Abstract
Streptococcus gordonii is an early colonizer of the human oral cavity and an abundant constituent of oral biofilms. Two tandemly arranged gene clusters, designated lac and gal, were identified in the S. gordonii DL1 genome, which encode genes of the tagatose pathway (lacABCD) and sugar phosphotransferase system (PTS) enzyme II permeases. Genes encoding a predicted phospho-β-galactosidase (LacG), a DeoR family transcriptional regulator (LacR), and a transcriptional antiterminator (LacT) were also present in the clusters. Growth and PTS assays supported that the permease designated EIILac transports lactose and galactose, whereas EIIGal transports galactose. The expression of the gene for EIIGal was markedly upregulated in cells growing on galactose. Using promoter-cat fusions, a role for LacR in the regulation of the expressions of both gene clusters was demonstrated, and the gal cluster was also shown to be sensitive to repression by CcpA. The deletion of lacT caused an inability to grow on lactose, apparently because of its role in the regulation of the expression of the genes for EIILac, but had little effect on galactose utilization. S. gordonii maintained a selective advantage over Streptococcus mutans in a mixed-species competition assay, associated with its possession of a high-affinity galactose PTS, although S. mutans could persist better at low pHs. Collectively, these results support the concept that the galactose and lactose systems of S. gordonii are subject to complex regulation and that a high-affinity galactose PTS may be advantageous when S. gordonii is competing against the caries pathogen S. mutans in oral biofilms.
INTRODUCTION
The human mouth accommodates a diverse and dynamic microbial ecosystem composed of hundreds of bacterial species, and the stability of these communities is associated with an array of antagonistic and cooperative interactions between constituents of the biofilms (26). Of particular importance to oral health and diseases are the viridans group streptococci, Gram-positive bacteria that are classified into the anginosus, mitis, salivarius, bovis, and mutans groups (25). The members of the mitis group, which includes Streptococcus gordonii, Streptococcus mitis, Streptococcus oralis, Streptococcus parasanguinis, and Streptococcus sanguinis (25), are among the first organisms to establish in the oral cavity and remain abundant members of the biofilms that colonize the hard and soft tissues of the mouth throughout the lifetime of the host. Many members of the mitis group, including the organism that is the focus of this study, S. gordonii, are frequently associated with dental health. In contrast, Streptococcus mutans, a member of the mutans group of streptococci and certain other lactic acid bacteria (e.g., lactobacilli), is generally regarded as the primary etiological agent of dental caries. In some cases, an inverse relationship of the proportions of S. mutans and S. gordonii (16, 27) in human plaque samples has been observed, consistent with the observation that S. gordonii can potently antagonize the growth of S. mutans in vitro (31).
The oral microbiome functions as a cooperative community to degrade complex macromolecules and to release a spectrum of compounds that can be readily transported and metabolized (40). Viridans group streptococci generate energy almost exclusively from the fermentation of carbohydrates that are obtained from the host diet, from other bacteria in the oral microbiome, from sloughed host cells, and from saliva (22). Saliva is a primary source of nutrients provided nearly continuously to the oral microbiome. Salivary glycoconjugates, particularly O- and N-linked oligosaccharides of glycoproteins, serve as the preferred carbon and energy sources for many of the more abundant members of the oral microbiome. Many bacteria produce secreted or surface-associated glycosidases that cleave glycoconjugates found in saliva (4, 11). Due to the intermittent eating patterns of most humans, the ability to scavenge and efficiently metabolize the collection of carbohydrates present in the oral cavity is critical for the persistence of the organisms and to the ecology of the biofilms (37). Notably, when carbohydrates are present in excess in the diet, a low-pH environment can be created, which favors the growth and persistence of acid-tolerant pathogens, like S. mutans, at the expense of less aciduric commensal organisms.
S. mutans encodes 14 putative sugar:phosphotransferase systems (PTSs) (2), including one for lactose (EIILac), while the genome of S. gordonii DL1 harbors a similar number of putative PTS gene clusters (http://oralgen.lanl.gov/). Lactose is transported and concomitantly phosphorylated by the PTS before it is processed by phospho-β-galactosidase (LacG) into galactose-6-phosphate (Gal-6-PO4) and glucose (18, 46). The metabolism of Gal-6-PO4 then occurs through the tagatose-6-phosphate (Tag-6-PO4) pathway consisting of the LacABCD proteins (23). Galactose can also be catabolized by the Leloir pathway (3, 20), which usually involves a non-PTS transporter, such as GalP (18). Although the enzymes for the tagatose and Leloir pathways are expressed in S. mutans, this organism lacks a GalP homologue and a high-affinity PTS transporter for galactose (46). Instead, galactose is transported at a relatively low efficiency by PTS porters that have other carbohydrates as their cognate substrates and is subsequently processed through the tagatose pathway in S. mutans (46). Not surprisingly then, S. mutans needs relatively high concentrations of galactose to support growth, and the organism grows more slowly on galactose than on carbohydrates for which a higher-affinity uptake system is present.
Given the abundance of galactose and lactose entering the oral cavity in salivary secretions (12) and in the diet, respectively, we hypothesized that commensal streptococci may hold a selective advantage over the caries pathogen S. mutans by virtue of their ability to better utilize one or both of these carbohydrates. Accordingly, we identified and characterized two tandemly arranged gene clusters encoding tagatose pathway enzymes, two PTS enzyme II permeases, and two regulatory proteins. These gene products were shown to dominantly control galactose and lactose assimilation by S. gordonii DL1. Furthermore, competition assays provided evidence that the more efficient utilization of galactose by S. gordonii may confer a selective advantage over S. mutans UA159, particularly during periods of fasting by the host. The results reveal important differences in the regulation of, and capacity for, galactose and lactose metabolism between a strongly cariogenic isolate of S. mutans and this abundant oral commensal.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. gordonii and S. mutans cells were maintained on brain heart infusion (BHI) agar plates (Difco Laboratories, Detroit, MI), and broth cultures were routinely grown in BHI or in tryptone-vitamin (TV) medium (9); the latter medium supports the growth of S. gordonii or S. mutans only if it is supplemented with metabolizable carbohydrates. Unless stated otherwise, cultures were incubated at 37°C in a 5% CO2 aerobic environment. Antibiotic-resistant strains were selected and maintained on BHI agar containing erythromycin (Em) (10 μg/ml) (BHI-Em agar), kanamycin (Km) (1 mg/ml), or spectinomycin (Sp) (1 mg/ml). BHI broth was supplemented with antibiotics at half those concentrations, when needed, or with 10% horse serum (Sigma-Aldrich, St. Louis, MO) to induce natural competence (35). Escherichia coli strain DH10B was used as a cloning host and was maintained on L agar with antibiotics added at the following concentrations: Em at 250 μg/ml, Km at 50 μg/ml, or Sp at 50 μg/ml.
To monitor growth on particular carbohydrate sources, individual colonies were inoculated in triplicate into BHI broth and grown overnight (17 h). Strains were then diluted 1:20 into fresh TV medium containing 0.5% (wt/vol) glucose (TV-glucose medium), galactose, or lactose and incubated at 37°C in a 5% CO2 atmosphere. Growth was monitored hourly by measurements of the optical density at 600 nm (OD600).
DNA manipulations.
Coding sequences of the genes in the lac and gal clusters of S. gordonii were deleted, in whole or in part, and replaced with a nonpolar Em, Km, or Sp resistance cassette via allelic exchange (29). Briefly, two DNA fragments flanking the gene of interest were amplified via PCR using gene-specific primers with integrated restriction enzyme sites. These fragments, along with the appropriate antibiotic resistance determinant, were digested by using restriction enzymes and ligated together with T4 DNA ligase (New England BioLabs, Ipswich, MA). The ligation products were then used to transform genetically competent S. gordonii cells. Transformants were subjected to PCR verification and DNA sequencing before being used for phenotypic studies.
A cat (chloramphenicol acetyltransferase [CAT]) gene fusion to the lacA1 promoter (PlacA1-cat) was constructed by cloning a 385-bp DNA fragment containing the promoter region of the lacA1 gene into plasmid pYQ4, which was derived by modifying the integration vector pMJB8 to contain an Em resistance marker (47). The desired construct was then used to integrate a single copy of the gene fusion into the gtfG gene in the DL1 genome. To construct the PlacA2-cat fusion, a 308-bp sequence of the lacA2 promoter region was first fused with the promoterless cat gene in pJL84 (48), and the fusion was then subcloned into pMJB8 for integration into the chromosome. The PlacA2-cat fusion carries a Km resistance marker.
A point mutation was generated in the lacT gene to convert the Met8 codon into a UAG stop codon to create the lacT(M8stop) strain by using a previously reported protocol (45). Briefly, a 2-kbp DNA fragment containing the lacT(M8stop) mutation was generated by using recombinant PCR and then used to transform S. gordonii DL1, along with the promoter fusion PlacA2-cat plasmid, which carries a Km resistance cassette (44). After plating, Km-resistant colonies were screened for the lacT(M8stop) mutation by using allele-specific mismatch amplification mutation analysis (MAMA) PCR (13). The desired transformants were then verified by PCR and sequencing.
Real-time RT-PCR.
Total RNA was extracted from cultures of S. gordonii strains by using the RNeasy minikit (Qiagen, Germantown, MD), and specific mRNAs were quantified by using real-time reverse transcription (RT-PCR), as detailed previously (1). After the conversion of RNA into cDNA using random hexamers, the transcript levels of various genes were measured by using specific primers: 5′-GGT CAG GAT TTT GTT GAT GTG ACC C-3′ (forward) and 5′-GGA CCA GCC CCA TAA GCA TCG AT-3′ (reverse) for lacA1, 5′-GAC AGG CTA TGG AGA GGT CAA TC-3′ (forward) and 5′-TGG TGT ATC AAA GTG GTG AAG GG-3′ (reverse) for lacG, 5′-GGT GCA GAT GCT GCT GGA AAT-3′ (forward) and 5′-CAC CTC AGC TGC AAC TGC CAA T-3′ (reverse) for lacA2, 5′-GAT AAC AAC GGA GTA AGC CAA GG-3′ (forward) and 5′-TTG GAG CAT TTA GGA GGT CGT C-3′ (reverse) for the EIICGal gene, or 5′-CAC ACC GCC CGT CAC ACC-3′ (forward) and 5′-CAG CCG CAC CTT CCG ATA CG-3′ (reverse) for the 16S rRNA gene as a control.
CAT and sugar transport assays.
CAT assays were performed by using exponentially growing bacterial cells in TV medium according to a previously reported protocol (38). Assays of galactose transport by the bacterial sugar:phosphotransferase system were carried out according to previously reported protocols (30), with minor modifications. Briefly, the assay couples pyruvate generation from phosphoenolpyruvate (PEP)-dependent sugar phosphorylation by the PTS to lactate generation via lactate dehydrogenase, which concurrently oxidizes NADH to NAD+. However, the background level of spontaneous oxidation of NADH in wild-type (WT) S. gordonii cells in the absence of added PEP was unacceptably high. To circumvent this problem, we created strains of S. gordonii specifically for use in PTS assays which contained a deletion of the gene for NADH oxidase (encoded by nox [SGO_1167]), which reduced the background level of NAD generation (non-PEP dependent) in the PTS assay to acceptable levels (39). Cultures of S. gordonii that were grown overnight in BHI broth in a 5% CO2 atmosphere were diluted 1:25 into fresh TV-galactose medium and incubated in an anaerobic chamber maintained with 85% N2, 10% H2, and 5% CO2. The use of anaerobically grown cells further reduced the non-PEP-dependent NADH oxidation in the PTS assays. Cells were harvested at the mid-exponential phase, washed in phosphate buffer, and permeabilized by using toluene-acetone (1:9, vol/vol), and the PTS activity was measured and normalized to protein concentrations (30, 39).
Mixed-species liquid culture competition assay.
S. gordonii DL1 and S. mutans UA159 were modified by replacing the fruA gene with Em and Km resistance markers, respectively. Strains were cultured in triplicate overnight in BHI broth. The next morning, bacterial cultures were diluted 1:50 into 10 ml of fresh BHI broth and cultured for 3 h. After the cell density was measured, S. gordonii and S. mutans samples were mixed together in a 1:1 ratio of 1 × 108 CFU into 10 ml fresh TV broth supplemented with either 0.5% glucose or galactose, with or without 50 mM potassium phosphate buffer (pH 7.5). This time point (t) was designated 0 h. The samples were incubated for 6 h (t = 6 h) before being diluted 1:50 into fresh TV medium. The rediluted (for viability) and original (for persistence) mixed cultures were then incubated overnight (t = 22 h). Subsequently, the persistence cultures were kept as-is, while the viability cultures were rediluted 1:50 into fresh medium. These samples were incubated for an additional 8 h (t = 30 h). At each time point, the absorbance and pH were measured, and aliquots (100 μl) of the samples were removed, serially diluted, and plated onto BHI-Em and BHI-Km agar to enumerate S. gordonii and S. mutans bacteria, respectively. All plates were incubated for 2 days at 37°C in a 5% CO2 atmosphere before the colonies were counted.
RESULTS AND DISCUSSION
Identification of the lactose and galactose gene clusters.
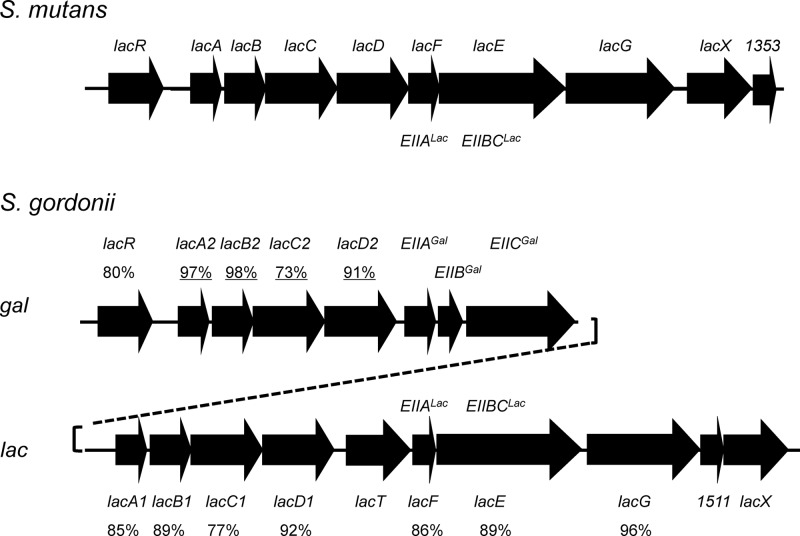
Lactose and galactose are carbohydrates that are frequently encountered by the bacterial communities in the oral cavity. To identify lactose- and galactose-utilizing enzymes in the genome of S. gordonii strain DL1 Challis, a BLAST search was performed by using protein sequences of the tagatose-6-phosphate (Tag-6-PO4) pathway (lac) of S. mutans UA159 (Fig. 1) (46). Two tandemly arranged gene clusters encoding the enzymes of the Tag-6-PO4 pathway were identified, and the duplicated gene products shared high degrees of homology (Fig. 1). For example, the A subunits (LacA1 and LacA2) of the galactose-6-phosphate isomerase enzyme, which converts Gal-6-PO4 into Tag-6-PO4, were 97% identical. Similar to that of S. mutans, one of the apparent operons contained genes for a predicted phospho-β-galactosidase (LacG) and a lactose-specific EII permease (EIILac). The other operon harbored the genes for a putative galactose-specific EII permease (EIIABCGal) encoded by the genes SGO_1520 to SGO_1522 (http://oralgen.lanl.gov/). For convenience, the operon containing LacG and EIILac is referred to as the lac operon, and that encoding EIIGal is referred to as the gal operon.
Fig 1.
Diagrams depicting the tagatose-6-phosphate pathway clusters in S. mutans and S. gordonii. Two sets (gal and lac) of predicted tagatose pathway genes (lacABCD) and putative regulatory genes (lacR and lacT) are present in S. gordonii DL1, as opposed to one set in S. mutans UA159. Additional genes encoding a galactose-PTS enzyme II permease (EIIABCGal) are also present in the gal cluster of S. gordonii. Numbers indicate the percent similarities among homologous genes; underlined numbers denote similarity between two homologues within the S. gordonii genome, and others represent similarity to homologues in the S. mutans genome. Other homologous open reading frames with unknown functions, SGO_1511 and lacX, are also indicated.
A gene for a transcriptional antiterminator (SGO_1515), designated here lacT, is present within the lac operon of S. gordonii but is absent in the S. mutans UA159 genome. LacT is a member of the BglG/SacY family of proteins commonly associated with the regulation of carbohydrate metabolism (15) and contains a coantiterminator (CoAT) RNA-binding domain (28) located in its amino terminus. CoAT domains facilitate binding to ribonucleic antiterminator (RAT) sequences in mRNA transcripts, preventing the formation of a terminator to allow RNA polymerase to continue the transcription of downstream genes (15). LacT also contains two PTS regulatory domains (PRDs), which serve as targets for phosphorylation by components of the PTS for the allosteric modulation of the DNA- or RNA-binding activity of the regulatory protein (42). Based on current models for the PTS-dependent regulation of antitermination (21), one would predict that when the PTS permease(s) for lactose or galactose is engaged in sugar transport, the PRD in LacT would be dephosphorylated, allowing for the antitermination of the lac or gal operon genes.
The transcriptional regulator 5′ to the gal gene cluster (Fig. 1) is designated LacR, based on similarities to known LacR proteins, and contains a helix-turn-helix domain of about 50 to 60 amino acids and a carboxy-terminal effector-binding domain. LacR-type proteins are present in multiple genera, including Streptococcus, Staphylococcus, Lactococcus, and Bacillus, and in many cases have been shown to negatively regulate sugar catabolism. Effector molecules for LacR-type regulators are often phosphorylated carbohydrates that are intermediates in the metabolic pathway controlled by the regulatory protein. In this case, Gal-6-PO4 and Tag-6-PO4 have each been suggested to bind to a LacR homologue to facilitate dissociation from its binding site (34, 41, 46). A protein alignment revealed 59% and 86% identities between LacR in S. gordonii and apparent homologues in S. mutans and S. sanguinis, respectively.
A survey of existing whole-genome sequences revealed that a number of other streptococci, lactococci, and lactobacilli retain various combinations of the gal and lac genes (5, 14, 32, 33, 36). For example, the gal and lac gene clusters of Streptococcus pyogenes strains closely resemble those of S. gordonii, encoding two sets of the tagatose enzymes, a lactose PTS, a predicted galactose PTS, and two homologous LacR proteins (32). Lactococcus lactis, Lactobacillus casei, S. sanguinis, S. mitis, Streptococcus pneumoniae, and Staphylococcus aureus also carry transporters that are similar to those in the S. gordonii lac and gal gene clusters.
Finally, it should be noted that the genes for the Leloir pathway (galKTE) for galactose utilization, as well as two predicted extracellular β-galactosidase enzymes, are present in the DL1 genome. However, as detailed below, the contribution of the Leloir pathway and these secreted enzymes to galactose or lactose catabolism appears to be nominal, so an analysis of the function or regulation of these genes was not pursued further in this study.
Growth phenotypes of the S. gordonii lac and gal gene mutants. (i) lac gene cluster.
Various mutant strains were created via allelic exchange using nonpolar antibiotic resistance genes. Growth in TV broth supplemented with 0.5% of the desired carbohydrates was monitored (Table 1). When grown in TV medium supplemented with 0.5% galactose, S. gordonii DL1 cells grew to a final OD600 of 1.2 with a doubling time of 68 ± 2 min, while in TV-lactose medium, the organism grew with a doubling time of 67 ± 4 min and reached a final OD600 of 1.0. All of the mutations in the lac gene cluster caused severe defects in growth on lactose, but the effects of the same mutations on growth on galactose varied (Table 1). For example, the ΔEIILac strain, in which the presumed lactose-specific transporter was deleted, failed to grow on lactose but achieved a final OD on galactose similar to that of strain DL1, with a doubling time of 70 ± 4 min. In contrast, the ΔlacA1B1 strain, lacking the subunits of the Tag-6-PO4 isomerase encoded by the lac operon, showed no growth on lactose but grew slowly (201 ± 18 min) and achieved a final OD600 of 0.2 when galactose was the growth carbohydrate. Similarly, the ΔlacG strain, which produces no phospho-β-galactosidase enzyme and could not grow at all on lactose, grew with a doubling time of 113 ± 8 min on galactose. Notably, a similar phenotype was observed previously for a lacG mutant of S. mutans (46) and was associated with a requirement of LacG for the induction of the operon (see below). The requirement for the induction of the lac operon of E. coli by β-galactosidase, which produces the cognate inducer of the operon, allolactose, is well established (10).
Table 1.
Doubling times and final optical densities of wild-type strain DL1 and various mutantsa
| Strain | Glucose |
Galactose |
Lactose |
|||
|---|---|---|---|---|---|---|
| Avg Td (min) ± SD | OD600 | Avg Td (min) ± SD | OD600 | Avg Td (min) ± SD | OD600 | |
| DL1 (wild type) | 57.4 ± 6.7 | 1.08 | 67.5 ± 2.1 | 1.17 | 67.1 ± 4.3 | 0.97 |
| ΔEIILac | 57.5 ± 4.4 | 1.07 | 70.0 ± 4.2 | 1.13 | ND | 0.15 |
| ΔlacG | 72.4 ± 2.6 | 1.07 | 113.2 ± 7.9 | 1.16 | ND | 0.09 |
| ΔlacA1B1 | 61.9 ± 1.2 | 1.03 | 201.0 ± 18.6 | 0.23 | ND | 0.20 |
| lacT(M8stop) | 64.8 ± 1.9 | 1.12 | 74.9 ± 0.4 | 1.16 | ND | 0.19 |
| ΔEIIGal | 68.5 ± 8.4 | 0.86 | 107.6 ± 26.2 | 1.15 | 94.2 ± 6.5 | 0.88 |
| ΔlacR | 57.8 ± 1.5 | 1.16 | 64.1 ± 0.9 | 1.23 | 60.9 ± 2.3 | 0.93 |
| ΔlacA2B2 | 57.0 ± 2.3 | 1.05 | 66.9 ± 2.5 | 1.27 | 69.4 ± 1.2 | 0.95 |
| ΔlacA1B1 ΔlacA2B2 | 55.7 ± 0.7 | 1.01 | ND | 0.10 | ND | 0.13 |
| ΔEIIABMan | 67.9 ± 1.7 | 0.89 | 109.7 ± 5.2 | 1.17 | 67.2 ± 3.0 | 0.77 |
| ΔEIIGal/ΔEIIABMan | 61.4 ± 1.2 | 0.90 | 254 ± 47 | 0.59 | 61.2 ± 5.1 | 0.76 |
Results (averages ± standard deviations) are each based on three manual growth curve analyses using TV medium containing the specified carbohydrate (0.5%) in the presence of 5% of CO2. ND (not determined) is denoted when a strain displayed minimal growth and when the doubling time (Td) was too large for accurate calculations.
Since the genome of S. gordonii also harbors two predicted extracellular β-galactosidases (BgaA [SGO_1486] and BgaC [SGO_0043]), the possibility exists that some of these enzymes could contribute to the growth of the bacteria on lactose by converting lactose into galactose and glucose in the supernatant fluid. In fact, a modest yet consistent growth defect on lactose was observed for a strain carrying deletions of both the bgaA and bgaC genes (data not shown). However, the phenotypes of the ΔEIILac and ΔlacG strains support that the predicted extracellular β-galactosidase enzymes are not sufficient to support growth on lactose, possibly reflecting that BgaA and BgaC may have relatively poor activity on lactose compared to, for example, galactose in glycoconjugates. Furthermore, when measured by using quantitative real-time RT-PCR, the transcript levels of both the bgaA and bgaC genes were relatively low in glucose-grown DL1 cells, and they remained so when cells were grown in lactose-based medium (data not shown). Also consistent with a lack of a significant contribution of BgaA or BgaC to lactose metabolism, when β-galactosidase assays were performed on intact cells of the wild type or the lacG mutant, using either 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) or o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate, significant levels of activity were not detected in either genetic background (data not shown). These findings are in agreement with data from a previous study which showed that an apparent homologue of the S. gordonii BgaA enzyme in S. pneumoniae is not responsible for the hydrolysis of lactose in the extracellular environment (43).
(ii) gal gene cluster.
Unlike the strain lacking EIILac, the growth of the EIIGal-deficient strain on galactose was clearly impaired compared to the growth of the wild-type strain, with a minimum doubling time of 108 ± 26 min. Thus, EIIGal appears to contribute significantly to the internalization of galactose. Consistent with these results, a PTS EII complex present in S. pneumoniae that has significant similarity to EIIGal of S. gordonii was shown previously to be required for growth on galactose (24). However, we previously reported that a mutation of the manL gene, encoding the AB domains of a glucose/mannose-PTS enzyme II complex (EIIMan), contributed to galactose utilization by S. gordonii (39). When both EIIMan and EIIGal were deleted from S. gordonii, the mutant displayed further reductions in levels of growth on galactose compared to the single mutants (Table 1). Therefore, in S. gordonii, EIILac, EIIMan, and EIIGal all appear to have the capacity to contribute to galactose uptake.
The deletion of the lacA2B2 genes (Fig. 1) did not significantly affect growth on galactose, but the mutant lacking lacA1B1 and lacA2B2 was unable to grow on galactose at all. These findings demonstrate that the tagatose pathway is the major pathway for S. gordonii to catabolize galactose for growth and that the Leloir pathway either is not involved in galactose catabolism or is regulated in such a way that it is not expressed in strains deficient in LacAB proteins. Consistent with these results, the deletion of the galK gene, encoding galactokinase in the Leloir pathway, had only a modest effect on growth on galactose (doubling time, 86 ± 4 min). Collectively, these data provide evidence that the GalKTE pathway is not a significant contributor to the catabolism of galactose by S. gordonii under the conditions tested and that the PTS is the main route of entry for galactose in this organism.
Consistent with the effects of the deletion of lacA2B2 on growth on galactose, there was little impact of this mutation on growth on lactose. However, when both sets of isomerase (lacAB) genes were deleted, no growth was observed on lactose, even in the presence of an intact lacG gene. Since LacG is expected to cleave lactose-6-PO4 to release Gal-6-PO4 and glucose inside the cells, we propose that the failure of the mutant lacking both lacAB gene sets to grow is due to the growth-inhibitory effects of the accumulation of Gal-6-PO4 in cells, as was observed previously for S. mutans and some other bacteria (46).
Interestingly, the mutant lacking the transcriptional antiterminator, lacT(M8stop), failed to grow on lactose and grew only slightly slower than the parental strain on galactose. The simplest interpretation of these results is that LacT functions in the antitermination of the genes for the EIILac permease. It appears less likely that LacT is required for the expression of the lacA1B1C1D1 genes, since the inactivation of lacA1B1, but not lacT, caused a severe growth defect on galactose (Table 1). It is also of interest that EIIGal has an influence on lactose utilization, perhaps reflecting a role in the regulation of LacT activity. Further analysis of the regulatory functions of LacT is under way in our laboratory. The loss of LacR, on the other hand, had relatively little influence on the growth of strains on lactose or galactose, consistent with its predicted role as a negative regulator of the tagatose pathway genes.
(iii) Effects of lac and gal mutations on growth on glucose.
When grown in TV broth supplemented with 0.5% glucose, wild-type strain DL1 grew to a final OD600 of 1.1 with a doubling time of 57 ± 7 min. With the exception of the ΔlacG and ΔEIIGal strains, which grew modestly slower, all other mutant strains exhibited similar growth rates and achieved final optical densities comparable to that of the wild-type strain when grown on glucose (Table 1). Also of note, the ΔlacR strain had a strong tendency to aggregate when grown in BHI broth, which contains 0.3% added glucose, and it formed much longer chains than the parental strain (about 15 to 20 cells per chain, compared to 2 to 5 cells per chain for the WT). Both LacR and LacG have been shown to play regulatory roles in the metabolism of galactose and lactose in S. mutans (46), and we propose that a perturbation of the cell wall biosynthetic pathways associated with aberrant galactose metabolism may account for the slower growth of the lacG mutant and the cell division defects of the lacR mutant. Consistent with this hypothesis was the observation that lac operon expression was altered in a lacG or lacR deletion strain grown on glucose (see below).
Transport of galactose via the PTS.
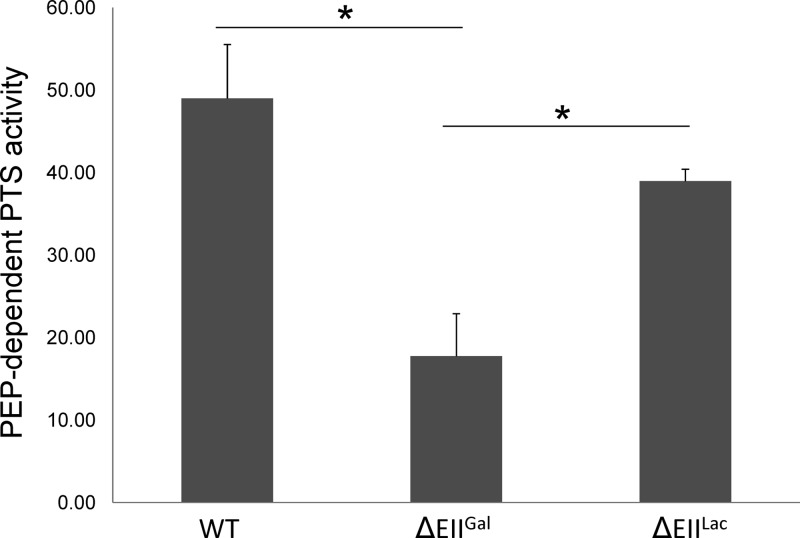
To further substantiate that EIIGal and, potentially, EIILac are the primary galactose transporters in S. gordonii, PTS assays were performed. Notably, S. gordonii appears to produce high levels of NADH oxidase activity when exposed to O2 or when grown on galactose (39), which interferes with accurate determinations of PTS activity in the coupled assay employed here (30). To circumvent this problem, PTS assays were conducted in the wild-type, ΔEIIGal, and ΔEIILac genetic backgrounds in strains carrying a deletion of the major NADH oxidase enzyme of S. gordonii (nox), as detailed in Materials and Methods. When grown anaerobically in 0.5% galactose, the ΔEIIGal strain had a significantly lower level of galactose-PTS activity than did strains with an intact EIIGal permease (Fig. 2). The strain lacking EIILac consistently had lower levels of PTS activity than those seen with the wild-type-background strain, although the differences were not statistically significant. However, the level of PTS activity in the strain lacking EIIGal was significantly lower than that in the strain without EIILac. These results provide additional support that, in addition to EIIMan (39), EIIGal is capable of transporting galactose in S. gordonii. We speculate that the EIILac complex is also involved in the transport of galactose albeit at levels too low to be demonstrated by our PTS assay in a statistically meaningful way.
Fig 2.
Galactose-PTS activities. In vitro PTS assays were performed on S. gordonii wild-type (WT) and ΔEIIGal and ΔEIILac strains grown anaerobically in TV medium supplemented with 0.5% galactose. All three strains contain a nox (NADH oxidase) deletion. Values were obtained from triplicate assays of three individual cultures, and the asterisks indicate a P value of less than 0.005 according to the Student t test.
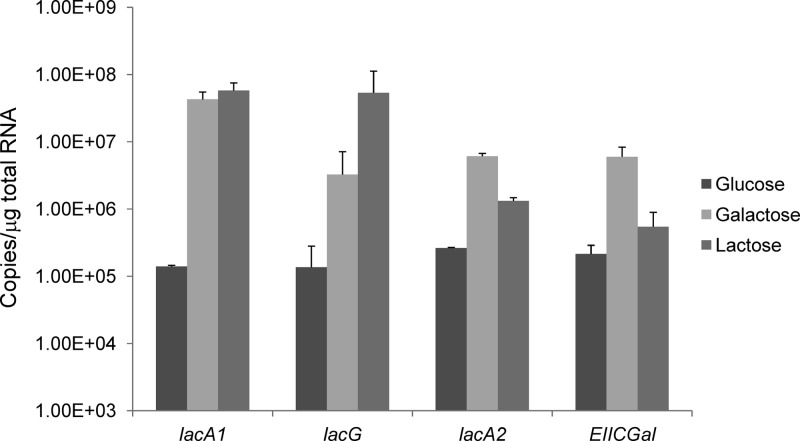
Operon-specific gene expression in response to carbohydrate.
Real-time RT-PCR was performed to evaluate the expressions of the lacA1 and lacG genes of the lac operon and the lacA2 and EIICGal genes of the gal operon, which were chosen based on their positions in each gene cluster (Fig. 1). Total RNA from strain DL1 was extracted from cells that were grown exponentially in TV medium supplemented with 0.5% glucose, galactose, or lactose and converted into cDNA by using random hexamers. The amount of the lacG transcript was 24-fold higher in cells grown on galactose (P = 0.04) than in glucose-grown cells (Fig. 3). Even higher expression levels were noted when the cells were grown on lactose, with a 390-fold induction compared to that of cells grown on glucose (P = 0.02). The greater increase in the lacG expression level in lactose-grown cells than in galactose-grown cells, together with the lack of growth of the lacT(M8stop) strain on lactose, supports the idea that the EIILac complex modulates LacT activity, since lacT and the downstream lacFEG genes are likely cotranscribed.
Fig 3.
Expression levels of the lacA1, lacG, lacA2, and EIICGal genes in cells grown on the indicated carbohydrates. Real-time quantitative RT-PCR was used to measure the transcript levels of each gene in DL1 cells grown in TV medium containing 0.5% glucose, galactose, or lactose. Results are the averages of data from three independent experiments, and the error bars represent standard deviations.
Interestingly, lacA1 transcript levels were elevated approximately 100-fold in cells grown on galactose or lactose, as opposed to glucose-grown cells. Additional tests of lacA1 expression in the lacT(M8stop) background also indicated that LacT is not required for lacA1 promoter activity (data not shown). On the other hand, the expression level of the gene for EIICGal in the gal gene cluster was only 2.5-fold higher in cells grown on lactose, compared with a 28-fold (P = 7.6 × 10−5) increase in cells grown on galactose over those measured in glucose-grown cells. A similar pattern of expression was found for the lacA2 gene, with increased transcript levels in cells grown on lactose and much higher levels in galactose-grown cells. These results are consistent with the observation that the lac genes appear to be dedicated mainly to lactose metabolism, whereas both the lac and gal operons were required for optimal galactose utilization.
In S. mutans, the inactivation of the apparent homologues of the S. gordonii lacR or lacG gene resulted in the aberrant expression of the lac gene cluster (46). To assess whether the loss of LacR or LacG impacted the expression of the lac or gal gene cluster in S. gordonii, promoter-cat fusions were constructed by using DNA fragments containing their respective promoters (PlacA1 for the lac operon and PlacA2 for the gal operon) and were integrated in a single copy into a distal site (gtfG) of the chromosome in both the wild-type and mutant strains (47). The loss of LacR led to a large increase in expression from the lacA1 promoter in cells grown on glucose, galactose, or lactose, whereas the loss of lacG led to clear reductions in lacA1 promoter activity in either glucose- or galactose-containing medium; lacG mutants cannot grow with lactose as the only carbohydrate. Interestingly, the deletion of lacR caused little change in the PlacA2-cat expression levels under the same conditions.
Further analysis of the promoter regions of the lac and gal gene clusters identified a conserved catabolite response element (cre), which is the binding site for catabolite control protein A (CcpA) (17), near the promoter of lacA2. Therefore, it was possible that the effect of the deletion of lacR on the lacA2 promoter was masked by catabolite repression. To exclude the involvement of CcpA in our genetic analyses, point mutations were introduced to disrupt cre in the PlacA2-cat promoter fusion, resulting in a PlacA2cre-cat fusion. As predicted, the PlacA2cre-cat fusion produced higher levels of CAT activity than did the PlacA2-cat gene fusion in the wild-type genetic background (Table 2). When assayed in cells grown on glucose or galactose (Table 2), PlacA2cre-cat also showed much higher expression levels in the lacR mutant background than those seen in the wild-type genetic background. Collectively, these data indicate that LacR plays a role in the repression of both the lac and gal operons. Moreover, the lacTFEG genes are likely regulated by the lactose PTS exerting its influence through LacT-dependent antitermination, whereas the gal operon is dominantly regulated by carbon catabolite repression (CCR) via the CcpA protein. In the in vitro studies described here, CcpA-mediated CCR is active in cells, since they were grown with relatively high concentrations of carbohydrate. However, under carbohydrate-limiting conditions, such as those seen during periods of fasting by the host, one would predict that the gal operon, including the EIIGal complex, could contribute significantly to the utilization of galactose in the human oral cavity, where carbohydrate is frequently a limiting nutrient and is present in concentrations that do not trigger CCR.
Table 2.
Expression levels of promoter-cat fusions in the wild type and lacR, lacG, and ccpA mutantsa
| Strain | Avg CAT sp act (nmol mg of protein−1 min−1) (SD) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| PlacA1-cat |
PlacA2-cat |
PlacA2cre-cat |
|||||||
| Glc | Gal | Lac | Glc | Gal | Lac | Glc | Gal | Lac | |
| DL1 | 83.6 (15) | 2,253 (165) | 1,886 (180) | 0.72 (0.2) | 41.0 (0.9) | 5.2 (0.5) | 43.5 (9.3) | 196 (6) | 231 (7) |
| lacR | 13,908 (6,531) | 14,431 (5,337) | 22,546 (5,679) | 7.4 (0.2) | 46.1 (1.2) | 4.6 (0.5) | 428 (22) | 305 (35) | 135 (29) |
| lacG | 45.1 (4) | 1,203 (387) | ND | ND | ND | ND | ND | ND | ND |
| ccpA | 43.6 (13.5) | 3,189 (138) | 1,724 (220) | 24.6 (5.1) | 129 (17) | 123 (30) | ND | ND | ND |
Cells were harvested from exponentially growing cultures in TV medium supplemented with 0.5% glucose (Glc), galactose (Gal), or lactose (Lac). The data are the averages (standard deviations) of the activities based on three individual cultures. CAT specific activities are expressed as nmol of chloramphenicol acetylated mg of protein−1 min−1. ND, not determined.
Comparison of growths of S. gordonii DL1 and S. mutans UA159 on galactose.
The possession of a dedicated high-affinity galactose transport system and redundant galactose metabolic pathways in S. gordonii, which are lacking in S. mutans UA159, may reflect a niche adaptation by the commensal that imparts a growth advantage over the caries pathogen S. mutans. To begin to test this hypothesis, wild-type S. gordonii and S. mutans strains were grown in TV medium supplemented with either 0.5% glucose or galactose or 2% galactose (Table 3). When grown in TV medium supplemented with 0.5% glucose, both strains grew to similar final ODs, and S. gordonii had a doubling time of 57 ± 7 min, while S. mutans divided every 63 ± 1 min. When grown in TV medium supplemented with 0.5% galactose, S. gordonii grew to a final OD600 of 1.2 with a doubling time of 68 ± 2 min, whereas S. mutans grew to a final OD600 of 0.9 with a doubling time of 135 ± 11 min. Similarly, in TV medium with 2% galactose, S. gordonii again grew faster than S. mutans and achieved a higher final optical density. However, S. mutans cells grew at a significantly higher rate (doubling time of 84 ± 2 min) in 2% galactose than in 0.5% galactose.
Table 3.
Doubling times and final optical densities of cultures of S. mutans strain UA159 and S. gordonii strain DL1a
| Strain | 0.5% glucose |
0.5% galactose |
2% galactose |
|||
|---|---|---|---|---|---|---|
| Avg Td (min) ± SD | OD600 | Avg Td (min) ± SD | OD600 | Avg Td (min) ± SD | OD600 | |
| S. mutans UA159 | 62.9 ± 1.2 | 1.09 | 135.3 ± 10.6 | 0.93 | 83.5 ± 2.4 | 0.91 |
| S. gordonii DL1 | 57.4 ± 6.7 | 1.08 | 67.5 ± 2.1 | 1.17 | 70.3 ± 2.2 | 1.14 |
Strains were grown in TV-based medium supplemented with various carbohydrates. Results for the doubling times are the averages and standard deviations derived from three independent cultures.
Collectively, these data suggest that S. gordonii may be better equipped than S. mutans UA159 to utilize galactose, as evidenced by the higher growth rate, higher final yield, and apparently higher affinity for this hexose (46). The improved growth of S. mutans on 2% galactose compared with growth on 0.5% galactose likely reflects that it has a lower capacity and a lower affinity for the transport of this hexose (46). Given that galactose is probably present at low steady-state concentrations as it is liberated from host glycoproteins, the differences between these two organisms in their abilities to scavenge and efficiently catabolize galactose could have a significant impact on the ecological balance of dental biofilms, particularly during periods of fasting by the host.
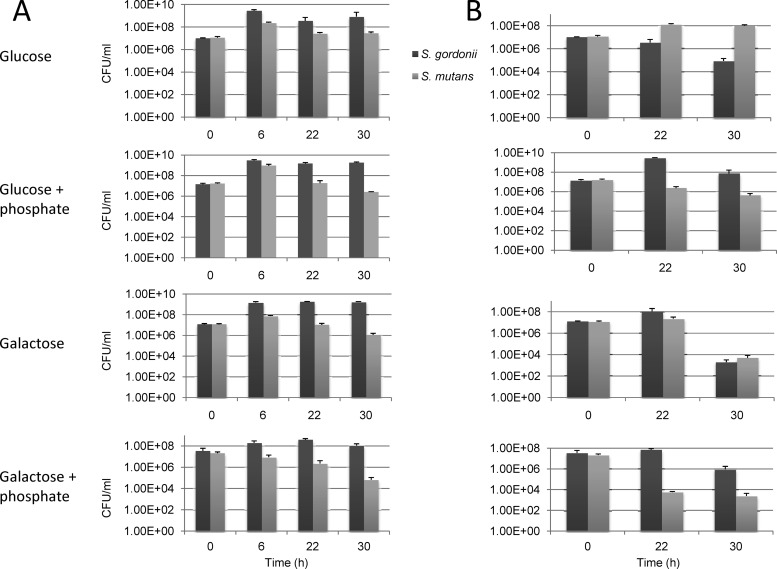
Mixed-species liquid culture competition assay.
While the production of bacteriocins and excess acid by S. mutans is detrimental to the survival of oral commensal bacteria, including S. gordonii, S. gordonii is capable of producing hydrogen peroxide, which is inhibitory to the growth of S. mutans (26). Here, a mixed-species liquid culture competition assay was used to assess the capacity of S. gordonii DL1 to compete with S. mutans UA159 in the presence of different carbohydrates. To be able to enumerate these two bacterial species accurately during plating, the S. gordonii fruA gene (39) was replaced with an Em resistance cassette, and the fruA gene of S. mutans (8, 48) was replaced with a Km resistance marker. FruA is an exo-β-fructosidase, and the loss of this enzyme affects growth only on sucrose or homopolymers of fructose (6–8, 39). Cells were grown to the mid-exponential phase to an OD600 of 0.5 and mixed together in a liquid culture in a 1:1 ratio (t = 0 h). All incubations were static in a 5% CO2 atmosphere at 37°C. At the 0-h time point and at the 6-, 22-, or 30-h time point, the OD600 and pH were measured, serial dilutions were plated for CFU enumerations, and the samples were subcultured 1:50 into fresh medium with or without 50 mM phosphate buffer to assess viability. The original mixed culture was also incubated continuously and subjected to all of the same measurements at the same time points, but the medium was never changed, and buffer was not added to the samples. This allowed for the comparison of the abilities of these cells to persist in the same environment over time. All conditions were tested in triplicate, and the results are presented in Fig. 4 (CFU) and Table 4 (pH).
Fig 4.
Mixed-species competition assay testing viability (A) and persistence (B) of S. gordonii and S. mutans. Exponentially growing cultures of DL1 and UA159, each containing an antibiotic marker (Em for DL1 and Km for UA159), were inoculated in a 1:1 ratio into TV medium supplemented with 0.5% glucose or galactose, with or without 50 mM potassium phosphate buffer (pH 7.5) (see Materials and Methods for details). At time points of 6, 22, and 30 h, the cultures for viability testing were diluted into fresh medium (excluding t = 30 h), while the pH was measured (Table 4), and CFU were determined by plating. Results are the averages of data from three independent experiments, and the error bars represent standard deviations.
Table 4.
pH measurements at various time points of the mixed-species liquid culturesa
| Assay and t (h) | Avg pH ± SD |
|||
|---|---|---|---|---|
| Glucose | Glucose + phosphate | Galactose | Galactose + phosphate | |
| Viability | ||||
| 6 | 4.9 ± 0.2 | 6.3 ± 0.6 | 6.0 ± 0 | 6.8 ± 0.2 |
| 22 | 4.6 ± 0.1 | 5.6 ± 0.3 | 4.8 ± 0.1 | 5.5 ± 0.2 |
| 30 | 6.7 ± 0.5 | 6.6 ± 0.6 | 6.0 ± 0.1 | 6.9 ± 0.1 |
| Persistence | ||||
| 22 | 4.5 ± 0.1 | 5.7 ± 0.3 | 4.7 ± 0.1 | 5.4 ± 0.2 |
| 30 | 4.5 ± 0.1 | 5.7 ± 0.3 | 4.7 ± 0 | 5.4 ± 0.2 |
S. gordonii and S. mutans cells were mixed at a 1:1 ratio in TV-glucose or TV-galactose medium supplemented with or without 50 mM potassium phosphate buffer (pH 7.5) and incubated continuously for 30 h (persistence assay) or diluted into fresh medium periodically (viability assay). Results are the averages and standard deviations from three individual experiments.
In the viability aspects of the competition assays, S. gordonii outcompeted S. mutans under all conditions tested. As shown in Fig. 4, both species reached their highest cell densities at 6 h, but the proportion of S. gordonii cells in the mixed culture was up to 20-fold higher than that of S. mutans. At subsequent time points, S. gordonii maintained largely similar viable counts, whereas the proportion of S. mutans cells consistently declined. When cultured without phosphate buffer, S. mutans maintained higher CFU counts throughout the test when grown in glucose than when grown in galactose, whereas S. gordonii showed little preference between these two sugars when viability was assessed. The addition of 50 mM phosphate buffer had little impact on the viability of S. gordonii in TV-glucose medium but resulted in a slight reduction in CFU in TV-galactose medium. Conversely, the buffering of the TV-glucose medium appeared to lead to a faster decline in the numbers of S. mutans cells than in nonbuffered medium, perhaps associated with an enhanced antagonism by S. gordonii against S. mutans in a less acidic environment.
Without phosphate buffer, both glucose- and galactose-containing media were acidified overnight to pH values of 4.6 and 4.8, respectively (Table 4). However, the next stage of culturing saw the pH of the cultures stabilize and rise above 6.0 by the end of the assay, a strong indication that a mixed culture dominated by S. gordonii could maintain a less acidic environment than S. mutans alone. The elevated pH in the S. gordonii-enriched cultures was almost assuredly due to the hydrolysis of arginine by the arginine deiminase pathway of S. gordonii (19), which is derepressed in galactose cultures compared with glucose cultures.
In the persistence portion of the competition assays, S. mutans was found at higher levels than S. gordonii under both galactose and glucose conditions without the addition of phosphate buffer. Specifically, in TV medium containing galactose alone, S. gordonii managed to maintain a slight dominance at the 22-h time point, but S. mutans was present in 2.5-fold-higher numbers at the 30-h time point (Fig. 4). In TV-glucose medium, S. mutans cells outnumbered S. gordonii cells by 38-fold at the 22-h time point and by 3 logs after 30 h of continuous incubation. When supplemented with 50 mM phosphate buffer, however, S. gordonii was able to maintain high levels of viability and to survive much better than S. mutans. This was especially clear in galactose cultures with phosphate buffer, where at end of the 30-h mixed coincubation, S. mutans was barely detectable. Thus, S. gordonii is able to outcompete S. mutans when galactose is present and when the environment is not extremely acidic.
Summary and conclusions.
The genome of S. gordonii strain DL1 contains more than 16 genes dedicated to the metabolism of lactose and galactose, carbohydrates that are commonly found in the oral environment. The growths and PTS activities of various strains constructed for this study indicate that galactose and lactose are transported by PTS permeases and catabolized primarily through the tagatose pathway. Although S. gordonii has a galKTE pathway, this pathway either is not sufficiently active or is cross-regulated by the lac and gal systems in a way that does not allow for compensatory growth when certain mutations are present or under the particular conditions tested here. A quantitative assessment of gene expression showed that the lac operon, and lacG in particular, is strongly induced by lactose but less effectively induced by galactose. In contrast, the induction of the gal operon occurs mainly in the presence of galactose and when CCR is relieved. Growth comparisons and a mixed-species competition assay highlight a potentially important role for the gal operon in the ability of S. gordonii to compete for galactose with the caries pathogen S. mutans. Thus, the ability of S. gordonii to utilize galactose more efficiently than S. mutans may represent an ecological advantage for these, and possibly other, commensal bacteria. It is also notable that the production of hydrogen peroxide by S. gordonii is strongly inhibitory to the growth of S. mutans and that one of the major pathways for H2O2 production, pyruvate oxidase, is repressed by high glucose concentrations but is highly expressed in cells grown in galactose (49). Therefore, during fasting periods, when galactose is a primary carbohydrate for growth, S. gordonii may gain a selective advantage over S. mutans not only by growing more efficiently but also through more effective antagonism. Possibly, then, galactose or its derivatives may prove effective at promoting an oral flora that is less cariogenic.
ACKNOWLEDGMENT
This study was supported by grant DE12236 from the NIDCR.
Footnotes
Published ahead of print 1 June 2012
REFERENCES
- 1. Ahn SJ, Lemos JA, Burne RA. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187:3028–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ajdic D, et al. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ajdic D, Sutcliffe IC, Russell RR, Ferretti JJ. 1996. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene 180:137–144 [DOI] [PubMed] [Google Scholar]
- 4. Beighton D, Whiley RA. 1990. Sialidase activity of the “Streptococcus milleri group” and other viridans group streptococci. J. Clin. Microbiol. 28:1431–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breidt F, Jr, Hengstenberg W, Finkeldei U, Stewart GC. 1987. Identification of the genes for the lactose-specific components of the phosphotransferase system in the lac operon of Staphylococcus aureus. J. Biol. Chem. 262:16444–16449 [PubMed] [Google Scholar]
- 6. Burne RA, Penders JE. 1992. Characterization of the Streptococcus mutans GS-5 fruA gene encoding exo-β-D-fructosidase. Infect. Immun. 60:4621–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burne RA, Penders JE. 1994. Differential localization of the Streptococcus mutans GS-5 fructan hydrolase enzyme, FruA. FEMS Microbiol. Lett. 121:243–249 [DOI] [PubMed] [Google Scholar]
- 8. Burne RA, Schilling K, Bowen WH, Yasbin RE. 1987. Expression, purification, and characterization of an exo-β-D-fructosidase of Streptococcus mutans. J. Bacteriol. 169:4507–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burne RA, Wen ZT, Chen YY, Penders JE. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burstein C, Cohn M, Kepes A, Monod J. 1965. Role of lactose and its metabolic products in the induction of the lactose operon in Escherichia coli. Biochim. Biophys. Acta 95:634–639 [PubMed] [Google Scholar]
- 11. Byers HL, Tarelli E, Homer KA, Beighton D. 1999. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human α1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9:469–479 [DOI] [PubMed] [Google Scholar]
- 12. Caldwell RC, Pigman W. 1966. Changes in protein and glycoprotein concentrations in human submaxillary saliva under various stimulatory conditions. Arch. Oral Biol. 11:437–450 [DOI] [PubMed] [Google Scholar]
- 13. Cha RS, Zarbl H, Keohavong P, Thilly WG. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 2:14–20 [DOI] [PubMed] [Google Scholar]
- 14. Chassy BM, Thompson J. 1983. Regulation and characterization of the galactose-phosphoenolpyruvate-dependent phosphotransferase system in Lactobacillus casei. J. Bacteriol. 154:1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Declerck N, Vincent F, Hoh F, Aymerich S, van Tilbeurgh H. 1999. RNA recognition by transcriptional antiterminators of the BglG/SacY family: functional and structural comparison of the CAT domain from SacY and LicT. J. Mol. Biol. 294:389–402 [DOI] [PubMed] [Google Scholar]
- 16. De Stoppelaar JD, Van Houte J, Backer DO. 1970. The effect of carbohydrate restriction on the presence of Streptococcus mutans, Streptococcus sanguis and iodophilic polysaccharide-producing bacteria in human dental plaque. Caries Res. 4:114–123 [DOI] [PubMed] [Google Scholar]
- 17. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 18. de Vos WM, Vaughan EE. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217–237 [DOI] [PubMed] [Google Scholar]
- 19. Dong Y, Chen YY, Snyder JA, Burne RA. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68:5549–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fridovich-Keil JL. 2006. Galactosemia: the good, the bad, and the unknown. J. Cell. Physiol. 209:701–705 [DOI] [PubMed] [Google Scholar]
- 21. Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73:245–259 [DOI] [PubMed] [Google Scholar]
- 22. Hojo K, Nagaoka S, Ohshima T, Maeda N. 2009. Bacterial interactions in dental biofilm development. J. Dent. Res. 88:982–990 [DOI] [PubMed] [Google Scholar]
- 23. Jagusztyn-Krynicka EK, et al. 1992. Streptococcus mutans serotype c tagatose 6-phosphate pathway gene cluster. J. Bacteriol. 174:6152–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaufman GE, Yother J. 2007. CcpA-dependent and -independent control of β-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon. J. Bacteriol. 189:5183–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406–408 [DOI] [PubMed] [Google Scholar]
- 26. Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuramitsu HK, Wang BY. 2006. Virulence properties of cariogenic bacteria. BMC Oral Health 6(Suppl 1):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langbein I, Bachem S, Stulke J. 1999. Specific interaction of the RNA-binding domain of the Bacillus subtilis transcriptional antiterminator GlcT with its RNA target, RAT. J. Mol. Biol. 293:795–805 [DOI] [PubMed] [Google Scholar]
- 29. Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205 [DOI] [PubMed] [Google Scholar]
- 30. LeBlanc DJ, Crow VL, Lee LN, Garon CF. 1979. Influence of the lactose plasmid on the metabolism of galactose by Streptococcus lactis. J. Bacteriol. 137:878–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu J, Wu C, Huang IH, Merritt J, Qi F. 2011. Differential response of Streptococcus mutans towards friend and foe in mixed-species cultures. Microbiology 157:2433–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loughman JA, Caparon MG. 2007. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 64:269–280 [DOI] [PubMed] [Google Scholar]
- 33. Neves AR, et al. 2010. Towards enhanced galactose utilization by Lactococcus lactis. Appl. Environ. Microbiol. 76:7048–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oskouian B, Stewart GC. 1990. Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J. Bacteriol. 172:3804–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petersen FC, Scheie AA. 2010. Natural transformation of oral streptococci. Methods Mol. Biol. 666:167–180 [DOI] [PubMed] [Google Scholar]
- 36. Rosey EL, Stewart GC. 1992. Nucleotide and deduced amino acid sequences of the lacR, lacABCD, and lacFE genes encoding the repressor, tagatose 6-phosphate gene cluster, and sugar-specific phosphotransferase system components of the lactose operon of Streptococcus mutans. J. Bacteriol. 174:6159–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scannapieco FA. 1994. Saliva-bacterium interactions in oral microbial ecology. Crit. Rev. Oral Biol. Med. 5:203–248 [DOI] [PubMed] [Google Scholar]
- 38. Shaw WV. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737–755 [DOI] [PubMed] [Google Scholar]
- 39. Tong H, Zeng L, Burne RA. 2011. The EIIABMan PTS permease regulates carbohydrate catabolite repression in Streptococcus gordonii. Appl. Environ. Microbiol. 77:1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van der Hoeven JS, Camp PJ. 1991. Synergistic degradation of mucin by Streptococcus oralis and Streptococcus sanguis in mixed chemostat cultures. J. Dent. Res. 70:1041–1044 [DOI] [PubMed] [Google Scholar]
- 41. van Rooijen RJ, Dechering KJ, Niek C, Wilmink J, de Vos WM. 1993. Lysines 72, 80 and 213 and aspartic acid 210 of the Lactococcus lactis LacR repressor are involved in the response to the inducer tagatose-6-phosphate leading to induction of lac operon expression. Protein Eng. 6:201–206 [DOI] [PubMed] [Google Scholar]
- 42. van Tilbeurgh H, Declerck N. 2001. Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr. Opin. Struct. Biol. 11:685–693 [DOI] [PubMed] [Google Scholar]
- 43. Zahner D, Hakenbeck R. 2000. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 182:5919–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeng L, Burne RA. 2008. Multiple sugar:phosphotransferase system permeases participate in catabolite modification of gene expression in Streptococcus mutans. Mol. Microbiol. 70:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zeng L, Das S, Burne RA. 2011. Genetic analysis of the functions and interactions of components of the LevQRST signal transduction complex of Streptococcus mutans. PLoS One 6:e17335 doi:10.1371/journal.pone.0017335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zeng L, Das S, Burne RA. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J. Bacteriol. 192:2434–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeng L, Dong Y, Burne RA. 2006. Characterization of cis-acting sites controlling arginine deiminase gene expression in Streptococcus gordonii. J. Bacteriol. 188:941–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeng L, Wen ZT, Burne RA. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol. Microbiol. 62:187–200 [DOI] [PubMed] [Google Scholar]
- 49. Zheng L, Itzek A, Chen Z, Kreth J. 2011. Environmental influences on Streptococcus gordonii competitive hydrogen peroxide production. Appl. Environ. Microbiol. 77:4318–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]