal amounts of cell lysates were analyzed with a 12% SDS-polyacrylamide gel, and protein bands were visualized by Coomassie blue R-250 staining. The position of the recombinant EstA proteins is indicated with an arrowhead.
As shown in Fig. 2B, A major band of recombinant protein appeared at the predicted size in the sample of each transformant grown in ACVy, indicating that all tested promoters could drive high-level EstA expression in S. islandicus. When grown in GCVy, essentially no recombinant protein was detectable for the cells transformed with pSeTL1-estA, although a significant amount of recombinant protein was synthesized from pSeTL2-estA and pSeSD-estA (Fig. 2B). Because the araS promoter (−55, +4) retained 19% residual activity under a noninducible condition, the essentially inactive nature of ParaS-TL1 was indicative of an additional negative regulatory element in the araS N-terminal coding sequence. Importantly, ParaS-SD conferred the highest level of protein expression among the araS-derived promoters.
Translational start from the ATG codon in front of the hexahistidine codons required a preceded RBS motif.
To systematically study tagged protein expression from ParaS2 and from ParaS-SD, we designed fusion genes encoding recombinant β-glycosidases carrying N-terminal, C-terminal, or N-terminal plus C-terminal His tags (Fig. 3A). The expression of the recombinant proteins in Sulfolobus cells was assessed by the β-galactosidase assay and by SDS-PAGE. High levels of β-galactosidase activity were obtained from expression of N-terminal and C-terminal 6×His-tagged β-galactosidase for all four constructs either from pEXA or from pSeSD. However, there were also important differences in their activities. The activity from pSeSD-C-lacS was 2-fold of that from pSeSD-N-lacS and ca. 3-fold of the activities from the pEXA-lacS expression plasmids (Fig. 3B).
We also studied the synthesis of recombinant β-glycosidase from all three pSeSD-lacS constructs by SDS-PAGE analysis of the total proteins of their transformants. While a relatively high level of recombinant protein was already expressed from pSeSD-C-lacS in GCVy, no clear bands were observed for the recombinant proteins expressed from the other two expression plasmids in the same medium. However, protein bands of a high intensity were present in the total proteins after arabinose induction for all three pSeSD constructs, which is indicative of high levels of induction of protein expression, at least for pSeSD-N-lacS and pSeSD-N/C-lacS (Fig. 3C).
More strikingly, when cell extracts of these strains were used for tag-affinity purification of recombinant protein, His-tagged recombinant protein was not obtainable for pEXA-N-lacS, suggesting that the recombinant protein produced from the latter expression plasmid could be missing the designed His tag at the N terminus. To study this possibility, the C-terminal His-tagged and the N-terminal plus C-terminal His-tagged recombinant forms of β-glycosidase were purified by histidine-affinity purification. The purified proteins were analyzed on an SDS-polyacrylamide gel, and proteins with sizes of 50 to 60 kDa were obtained. Both proteins were sent for N-terminal amino acid sequencing, and the sequencing results obtained are summarized in Table 2.
Table 2.
Translational initiation of recombinant proteins from the pEXA vector
| Cycle | Residue no. | pEXA-C-lacS |
pEXA-N/C-lacS |
||
|---|---|---|---|---|---|
| Deduced amino acid | Sequencing result | Deduced amino acid | Sequencing result(s) | ||
| 1 | Blank | ||||
| 2 | Standard | ||||
| 3 | 1 | M | M | M | S/A/T (M/Y/G/K/V/L/I) |
| 4 | 2 | Y | Y | H | ? |
| 5 | 3 | S | S | H | (P/Y) |
| 6 | 4 | F | F | H | (L) |
| 7 | 5 | P | P | H | (Y/A/P/V/I) |
| 8 | 6 | N | N | H | (W/A/Y/P) |
While the C-terminus-tagged β-glycosidase protein was expressed as the designed fusion protein because the amino acid sequence obtained at the N terminus was identical to its coding sequence, the N-terminal sequence of the recombinant protein with both N- and C-terminal tags could not be resolved because signals of different amino acids were observed from each round of amino acid determination. This indicates that protein synthesis must have initiated at multiple sites downstream of the designed His tag codons, suggesting that the AUG codon preceding the synthetic His tag-coding sequence failed to specify translation initiation. Taken together, our results unravel for the first time that the N-terminal coding sequence of β-glycosidase could contain a signal(s) that specifies translational initiation.
Genomics analyses of several archaeal genomes have revealed that archaeal genes belong to two groups: group 1 includes all single genes and first genes of operons, while group 2 genes include those lying downstream from the first genes of operons (25, 28). In Sulfolobus genomes, group 1 genes produce either leaderless or short-leadered transcripts, all of which lack an RBS motif, while group 2 genes are often preceded by an SD motif (28, 29). An early study with a Sulfolobus cell-free translation system revealed that the SD motif was not essential to archaeal translation and deletion of the element increased the reporter gene activity (7). Here we found that insertion of hexahistidine codons immediately after the AUG start codon caused the shift of translation initiation from the start codon to multiple sites downstream of the hexahistidine codons, i.e., within the coding region of the N-terminal sequence, and the proteins yielded were still very active. Taken together, both in vivo and in vitro data suggested that, for the genes lacking an RBS site, their N-terminal coding sequences could have the ability to confer translation initiation. Conversely, when a gene already possesses an RBS, its coding sequence may lack such a function. Thus, by inserting an RBS motif into the araS promoter, the resulting synthetic promoter should function for both categories of genes, and this feature could account for the application of pSeSD as a more successful vector in expressing recombinant proteins in Sulfolobus than all other existing expression vectors.
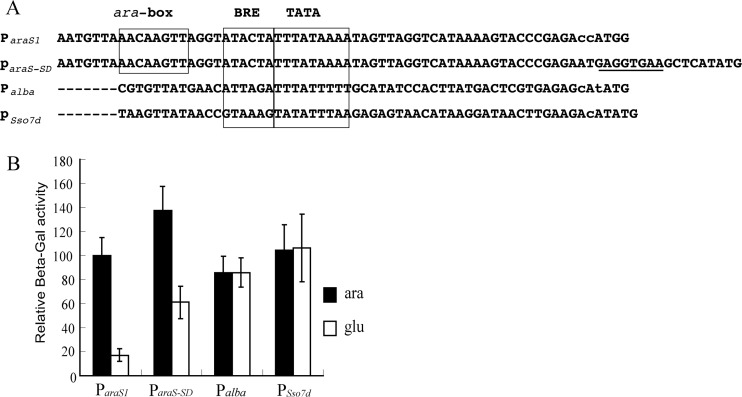
Demonstration of ParaS-SD as the strongest promoter in driving protein expression in S. islandicus known to date.
The Sulfolobus chromosomal proteins Sac7d/Sso7d and Alba proteins were found to be very abundant in the Sulfolobus cells (4, 5, 18), and consequently, their gene promoters are regarded as very strong promoters. To compare the activity of these promoters with that of ParaS-SD, the expression plasmids p10b-C-lacS and p7d-C-lacS were constructed. Again, the reporter gene plasmid with ParaS2 was used as a reference. Interestingly, the alba (Sso10b, Sso6877) promoter showed 87% relative activity both in ACVy and in GCVy, whereas the Sso7d (Sso9180) promoter showed 105% and 108% activity in these two media, respectively (Fig. 4). Strikingly, however, the relative activity of the artificial arabinose-inducible promoter ParaS-SD was the highest among the promoters tested, exhibiting 138% relative activity in ACVy, and its activity was also very high in GCVy, retaining 60% of it activity in the absence of arabinose, indicating that araS-SD could confer much higher constitutive gene expression than ParaS. These results are also in good agreement with those of the SDS-PAGE analysis of total proteins expressed from pSeSD-C-lacS (Fig. 3C), where a large amount of recombinant protein was synthesized in the absence of arabinose.
Fig 4.
Comparison of the promoter activities of ParaS-SD and other known strong promoters. (A) Sequence comparison of ParaS1, ParaS-SD, Palba, and Psso7d. Promoter elements BRE (TFB recognition element) and TATA (TATA-binding motif), identified for the S. solfataricus araS promoter, are boxed along with the ara box element, responsible for arabinose-inducible expression. Dashes indicate sequence gaps. The engineered ribosome-binding site in ParaS-SD is underlined. (B) Relative β-galactosidase activity of different promoter constructs. Cells were cultured in ACVy (ara) or GCVy (glu) and were harvested at an OD600 of ca. 0.2. Crude cell extracts were prepared and assayed for β-galactosidase activity. All assays were conducted in triplicate.
We also noticed that the β-galactosidase activity from p10b-C-lacS was 87% of that from pRp (Fig. 4), but this does not simply imply that ParaS1 could be more active than Palba. The recombinant β-glycosidase proteins expressed from the two plasmids are in different forms. While p10b-C-lacS expresses a β-galactosidase with a 6×His tag at the C terminus, the enzyme expressed from pRp is devoid of any peptide tag. This fusion β-glycosidase could exhibit lower specific activity than the tag-free form. Indeed, ParaS1 conferred ca. 80% of the activity of ParaS2 in a reporter gene assay, but the activity of untagged β-glycosidase from pRp (ParaS1) was about 2-fold of the activity of C-terminal 6×His-tagged β-glycosidase produced from pEXA-C-lacS (Fig. 2B).
To date, the usefulness of the pSeSD expression vector has been documented worldwide both in S. islandicus and in S. solfataricus. This includes usefulness for (i) expression of viral protein c92 and demonstration of its function in the formation of pyramid-like cellular lysis structures in S. solfataricus (26); (ii) expression of Csa2 (a CRISPR-associated protein in the same host has led to the copurification of its interaction partners and the subsequent identification of the first putative CRISPR-associated complex for antiviral defense complex in archaea [16]); and (iii) retrieval of large amounts of the S. islandicus enzymes EstA and HerA (19, 34) for biochemical characterization of the recombinant enzyme, which has revealed exceptional biochemical properties for the EstA enzyme. Currently, we employ this expression system to obtain large amounts of homologous recombinant proteins for crystal structural analysis. A wider range of application of this versatile cloning vector for producing recombinant hyperthermophilic proteins will further facilitate research in archaeal biology and biotechnology.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the Danish Council for Independent Research/FTP (09-062332 and 11-106683) and by grants from the National Natural Science Foundation of China (no. 31100050, no. 31100096, and no. 31128011).
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Albers SV, Driessen AJ. 2008. Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea 2:145–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albers SV, et al. 2006. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 72:102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allers T, Barak S, Liddell S, Wardell K, Mevarech M. 2010. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76:1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baumann H, Knapp S, Karshikoff A, Ladenstein R, Hard T. 1995. DNA-binding surface of the Sso7d protein from Sulfolobus solfataricus. J. Mol. Biol. 247:840–846 [DOI] [PubMed] [Google Scholar]
- 5. Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. 2002. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148–151 [DOI] [PubMed] [Google Scholar]
- 6. Berkner S, Wlodkowski A, Albers SV, Lipps G. 2010. Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles 14:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Condo I, Ciammaruconi A, Benelli D, Ruggero D, Londei P. 1999. Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 34:377–384 [DOI] [PubMed] [Google Scholar]
- 8. Contursi P, et al. 2006. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10:615–627 [DOI] [PubMed] [Google Scholar]
- 9. Deng L, Zhu H, Chen Z, Liang YX, She Q. 2009. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13:735–746 [DOI] [PubMed] [Google Scholar]
- 10. Garrett RA, Klenk HP. (ed). 2007. Archaea: evolution, physiology, and molecular biology. Blackwell Publishing, Malden, MA [Google Scholar]
- 11. Garrett RA, et al. 2011. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem. Soc. Trans. 39:51–57 [DOI] [PubMed] [Google Scholar]
- 12. Gudbergsdottir S, et al. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 79:35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo L, et al. 2011. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 193:1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jonuscheit M, Martusewitsch E, Stedman KM, Schleper C. 2003. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol. Microbiol. 48:1241–1252 [DOI] [PubMed] [Google Scholar]
- 15. Leigh JA, Albers SV, Atomi H, Allers T. 2011. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35:577–608 [DOI] [PubMed] [Google Scholar]
- 16. Lintner NG, et al. 2011. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 286:21643–21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lubelska JM, Jonuscheit M, Schleper C, Albers SV, Driessen AJ. 2006. Regulation of expression of the arabinose and glucose transporter genes in the thermophilic archaeon Sulfolobus solfataricus. Extremophiles 10:383–391 [DOI] [PubMed] [Google Scholar]
- 18. Mai VQ, Chen X, Hong R, Huang L. 1998. Small abundant DNA binding proteins from the thermoacidophilic archaeon Sulfolobus shibatae constrain negative DNA supercoils. J. Bacteriol. 180:2560–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mei Y, et al. 2012. Exceptional thermal stability and organic solvent tolerance of an esterase expressed from a thermophilic host. Appl. Microbiol. Biotechnol. 93:1965–1974 [DOI] [PubMed] [Google Scholar]
- 20. Peng N, Ao X, Liang YX, She Q. 2011. Archaeal promoter architecture and mechanism of gene activation. Biochem. Soc. Trans. 39:99–103 [DOI] [PubMed] [Google Scholar]
- 21. Peng N, Xia Q, Chen Z, Liang YX, She Q. 2009. An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol. Microbiol. 74:928–939 [DOI] [PubMed] [Google Scholar]
- 22. Santangelo TJ, Cubonova L, Reeve JN. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74:3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. She Q, et al. 2008. Host-vector systems for hyperthermophilic archaeon Sulfolobus, p 151–156 In Liu S-J, Drake HL. (ed), Microbes and the environment: perspective and challenges. Science Press, Beijing, China [Google Scholar]
- 24. She Q, et al. 2009. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 37:92–96 [DOI] [PubMed] [Google Scholar]
- 25. Slupska MM, et al. 2001. Leaderless transcripts of the crenarchaeal hyperthermophile Pyrobaculum aerophilum. J. Mol. Biol. 309:347–360 [DOI] [PubMed] [Google Scholar]
- 26. Snyder JC, Brumfield SK, Peng N, She Q, Young MJ. 2011. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J. Virol. 85:6287–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stedman KM, Schleper C, Rumpf E, Zillig W. 1999. Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: construction and testing of viral shuttle vectors. Genetics 152:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tolstrup N, Sensen CW, Garrett RA, Clausen IG. 2000. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4:175–179 [DOI] [PubMed] [Google Scholar]
- 29. Torarinsson E, Klenk HP, Garrett RA. 2005. Divergent transcriptional and translational signals in Archaea. Environ. Microbiol. 7:47–54 [DOI] [PubMed] [Google Scholar]
- 30. Waege I, Schmid G, Thumann S, Thomm M, Hausner W. 2010. Shuttle vector-based transformation system for Pyrococcus furiosus. Appl. Environ. Microbiol. 76:3308–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagner M, et al. 2009. Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem. Soc. Trans. 37:97–101 [DOI] [PubMed] [Google Scholar]
- 32. Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338 [DOI] [PubMed] [Google Scholar]
- 33. Zhang C, et al. 2010. Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology 156:3386–3397 [DOI] [PubMed] [Google Scholar]
- 34. Zheng T, et al. 2012. Development of a simvastatin selection marker for a hyperthermophilic acidophile, Sulfolobus islandicus. Appl. Environ. Microbiol. 78:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.