Abstract
Psychrotolerant sporeformers, specifically Paenibacillus spp., are important spoilage bacteria for pasteurized, refrigerated foods such as fluid milk. While Paenibacillus spp. have been isolated from farm environments, raw milk, processing plant environments, and pasteurized fluid milk, no information on the number of Paenibacillus spp. that need to be present in raw milk to cause pasteurized milk spoilage was available. A real-time PCR assay targeting the 16S rRNA gene was designed to detect Paenibacillus spp. in fluid milk and to discriminate between Paenibacillus and other closely related spore-forming bacteria. Specificity was confirmed using 16 Paenibacillus and 17 Bacillus isolates. All 16 Paenibacillus isolates were detected with a mean cycle threshold (CT) of 19.14 ± 0.54. While 14/17 Bacillus isolates showed no signal (CT > 40), 3 Bacillus isolates showed very weak positive signals (CT = 38.66 ± 0.65). The assay provided a detection limit of approximately 3.25 × 101 CFU/ml using total genomic DNA extracted from raw milk samples inoculated with Paenibacillus. Application of the TaqMan PCR to colony lysates obtained from heat-treated and enriched raw milk provided fast and accurate detection of Paenibacillus. Heat-treated milk samples where Paenibacillus (≥1 CFU/ml) was detected by this colony TaqMan PCR showed high bacterial counts (>4.30 log CFU/ml) after refrigerated storage (6°C) for 21 days. We thus developed a tool for rapid detection of Paenibacillus that has the potential to identify raw milk with microbial spoilage potential as a pasteurized product.
INTRODUCTION
Despite advances in food preservation techniques, bacterial spoilage remains a leading cause of global food loss (14). Nearly one-third of all food produced worldwide is estimated to be lost postharvest, and much of this loss can be attributed to microbial spoilage (16). Dairy products constitute one of the leading sectors impacted by food loss in the United States, as nearly 20% of conventionally pasteurized (high temperature, short time [HTST]) fluid milk is discarded prior to consumption each year (23). In the United States, the shelf life of fluid milk ranges from approximately 1 to 3 weeks. Most consumer complaints result from the growth of psychrotolerant bacteria, typically, either non-spore-forming Gram-negative rods or Gram-positive spore-forming bacteria (12, 18, 19, 28, 34, 39). The presence of psychrotolerant, non-spore-forming bacteria (e.g., Pseudomonas) in pasteurized milk indicates either inadequate heating of the milk or, more commonly, postpasteurization contamination (6). Therefore, pasteurized milk contamination with Pseudomonas and other non-spore-forming bacteria can be controlled or eliminated by adhering to pasteurization specifications for minimum time and temperature combinations (8) and by adhering to proper sanitation and equipment maintenance protocols, particularly with respect to milk filler sites (33). Conversely, Gram-positive psychrotolerant sporeformers can survive pasteurization as spores, germinate, and then grow during refrigerated storage to numbers capable of causing off flavors or curdling of milk (5, 20, 34, 36).
The predominant Gram-positive spore-forming bacteria isolated from milk are Bacillus spp. and Paenibacillus spp. Both Bacillus spp. and Paenibacillus spp. have been isolated from farm environments (e.g., soil, water, and feed), raw milk, dairy-processing plants, and pasteurized milk (5, 19, 38, 42). In HTST pasteurized milk, when postpasteurization contamination is excluded, Bacillus spp. represent the predominant bacteria found early in the shelf life (<7 days). However, during refrigerated storage of pasteurized milk, Paenibacillus spp. become the predominant spoilage organisms, typically representing over 95% of the bacterial population identified late in shelf life (>10 days) (35). Paenibacillus spp. are generally present in very low numbers in raw milk and early in pasteurized milk shelf life, yet they can reproduce to high numbers during cold storage. Numerous microbiological tests have been applied to raw milk with the goal of predicting the shelf life performance of the milk, but none are adequately predictive of HTST pasteurized fluid milk shelf life (25). This, in part, is likely due to the inability of traditional microbiological tests to identify or quantify low levels (<10 spores/ml) of Paenibacillus spp. Currently, only limited phenotypic methods are available to differentiate between Bacillus spp. and closely related Paenibacillus spp., including cold growth, which requires 7 to 10 days of incubation, and lactose utilization, which can be difficult to interpret and is not a consistent indicator of sporeformer genus (21).
The aim of this study was to develop a novel PCR assay targeting the 16S rRNA gene so that specific identification of Paenibacillus spp. could be performed rapidly. The objectives of this study were to (i) design primers and a probe for detection of Paenibacillus spp. while limiting nonspecific detection of closely related Bacillus spp., (ii) validate the primers and probe using a real-time PCR assay on select Paenibacillus and Bacillus isolates from a collection of over 1,200 isolates from fluid milk and dairy environments, and (iii) develop a systematic approach to aid in identification of Paenibacillus spp. from raw milk. The results of this study will provide the food industry with an assay to monitor the quality of raw milk. This assay may even be adapted to aid in the development of strategies to limit spoilage of other pasteurized, refrigerated foods like vegetable purees (2, 15) and fermented beverages (17). Finally, our assay has potential for use as a screening tool to isolate novel enzyme-producing Paenibacillus spp. from other foods (31) and the natural environment (37), as previous identification of Paenibacillus strains has led to the discovery of many compounds with promising applications in agriculture and medicine (29).
MATERIALS AND METHODS
TaqMan probe and primer design.
rpoB and 16S rRNA gene alignments were performed in the MegAlign program (DNAStar, Inc. Madison, WI). rpoB sequences (632 bp) from a total of 1,288 isolates representing the Paenibacillus genus (n = 737), the Bacillus genus (n = 467), and genera formerly classified as Bacillus (e.g., Viridibacillus; n = 84) collected from farm environments, raw milk, fluid milk-processing plants, and HTST fluid milk products were analyzed to identify unique subtypes (21). rpoB sequences lacked sufficient conservation for design of TaqMan primers and probes that could detect all 737 Paenibacillus sequences represented in this collection. Therefore, alignments of partial (>600-bp) 16S rRNA gene sequences representing each of the 283 rpoB subtypes identified among these Bacillus and Paenibacillus spp. were used to create consensus sequences for (i) all Paenibacillus rRNA gene sequences and (ii) all non-Paenibacillus rRNA gene sequences (which includes sequences for Bacillus, Lysinibacillus, Oceanobacillus, Psychrobacillus, Solibacillus, and Viridibacillus). The consensus sequences were exported to Primer Express software (version 2.0.0; Applied Biosystems, Foster City, CA) for primer-probe design. Primers were designed to detect a conserved region within the Paenibacillus genus, while excluding Bacillus spp. and other closely related genera. The designed amplicon was 158 bp and included a 24-bp probe located 34 bp downstream from the 5′ end of the forward primer (see Table 1 for primers and probe). The probe was labeled on the 5′ end with 6-carboxyfluorescein (FAM) and the 3′ end with tetramethylrhodamine (TAMRA). Detailed information on all isolates used in this study, including 16S rRNA and rpoB sequences, can be accessed at www.pathogentracker.net.
Table 1.
TaqMan primers and probe designed for detection of Paenibacillus spp. 16S rRNA gene
| Primer or probe | Sequence (5′–3′) | Denaturation temp (°C)a |
|---|---|---|
| Primers | ||
| MR-18_16S F | AAA TCA TCA TGC CCC TTA TG | 61.1 |
| MR-19_16S R | CGA TTA CTA GCA ATT CCG ACT | 59.8 |
| MR-21_16S probe | CGT ACT ACA ATG GCC GGT ACA ACG | 69.6 |
Denaturation temperatures were calculated using the Sigma-Aldrich DNA calculator (Sigma-Aldrich, St. Louis, MO).
TaqMan conditions.
Real-time PCR was conducted in a 12.5-μl reaction mixture containing 6.25 μl of 2× TaqMan universal master mix (Applied Biosystems), 900 nM each forward and reverse primer (MR-18_16S F, MR-19_16S R), 250 nM TaqMan probe (MR-21_16S probe), and 1.375 μl water (Table 1). Each reaction mixture also contained 1.25 μl of 10× exogenous internal positive control (IPC) mix and 0.25 μl of 50× exogenous IPC DNA (PE Applied Biosystems). Finally, 1.0 μl of DNA template was added to each reaction mixture.
Real-time PCR was performed as follows: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, and 40 cycles of denaturation at 95°C for 15 s, followed by extension and annealing at 60°C for 1 min. Threshold cycle (CT) values represent the fractional PCR cycle in which fluorescence first passed a defined threshold for each sample amplification plot.
Bacterial isolate selection and assay validation.
To validate the primers and probe, Paenibacillus and closely related Bacillus strains (n = 9 for each genus) were selected to represent the rpoB allelic types (ATs; i.e., those isolated ≥10 times) most frequently found among a collection of over 1,200 isolates collected from dairy farms, processing plants, raw milk, and pasteurized fluid milk (21). An additional 8 Bacillus strains (or strains of the closely related genera Lysinibacillus, Oceanobacillus, and Viridibacillus) and 7 Paenibacillus strains were included to represent genetic diversity (Table 2).
Table 2.
Bacterial isolates used to evaluate the specificity of a real-time PCR assay for detection of Paenibacillus spp.
| Isolate | ATa | Group IDb | No. of isolates in ATc | Mean CTd |
|---|---|---|---|---|
| FSL R5-510 | 1 | Bacillus licheniformis sensu lato 1 | 134 | >40 |
| FSL H7-687 | 3 | Bacillus weihenstephanensis | 19 | >40 |
| FSL R5-450 | 6 | Bacillus licheniformis sensu lato 1 | 35 | 39.53 |
| FSL R5-213 | 17 | Viridibacillus spp. | 24 | >40 |
| FSL H7-346 | 20 | Bacillus pumilus | 24 | >40 |
| FSL H7-608 | 59 | Bacillus cereus sensu lato | 26 | >40 |
| FSL R5-280 | 73 | Viridibacillus spp. | 18 | >40 |
| FSL H8-103 | 75 | Bacillus weihenstephanensis | 23 | >40 |
| FSL R5-860 | 158 | Bacillus cereus sensu lato | 137 | >40 |
| FSL H3-288 | 34 | Lysinibacillus spp. | 3 | >40 |
| FSL H7-305 | 55 | Bacillus clausii | 2 | >40 |
| FSL H7-431 | 64 | Bacillus sp. 2 | 2 | >40 |
| FSL H7-432 | 65 | Bacillus subtilis sensu lato 1 | 6 | >40 |
| FSL H7-719 | 84 | Oceanobacillus chironomi | 1 | >40 |
| FSL H7-729 | 85 | Bacillus flexus | 1 | 38.65 |
| FSL H8-493 | 135 | Bacillus aerophilus sensu lato | 9 | >40 |
| FSL R5-231 | 140 | Bacillus safensis | 6 | 38.22 ± 0.34 |
| FSL F4-077 | 2 | Paenibacillus odorifer 1 | 52 | 19.06 ± 0.06 |
| FSL F4-126 | 13 | Paenibacillus odorifer 1 | 21 | 18.31 ± 0.25 |
| FSL H7-592 | 15 | Paenibacillus odorifer 1 | 112 | 19.09 ± 0.11 |
| FSL F4-190 | 21 | Paenibacillus odorifer 3 | 28 | 18.62 ± 0.18 |
| FSL H7-689 | 23 | Paenibacillus amylolyticus sensu lato | 35 | 18.88 ± 0.04 |
| FSL F4-242 | 25 | Paenibacillus odorifer 1 | 19 | 18.40 ± 0.08 |
| FSL F4-248 | 27 | Paenibacillus odorifer 1 | 79 | 19.01 ± 0.10 |
| FSL R5-925 | 30 | Paenibacillus odorifer 3 | 12 | 20.28 ± 0.22 |
| FSL H3-442 | 32 | Paenibacillus odorifer 1 | 16 | 19.43 ± 0.10 |
| FSL F4-100 | 8 | Paenibacillus lautus | 3 | 18.92 ± 0.21 |
| FSL H3-318 | 41 | Paenibacillus sp. 1 | 3 | 19.87 ± 0.28 |
| FSL R7-277 | 45 | Paenibacillus graminis 1 | 3 | 19.02 ± 0.13 |
| FSL H7-331 | 58 | Paenibacillus sp. 10 | 5 | 19.95 ± 0.06 |
| FSL H8-287 | 100 | Paenibacillus cf. xylanilyticus | 9 | 18.94 ± 0.04 |
| FSL H8-551 | 157 | Paenibacillus cf. peoriae | 8 | 19.38 ± 0.23 |
| FSL R5-978 | 163 | Paenibacillus graminis 2 | 9 | 19.01 ± 0.14 |
Bacillus AT1, AT6, and AT17 and Paenibacillus AT2, AT15, AT23, and AT27 represent ATs commonly isolated from HTST milk produced in plants throughout the United States (34). AT1, AT15, AT21, and AT27 also represent ATs commonly isolated throughout the dairy-processing continuum (i.e., dairy farm environment, tank trucks, plant storage silos, and pasteurized milk) in New York State (19).
Group identifier based on previously described phylogenetic comparison (21); cf. denotes closely related species according to 16S rRNA sequence analysis.
Numbers are based on a total of 737 Paenibacillus and 551 non-Paenibacillus (i.e., Bacillus, Lysinibacillus, Oceanobacillus, Viridibacillus) dairy-associated isolates characterized by rpoB sequence-based subtyping (21). ATs isolated >10 times were considered predominant and used to test assay specificity; all other isolates were included to represent unique phylogenetic clades based on partial rpoB sequence comparison.
Samples not detected in 40 cycles were assigned a value of >40. Samples without SDs were detected in only one of two replicates.
Pure bacterial cultures, stored in 15% glycerol at −80°C, were streaked onto brain heart infusion (BHI) agar (Difco, BD Diagnostics, Franklin Lakes, NJ) and grown for 18 to 24 h at 32°C. A single colony from plates that confirmed a pure culture was inoculated into 5 ml of BHI broth (Difco) and grown for 18 to 24 h at 32°C. Total genomic DNA was extracted from 1 ml of overnight culture according to the QIAamp DNeasy kit instructions (Qiagen Inc., Valencia, CA). Purified DNA concentrations were determined using Hoechst dye assay (Thermo Fisher Scientific, Wilmington, DE) and standardized to 105 genomes/μl.
To determine amplification efficiency, genomic DNA from Paenibacillus odorifer isolate FSL H7-592, representing the predominant spoilage allelic type (AT15), was serially diluted (107 to 101 genomes/ml) to produce a standard curve. Amplification efficiency (E) was calculated using the following equation: [10(−1/slope)] − 1.
Detection limit and raw milk sample testing.
To determine the detection limit for Paenibacillus in the presence of other bacteria in a complex matrix, raw milk was obtained from the Cornell Teaching and Research Center (Dryden, NY). An overnight culture of Paenibacillus odorifer (FSL H7-592; AT15) was grown in BHI broth (Difco) and then centrifuged at 10,000 × g (5417C; Eppendorf, Hamburg, Germany) and resuspended in phosphate-buffered saline solution (Weber Scientific, Hamilton, NJ) before serial dilution into the raw milk; final Paenibacillus concentrations of 105, 104, 103, 102, and 101 CFU/ml of milk were achieved. A negative control containing no added Paenibacillus DNA was also included. To test the sensitivity of the PCR assay with a high background flora of mesophilic spore-forming bacteria typically found in milk, 100 ml of raw milk was heated to 80°C and held for 12 min, cooled, and then incubated at 32°C for 18 h before inoculation with Paenibacillus odorifer to achieve final Paenibacillus DNA concentrations of 105, 104, 103, 102, and 101 CFU/ml of enriched milk. A Norgen milk bacterial DNA isolation kit (Norgen Biotek Corp., Ontario, Canada) was used according to the manufacturer's instructions to extract DNA from 1 ml of all milk samples, and a final elution volume of 100 μl was obtained.
Paenibacillus assay testing of raw milk samples.
Approximately 400 ml of raw milk was collected from 10 different farms across upstate New York from March to May of 2011. Bulk tank raw milk samples (n = 24) were shipped on ice to the Cornell University Milk Quality Improvement Laboratory (Ithaca, NY). Upon receipt, raw milk was spore shocked (80°C for 12 min) to eliminate vegetative cells and activate spores (11). Approximately 150 ml of milk was aliquoted to 4 sterile 250-ml screw-cap Pyrex containers for aerobic plate count (APC) determination on the initial day of heat treatment and at days 7, 14, and 21 of storage at 6°C; APCs were performed according to Standard Methods for the Examination of Dairy Products (11). An additional 25 ml of spore-shocked milk was aliquoted into a sterile vial; this sample was incubated at 13°C for 48 h to encourage growth of Paenibacillus spp. while limiting Bacillus sp. growth. Bacterial counts in the 13°C enrichment were monitored immediately following the spore shock, at 24 h after spore shock, and 48 h after spore shock; bacterial counts were determined by plating 1 ml of milk over 5 BHI medium plates (200 μl per plate) supplemented with bromo-chloro-indolyl galactopyranoside (X-Gal; 100 mg/liter; Gold Biotechnology, St. Louis, MO). Plates were incubated at 32°C for 24 h before enumeration. Plating onto BHI agar supplemented with X-Gal allowed simultaneous APC determination and identification of β-galactosidase (β-Gal)-positive spore-forming bacteria, which, in milk, have generally been found to be Paenibacillus spp. (21). From APC plates, both X-Gal-positive and -negative colony counts were recorded. Up to 5 isolates representing colonies with unique morphologies and including colonies with both β-Gal-positive and -negative activity were selected from each plate for Paenibacillus TaqMan PCR; crude lysates were prepared by touching a single colony with a sterile toothpick, transferring the cells into 100 μl of sterile water in a 1.5-ml Eppendorf tube (Eppendorf, Hamburg, Germany), vortexing briefly, and then microwaving on high for 4 min. TaqMan PCR results from colony lysates were interpreted as positive for Paenibacillus if the CT value was <36.71; this cutoff value was the mean CT for the non-Paenibacillus isolates (38.66 ± 0.65) that were used to evaluate assay specificity (Table 2) minus 3 standard deviations (SDs; to limit false-positive detection). An isolate representing each colony was also characterized to the genus and species levels by 16S rRNA gene or rpoB sequence-based subtyping, as previously described (20).
In addition to direct testing of colonies, total genomic DNA was isolated from milk, after incubation of the spore-shocked milk at 13°C for 48 h, using the Norgen milk bacterial DNA isolation kit. Final elution volumes of 100 μl were collected and used in the TaqMan PCR reported here to test for the presence of Paenibacillus.
To test for an association between the detection of Paenibacillus colonies in raw milk samples (after heat shock of milk, 48 h of incubation at 13°C, and plating onto BHI agar supplemented with X-Gal) and the final bacterial count in heat-treated milk samples stored for 21 days at 6°C, Fisher's exact tests were performed (JMP, version 8.0; SAS Institute Inc., Cary, NC). Paenibacillus assay results were coded as present (≥1 Paenibacillus colony confirmed by TaqMan PCR) or absent (no detectable Paenibacillus colonies), depending on TaqMan colony PCR results. For statistical analysis, final bacterial counts at day 21 were used to assign milk samples into one of two groups (≤2 × 104 or >2 × 104 CFU/ml), based on the Pasteurized Milk Ordinance (8) bacterial count limit of 2 × 104 CFU/ml for grade A pasteurized fluid milk. For descriptive analysis, milk samples with day 21 bacterial counts of >2 × 104 CFU/ml were separated into intermediate (>2 × 104 and ≤1 × 106 CFU/ml) and high (>1 × 106 CFU/ml) categories, while day 21 bacterial counts of ≤2 × 104 remained designated low. P values of less than 0.05 were considered significant.
RESULTS
TaqMan PCR allows specific detection of Paenibacillus spp.
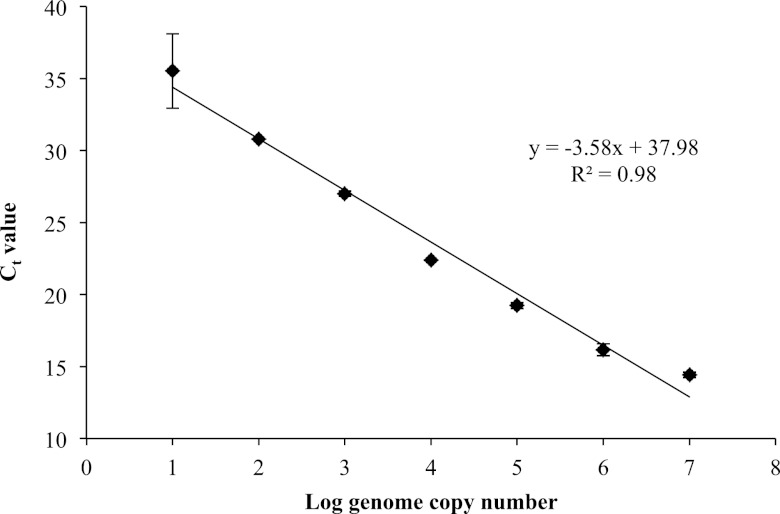
The TaqMan primers and probes designed here (Table 1) were first used to generate a standard curve based on mean CT values from assays performed in duplicate with Paenibacillus DNA at levels representing 107 to 101 log genome copies (Fig. 1). The linear regression line relating log genome copy number to CT values was y = −3.58x + 37.98, and the R2 value for the linear equation was 0.98. The amplification efficiency for real-time PCR amplification was determined to be 90.11%.
Fig 1.
Standard curve for determination of amplification efficiency. Error bars indicate ±1 standard deviation for duplicate tests of each genome copy number. The average efficiency for real-time amplification was 90.11%.
The specificity of the primers and probe for detection of Paenibacillus spp. was evaluated using 105 copies of genomic DNA isolated from 16 Paenibacillus isolates. All 16 Paenibacillus isolates were detected with the assay, and the mean CT value was 19.14 ± 0.54 (Table 2). The 16 isolates tested represented 16 rpoB ATs. These rpoB ATs represent over 56% (414/737) of Paenibacillus isolates previously collected from each of the four fundamental steps in dairy processing (i.e., from dairy farms [feed, bedding materials, manure, soil and milking parlor wash water], tank trucks, plant storage silos, and pasteurized milk). These ATs also represent five of the predominant rpoB ATs identified among sporeformer isolates obtained from HTST pasteurized milk processed in different geographical regions throughout the United States (AT2, AT15, AT23, and AT27; representing milk processed in the northeastern, midwestern, western, southern, and southeastern United States) (34).
A total of 17 isolates representing Bacillus and other genera closely related to Bacillus (i.e., Viridibacillus, Lysinibacillus, and Oceanobacillus) were also tested with the TaqMan PCR. These isolates represented 17 unique rpoB ATs, including 9 common ATs (i.e., ATs that represented ≥10 isolates among a total of 551 non-Paenibacillus isolates). In total, >85% (470/551) of non-Paenibacillus isolates collected and characterized from the fluid milk-processing continuum, including dairy farm environments, tank trucks, plant storage silos, and raw and pasteurized milk, were classified into the 17 ATs tested here. Overall, 14 isolates were negative in the TaqMan PCR (CT > 40), including 8/9 predominant Bacillus ATs found in fluid milk- or dairy-processing environments. The remaining three isolates (FSL R5-450, FSL H7-729, and FSL R5-231) yielded weakly positive results in the TaqMan PCR (i.e., CT values of ≥38.22). Isolate FSL R5-450, which represents a common AT (i.e., AT6; Table 2), was negative in one replicate and weakly positive in the other replicate (CT = 39.53). FSL H7-729 (AT85, an isolate included for genetic diversity; Table 2) was also negative in one replicate and weakly positive in the other replicate (CT = 38.65). Bacillus strain FSL R5-231 (AT140, an AT isolated only 6 times) was the only non-Paenibacillus strain that yielded a positive result in both TaqMan PCR replicates (CT = 38.22 ± 0.34).
Detection limit for vegetative Paenibacillus cells in raw milk is 3.25 × 101 CFU/ml.
The ability of the assay to detect vegetative Paenibacillus cells in whole raw milk with and without spore enrichment was tested. Detection of Paenibacillus in raw milk (no enrichment) inoculated with Paenibacillus isolate FSL H7-592 (AT15) was possible at concentrations ranging from 3.25 × 105 ± 0.21 × 105 CFU/ml (CT = 26.14 ± 0.78) to as few as 3.25 × 101 ± 0.21 × 101 Paenibacillus CFU/ml (CT = 39.15; only one of two replicates had a CT value of <40) (Table 3); background flora in the raw milk was present at 3.85 × 103 ± 1.91 × 103 CFU/ml (Table 3). The negative control was not detected in two biological replicates (CT > 40).
Table 3.
Sensitivity of Paenibacillus detection using real-time PCR
| Paenibacillus count (no. of CFU/ml) |
CT after: |
|
|---|---|---|
| Paenibacillus inoculated into raw milka | Paenibacillus inoculated into heat-shocked and enriched raw milkb | |
| 3.25 × 105 ± 0.21 × 105 | 26.14 ± 0.78 | 26.73 ± 0.09 |
| 3.25 × 104 ± 0.21 × 104 | 29.47 ± 0.40 | 30.80 ± 0.50 |
| 3.25 × 103 ± 0.21 × 103 | 31.76 ± 1.20 | 38.22 ± 0.06 |
| 3.25 × 102 ± 0.21 × 102 | 35.61 ± 0.95 | 39.46c |
| 3.25 × 101 ± 0.21 × 101 | 39.15c | >40d |
| Negative control | >40 | >40 |
Mean aerobic plate count of raw milk, 3.85 × 103 ± 1.91 × 103 CFU/ml; mean aerobic plate count postenrichment, 4.65 × 107 ± 0.21 × 107 CFU/ml.
Milk was incubated at 32°C for 18 h to achieve high levels of competitive microflora.
Only one of two sample replicates was detected in 40 cycles.
Samples not detected in 40 cycles were assigned a CT value of >40.
The detection of Paenibacillus cells inoculated into spore-activated and enriched raw milk ranged from 3.25 × 105 ± 0.21 × 105 Paenibacillus CFU/ml (CT = 26.73 ± 0.09) to 3.25 × 102 ± 0.21 × 105 Paenibacillus CFU/ml (CT = 39.46; only one of two replicates detected). Paenibacillus was not detected (CT > 40) in the enriched milk sample containing 3.25 × 105 ± 0.21 × 105 CFU/ml or the negative control. While the CT values at higher Paenibacillus concentrations (3.25 × 104 and 3.25 × 105 CFU/ml) were similar for both raw milk and spore-enriched raw milk, at lower Paenibacillus concentrations (3.25 × 101, 3.25 × 102, and 3.25 × 103 CFU/ml), the CT values were higher for heat-shocked and enriched samples. The sensitivity of detection for Paenibacillus was approximately 10-fold lower when Paenibacillus was inoculated in the nonenriched raw milk (with a mean background flora of 3.85 × 103 ± 1.91 × 103 CFU/ml) than when Paenibacillus was inoculated in the enriched milk samples, which showed a background flora of 4.65 × 107 ± 0.21 CFU/ml. A high concentration of a mesophilic spore-forming (i.e., Bacillus) bacterial 16S rRNA gene may have contributed to the decreased Paenibacillus sensitivity observed in the enriched milk samples.
Assay detects low levels of Paenibacillus spores capable of germination and outgrowth to spoilage levels in milk.
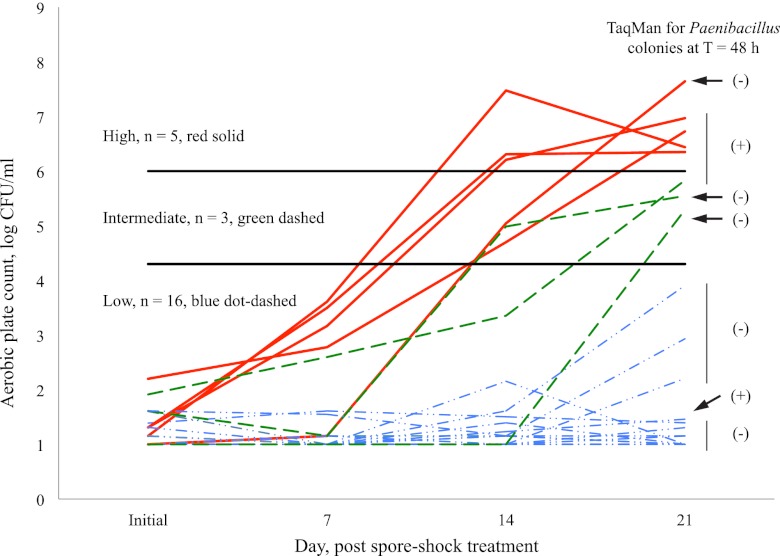
In order to evaluate the utility of the Paenibacillus TaqMan colony PCR, we also compared results from Paenibacillus detection in raw milk by TaqMan colony PCR to bacterial counts of milk stored at 6°C after heat treatment. Briefly, 24 raw milk samples collected from farm bulk tanks were (i) evaluated by the TaqMan colony PCR and (ii) subjected to simulated HTST pasteurization, followed by monitoring of bacterial numbers in the HTST-treated milk over a simulated shelf life of 21 days (i.e., incubation at 6°C) (Fig. 2). While initial-day counts for all 24 milk samples were below 2 × 102 spores/ml and ranged from <1 spore/ml to 117 spores/ml (mean, 11 spores/ml), subsequent bacterial outgrowth varied. At day 21 after spore shock treatment, bacterial numbers in the milk samples ranged from <10 CFU/ml (8 samples) to 4.37 × 107 CFU/ml (Table 4, sample D-3). Bacterial numbers after storage at 6°C for 21 days were categorized: 5 samples had bacterial counts of >1 ×106 CFU/ml (high), 16 samples had counts that remained at <2 × 104 CFU/ml (low), and 3 had counts of between 2 × 104 and 1 × 106 CFU/ml (intermediate). In 4/5 milk samples that reached bacterial numbers over 1 × 106 CFU/ml by day 21, Paenibacillus was detected by applying the Paenibacillus TaqMan assay to β-Gal-positive colonies recovered from raw milk after a 48-h enrichment at 13°C (48-h assay result, positive [+]; Fig. 2). For example, for sample D-4, total bacterial counts were 4, 15, and 153 CFU/ml after enrichment at 13°C for 0, 24, and 48 h, respectively. Of these counts, 0 β-Gal-positive CFU/ml were identified at time zero, 5 β-Gal-positive CFU/ml were identified at 24 h, and 114 β-Gal-positive CFU/ml were identified at 48 h. Representative blue colonies selected from 24 h (FSL R7-693) and 48 h (FSL R7-708) were identified as Paenibacillus by the TaqMan colony PCR reported here (CT values = 22.56 and 21.69, respectively). Confirmation of genus and species was performed by rpoB or 16S rRNA gene sequence-based characterization, and isolates FSL R7-693 and FSL R7-708 were determined to be Paenibacillus peoriae and Paenibacillus polymyxa, respectively. By day 21, milk sample D-4 reached a bacterial count of 9.33 × 106 CFU/ml. The predominant spoilage bacteria identified in the heat-treated milk stored at 6°C for 21 days were also determined to be Paenibacillus.
Fig 2.
Aerobic plate counts of spore-shocked milk stored at 6°C for 21 days. For each milk sample (n = 24), the Paenibacillus TaqMan assay was applied to individual colonies following heat treatment (80°C for 12 min), enrichment (13°C for 48 h), and plating of raw milk samples. Assay results indicate the presence (+) or absence (−) of one or more Paenibacillus colonies. The horizontal line at 4.3 log CFU/ml indicates the maximum permissible bacterial count in high-temperature, short-time-pasteurized milk in the United States (8). The horizontal line at 6 log CFU/ml indicates the maximum bacterial count typically associated with sensory scores of 8 and above (good flavor) on a 10-point scale (1).
Table 4.
Summary of assay results for raw milk samples that showed evidence of bacterial spoilage after heat treatment and subsequent incubation at 6°C
| Spoilage growth categoryb | Milk sample | No. of CFU/ml (no. of β-Gal-positive CFU/ml) after 13°C milk enrichment time ofa: |
Colony screening results after 13°C milk enrichment time of: |
Milk assessment following 21 days at 6°C |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | 0 h |
24 h |
48 h |
|||||||||||||
| Isolate | β-Galc | TaqMan PCR result (CT)d | rpoB-based genus | Isolate | β-Gal | TaqMan PCR result (CT) | rpoB-based genus | Isolate | β-Gal | TaqMan PCR result (CT) | rpoB-based genus | Bacterial count (CFU/ml) | Predominant spoilage bacteriae | |||||
| High | D-3 | 2 | 2 | <1 | FSL R7-644 | − | >40 | Bacillus | FSL R7-645 | − | >40 | Bacillus | NAf | 4.37 × 107 | Paenibacillus | |||
| D-4 | 4 | 15 (5) | 153 (114) | FSL R7-677 | − | >40 | Bacillus | FSL R7-692 | − | >40 | Bacillus | FSL R7-708 | + | 21.69 | Paenibacillus | 9.33 × 106 | Paenibacillus | |
| D-4 | FSL R7-678 | − | >40 | Bacillus | FSL R7-693 | + | 22.56 | Paenibacillus | FSL R7-709 | − | >40 | Bacillus | ||||||
| FSL R7-710 | − | >40 | Bacillus | |||||||||||||||
| C-4 | 30 | 29 | 313 (13) | FSL R7-674 | − | >40 | Bacillus | FSL R7-690 | − | >40 | Bacillus | FSL R7-704 | − | >40 | Bacillus | 5.37 × 106 | Paenibacillus | |
| FSL R7-675 | − | >40 | Bacillus | FSL R7-691 | − | >40 | Bacillus | FSL R7-705 | − | >40 | Bacillus | |||||||
| FSL R7-676 | − | >40 | Bacillus | FSL R7-706 | + | 25.18 | Paenibacillus | |||||||||||
| FSL R7-707 | + | 20.73 | Paenibacillus | |||||||||||||||
| C-3 | 27 | 23 | 197 (40) | FSL R7-646 | − | >40 | Bacillus | FSL R7-650 | − | >40 | Bacillus | FSL R7-652 | + | 18.9 | Paenibacillus | 2.75 × 106 | Paenibacillus | |
| FSL R7-651 | − | >40 | Bacillus | FSL R7-653 | − | >40 | Bacillus | |||||||||||
| FSL R7-654 | − | >40 | Bacillus | |||||||||||||||
| FSL R7-655 | − | >40 | Bacillus | |||||||||||||||
| FSL R7-656 | + | 17.9 | Paenibacillus | |||||||||||||||
| G-4 | 14 | 42 (1) | 530 (190) | FSL R7-679 | − | 18.37 | Paenibacillus | FSL R7-695 | − | >40 | Bacillus | FSL R7-711 | + | 21.8 | Paenibacillus | 2.24 × 106 | Paenibacillus | |
| FSL R7-680 | − | >40 | Bacillus | FSL R7-696 | − | >40 | Bacillus | FSL R7-712 | +/− | >40 | Bacillus | |||||||
| FSL R7-681 | − | >40 | Bacillus | FSL R7-697 | − | >40 | Bacillus | FSL R7-713 | − | >40 | Bacillus | |||||||
| FSL R7-682 | − | >40 | Bacillus | FSL R7-714 | − | >40 | Bacillus | |||||||||||
| Intermediate | C-5 | 117 (6) | 87 | 550 (10) | FSL R7-722 | − | >40 | Bacillus | FSL R7-733 | − | >40 | Bacillus | FSL R7-736 | − | >40 | Bacillus | 6.76 × 105 | Paenibacillus |
| FSL R7-723 | − | >40 | Bacillus | FSL R7-734 | − | >40 | Bacillus | FSL R7-737 | − | >40 | Bacillus | |||||||
| FSL R7-724 | − | >40 | Bacillus | FSL R7-735 | − | >40 | Bacillus | FSL R7-738 | − | >40 | Bacillus | |||||||
| FSL R7-725 | − | >40 | Bacillus | FSL R7-739 | +/− | 18.98 | Paenibacillus | |||||||||||
| FSL R7-726 | + | 21.5 | Paenibacillus | |||||||||||||||
| FSL R7-727 | + | 23.9 | Paenibacillus | |||||||||||||||
| D-5 | <1 | 3 | <1 | NA | FSL R7-740 | − | >40 | Bacillus | NA | 3.55 × 105 | B. weihenstephanensis | |||||||
| FSL R7-741 | − | >40 | Bacillus | |||||||||||||||
| FSL R7-742 | − | >40 | Bacillus | |||||||||||||||
| J-5 | <1 | 12 | 686 | NA | FSL R7-744 | − | >40 | Bacillus | FSL R7-745 | − | >40 | Bacillus | 1.95 × 105 | B. weihenstephanensis | ||||
Bacterial counts for the remaining 16 milk samples remained below 2 × 104 CFU/ml after heat shock and storage at 6°C for 21 days. See Table S1 in the supplemental material for complete summary of all 24 samples.
Growth category assigned on the basis of APCs following heat shock and storage of milk at 6°C for 21 days. High, APC > 1 × 106 CFU/ml; intermediate, APC ≤ 1 × 106 CFU/ml and >2 × 104 CFU/ml; low, APC ≤2 × 104 CFU/ml.
+, β-Gal-positive (blue) colonies; −, β-Gal-negative colonies; +/−,β-Gal weakly positive colonies (partial blue or light blue colony).
TaqMan CT values of <36.71 are interpreted as a positive Paenibacillus colony.
rpoB or 16S rRNA gene-based identification.
NA, not applicable.
Only one sample reached the high bacterial count category (>1 × 106 CFU/ml) after storage for 21 days at 6°C and did not contain detectable Paenibacillus after enrichment (sample D-3; Table 4). For the raw milk corresponding to this sample, the aerobic plate counts were 2, 2, and <1 CFU/ml following 0, 24, and 48 h of enrichment, respectively. The only colonies obtained at 0 and 24 h were determined to be Bacillus (Table 4). After 21 days of storage at 6°C, the bacterial count of sample D-3 reached 4.37 × 107 CFU/ml; the predominant organisms detected at this time were Paenibacillus, suggesting that very low levels (<1 spore/ml) of Paenibacillus are still capable of reaching high numbers in pasteurized products stored at refrigeration temperatures.
Among the 3 milk samples reaching intermediate bacterial counts by day 21 of cold storage (samples C-5, D-5, and J-5), only sample C-5 contained detectable Paenibacillus colonies during the 13°C enrichment and plating on BHI medium supplemented with X-Gal. Aerobic plate counts during enrichment of sample C-5 were 117 (6 β-Gal-positive colonies), 87, and 550 (10 β-Gal weakly positive [partial or light blue] colonies) at the 0-, 24-, and 48-h enrichment times, respectively. Two isolates, FSL R7-726 and FSL R7-727, from the time zero plating were β-Gal positive and were determined to be Paenibacillus by the TaqMan colony PCR (CT values = 21.5 and 23.9, respectively). Characterization by rpoB sequence analysis confirmed that both isolates were Paenibacillus. Plating at 24 h of enrichment yielded only Bacillus colonies (n = 3); however, one Paenibacillus colony was identified after 48 h of enrichment (FSL R7-739; CT = 18.98). After storage at 6°C for 21 days, the bacterial count for milk sample C-5 reached 6.76 × 105 CFU/ml, and the predominant bacteria identified were Paenibacillus. The other two milk samples (D-5 and J-5) in the intermediate count category contained no detectable Paenibacillus. Plating at 0, 24, and 48 h during sample enrichments yielded no β-Gal-positive colonies. Analysis of colonies using the Paenibacillus TaqMan assay determined colonies to be genera other than Paenibacillus (CT > 40). rpoB sequence-based characterization identified all 5 isolates collected from enrichment samples to be Bacillus pumilus or Bacillus licheniformis (Table 4). The predominant spoilage organism identified after storage of milk samples at 6°C for 21 days was determined to be cold-tolerant Bacillus weihenstephanensis. Final bacterial counts were 3.55 × 105 and 1.95 × 105 CFU/ml for samples D-5 and J-5, respectively.
A total of 16 raw milk samples had bacterial counts below 2 × 104 (4.30 log) CFU/ml after storage at 6°C for 21 days (see Table S1 in the supplemental material). During enrichment of those samples, 54 isolates were collected and only one sample (H-5) contained detectable Paenibacillus. After 48 h of enrichment, plating of sample H-5 resulted in 12 CFU/ml, 5 of which were weakly β-Gal positive. β-Gal weakly positive isolate FSL R7-747 was tested with the assay and determined to be Paenibacillus (CT = 20.37). rpoB-based characterization confirmed the identification of FSL H7-747 to be Paenibacillus. Following storage of milk sample H-5 for 21 days at 6°C, the bacterial count was 2.88 × 101 CFU/ml.
Results for the 24 milk samples were tested for a statistical association between detection of Paenibacillus (see Table S1 in the supplemental material; Paenibacillus colonies detected in 6 of 24 samples at 48 h) and the final APC after heat treatment and storage of raw milk samples for 21 days at 6°C. In raw milk samples where Paenibacillus was detected, there was a significant association with higher bacterial counts at day 21 (>2 × 104 CFU/ml; P = 0.0069).
Overall, a total of 109 bacterial isolates were collected during screening for Paenibacillus colonies by 13°C enrichment for 48 h and plating onto BHI medium supplemented with X-Gal. Of these, 97 isolates were β-Gal negative; 96/97 β-Gal-negative isolates were also negative in the TaqMan colony PCR (CT > 40; see Table S1 in the supplemental material). The only β-Gal-negative colony that yielded a positive signal with the TaqMan colony PCR (FSL R7-679, CT = 18.37) was confirmed to be Paenibacillus by rpoB sequence-based characterization. rpoB sequence-based identification identified the remaining 96 isolates as Bacillus (n = 92), Brevibacillus (n = 2), Oceanobacillus (n = 1), and Staphylococcus (n = 1). All 9 β-Gal-positive colonies were positive in the TaqMan colony PCR (mean CT = 21.57 ± 2.26). There were also 3 weakly β-Gal-positive (+/−; see Table S1 in the supplemental material) colonies. On the basis of rpoB characterization, 2/3 of these colonies were identified to be Paenibacillus and were detected with the TaqMan PCR (FSL R7-739 and FSL R7-747; CT values = 18.98 and 20.37, respectively). The remaining weakly β-Gal-positive colony (FSL R7-712) was determined to be Bacillus and was not detected by the TaqMan colony PCR (CT > 40).
In addition to testing individual colonies, total genomic DNA was collected from each of the 24 raw milk samples after 48 h of incubation at 13°C. Among these samples, only one milk sample was positive for Paenibacillus with the TaqMan PCR (sample G-4; 190 β-Gal-positive CFU/ml; CT = 34.49 ± 0.81). This suggests that Paenibacillus contamination in the raw milk is typically at levels below the detection limit of the TaqMan PCR when used on DNA directly extracted from milk (i.e., <3.25 × 101 ± 0.21 spores/ml).
DISCUSSION
Our real-time PCR-based approach represents an improved tool for identifying the predominant psychrotolerant spore-forming spoilage bacteria associated with pasteurized fluid milk stored at refrigerated temperatures. Based on a diverse collection of aerobic spore-forming bacteria, which included over 1,200 isolates collected from different segments of the dairy production continuum (21), we targeted Paenibacillus spp., the microbes that present the current biological limit to extension of pasteurized fluid milk shelf life. Our detection method requires heat treating raw milk at 80°C for 12 min to activate spores and eliminate vegetative bacterial cells, followed by a 48-h enrichment at 13°C to enrich for psychrotolerant bacteria. After enrichment, milk samples are plated onto BHI medium supplemented with X-Gal to allow direct screening of colonies, including β-Gal-positive colonies, which, in milk, generally represent Paenibacillus spp. Next, crude colony lysates are prepared for immediate testing of individual colonies using our TaqMan PCR, and final testing results (i.e., Paenibacillus or non-Paenibacillus spp.) can be obtained within a few hours. Overall, this colony-screening strategy combined with a TaqMan PCR presents a novel approach for detecting Paenibacillus in raw milk and for predicting psychrotolerant bacterial outgrowth in milk held at 6°C.
A Paenibacillus real-time PCR assay has potential applications for detection of psychrotolerant spore-forming bacteria in a variety of foods.
Few rapid, molecular-based detection methods targeting spore-forming bacteria responsible for food spoilage have been developed (9, 22, 24), and of these, none have focused on Paenibacillus, the psychrotolerant spore-forming genus associated with dairy spoilage. The absence of appropriate tools may reflect, in part, the fact that the bacterial ecology of pasteurized fluid milk has only recently been characterized at the molecular level, which led to identification of Paenibacillus as the predominant fluid milk spore-forming spoilage genera (12, 19, 21, 34). Rapid methods to detect spore-forming bacteria have primarily focused on food-borne pathogens, e.g., Bacillus cereus (13, 27, 43), that pose a significant health threat. However, the presence of spore-forming bacteria that can resist multiple processing hurdles and affect food product quality represents considerable economic and food security concerns. One commercial assay for the detection of spore-forming bacteria in food has been developed by Pall GeneSystems (32). However, when testing 34 food matrices, the authors reported that the detection system was unable to identify any Paenibacillus organisms. Conversely, when applying standard methods to the same 34 food matrices, researchers were able to identify Paenibacillus in sliced nuts and chocolate (32), which illustrates the difficulty of reliably identifying low levels of Paenibacillus in food. Other than this method, development of assays for Paenibacillus spp. to date has focused on Paenibacillus larvae (3, 26), an important honeybee pathogen. Thus, an assay targeting psychrotolerant Paenibacillus associated with milk spoilage represents a new and important tool for the dairy industry to identify Paenibacillus spp. in high-quality raw milk, as well as potential contamination sites at the farm and processing facility level. Sporeformers, including Paenibacillus, have the potential to form biofilms (44) and reside within processing facilities (20) and have been isolated from paperboard packaging (30). Thus, it is important to develop sensitive tools for detection of spoilage organisms and to apply them throughout the processing chain to identify entry points to enable development of control strategies to reduce spoilage and improve the quality of our foods. In the future, our assay could be extended to other refrigerated and pasteurized foods, including processed vegetables (2, 7, 15), where psychrotolerant Paenibacillus organisms are a potential spoilage concern.
Direct PCR-based detection of Paenibacillus in raw milk to predict shelf life is challenging due to the high sensitivity required.
Previous studies have demonstrated that low spore levels are typically found in raw milk. For example, sampling of raw milk from 43 processing plant silos in New York State yielded a mean aerobic spore count of 52 spores/ml (25). Additional studies in Europe reported similar findings, as mean counts of 131 mesophilic aerobic spores/ml (41) and <100 spores/ml of raw milk (42) were detected. Of these aerobic spores, only a small percentage are likely to be Paenibacillus, as a number of studies have found that Bacillus spp. comprise the majority of spores identified in raw and in recently heat-treated milk (4, 12, 19, 36). Consistent with this, only 12/109 (11%) isolates collected during our study represented Paenibacillus spp., and 9/12 of those Paenibacillus isolates were detected only after enrichment for 24 or 48 h at 13°C. Thus, due to the low levels of spores naturally present in raw milk, particularly spores of psychrotolerant Paenibacillus spp., an enrichment or concentration step is needed to improve assay sensitivity.
In addition to low levels of Paenibacillus spp., high levels of closely related Bacillus spp. further complicate detection, particularly for assays targeting the 16S rRNA gene. The detection limit for our assay increased nearly 10-fold when Paenibacillus was inoculated into heat-shocked and enriched (32°C for 18 h) raw milk. This reduction in sensitivity is likely due to high levels of closely related Bacillus spp. competing for primers and probe. Postollec and colleagues (32) encountered cross-reactivity when testing a commercial assay based on 16S rRNA gene primers and probes and reported Paenibacillus detection with Bacillus primers and vice versa. Many Bacillus and Paenibacillus spp. share over 99% identity, based on partial (632-bp) 16S rRNA gene analysis (21). Therefore, continued development of new assays, particularly through leveraging of full-genome sequencing technologies and concentration on definition of the characteristics of spore-forming bacteria, such as the differential presence of cold growth genes (10), will be critical to further improve detection capabilities.
PCR-based detection of individual colonies after enrichment and plating allows sensitive and specific detection of Paenibacillus spp.
Results from TaqMan PCR detection, performed on DNA extracted from milk samples, were predominantly negative due to low levels of Paenibacillus and competition from closely related Bacillus spp. Our observed detection limit for Paenibacillus inoculated into raw milk was 3.25 × 101 CFU/ml, which explains why only 1/24 raw milk samples tested positive for Paenibacillus. However, plating the same spore-shocked and enriched milk samples onto BHI medium supplemented with X-Gal allowed detection of Paenibacillus in 6/24 raw milk samples. Therefore, use of a TaqMan colony PCR following a short enrichment and plating on BHI medium supplemented with X-Gal greatly improves the reliability of the assay. Direct colony screening allowed us to lower the detection limit for Paenibacillus from 3.25 × 101 CFU/ml to 1 CFU/ml when a 1-ml sample was plated. In addition to improved sensitivity, the colony-screening method avoids the time and costs associated with genomic DNA purification steps.
The colony-screening method employs two important phenotypes that aid in distinguishing Paenibacillus from other sporeformers: cold growth and β-galactosidase activity. In general, Paenibacillus spp. are capable of growth at 6°C, whereas most Bacillus spp. are not; the most notable exception is Bacillus weihenstephanensis (21). By applying a 48-h incubation step for heat-shocked milk at 13°C, we were able to enrich for psychrotolerant Paenibacillus without promoting growth of mesophilic Bacillus spp., whose spores typically represent a higher proportion of spores in raw milk. However, two samples (C-5 and J-5) reached counts above the Pasteurized Milk Ordinance (8) limit for pasteurized milk (>20,000 CFU/ml) after storage at 6°C for 21 days and were not detected by our assay. The predominant spoilage organism in the two milk samples was determined to be B. weihenstephanensis. This outcome demonstrates the need for a detection system that utilizes genetic targets, such as cold-growth genes, shared by the psychrotolerant spoilage organisms of concern (i.e., Paenibacillus spp. and B. weihenstephanensis).
In addition to cold growth, β-galactosidase activity proved useful in identification of Paenibacillus. Previous work has shown that the majority of dairy-associated Paenibacillus subtypes are β-galactosidase positive, whereas the majority of Bacillus subtypes are not (5, 21). However, as some dairy-associated Bacillus isolates have expressed positive or weakly positive β-Gal activity, this phenotypic test cannot be completely relied upon to distinguish Paenibacillus from other sporeformers (21). In fact, we identified two β-Gal weakly positive isolates and one β-Gal-negative isolate as Paenibacillus by TaqMan colony PCR and rpoB sequence-based characterization. Thus, the combination of β-Gal screening and a 16S rRNA gene TaqMan assay proved necessary for accurate and sensitive detection of Paenibacillus spp. Application of this culture-dependent assay to screen for Paenibacillus spp. in nondairy environments could facilitate identification of strains with important metabolic capabilities (e.g., production of polymyxin, bioremediation, or nitrogen-fixing ability) of importance to agriculture, food processing, and medicine (29, 37, 40).
Conclusion.
We developed a sensitive and specific TaqMan assay that can detect psychrotolerant spore-forming Paenibacillus spp. associated with dairy spoilage. While the low levels of spores initially present in raw milk prevented direct detection of Paenibacillus in DNA extracted from raw milk or from enriched milk samples, an alternative colony-screening method proved feasible. A 16S rRNA gene-based TaqMan assay on crude colony lysates obtained from heat-shocked milk that had been enriched at 13°C for 48 h and plated on BHI medium supplemented with X-Gal provided fast and accurate identification of Paenibacillus. Overall, the assay provides an improved tool for the dairy industry to differentiate raw milk with the potential for lower postpasteurization bacterial outgrowth. Further development of rapid and effective detection methods for psychrotolerant sporeformers within a comprehensive farm-to-fork framework is needed for improved control of these important spoilage organisms in the food supply.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Milk Quality Improvement Program (MQIP) for their contributions to this research.
Research at the MQIP, including this project, is funded by the New York State Milk Promotion Board, New York State dairy farmers dedicated to the production, manufacture, and distribution of quality dairy products.
Footnotes
Published ahead of print 8 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Carey NR, Murphy SC, Zadoks RN, Boor KJ. 2005. Shelf lives of pasteurized fluid milk products in New York State: a ten-year study. Food Prot. Trends 25:102–113 [Google Scholar]
- 2. Carlin F, et al. 2000. Spore-forming bacteria in commercial cooked, pasteurised and chilled vegetable purées. Food Microbiol. 17:153–165 [Google Scholar]
- 3. Chagas SS, Vaucher RA, Brandelli A. 2010. Detection of Paenibacillus larvae by real-time PCR. Acta Sci. Vet. 38:251–256 [Google Scholar]
- 4. Coorevits A, et al. 2008. Comparative analysis of the diversity of aerobic spore-forming bacteria in raw milk from organic and conventional dairy farms. Syst. Appl. Microbiol. 31:126–140 [DOI] [PubMed] [Google Scholar]
- 5. De Jonghe V, et al. 2010. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int. J. Food Microbiol. 136:318–325 [DOI] [PubMed] [Google Scholar]
- 6. Eneroth A, Ahrne S, Molin G. 2000. Contamination routes of Gram-negative spoilage bacteria in the production of pasteurised milk, evaluated by randomly amplified polymorphic DNA (RAPD). Int. Dairy J. 10:325–331 [Google Scholar]
- 7. Fangio MF, Roura SI, Fritz R. 2010. Isolation and identification of Bacillus spp. and related genera from different starchy foods. J. Food Sci. 75:M218–M221 doi:10.1111/j.1750–3841.2010.01566.x [DOI] [PubMed] [Google Scholar]
- 8. FDA 2011. revision Grade “A” pasteurized milk ordinance, p 8–31 No. 229 Public Health Service, U.S. Department of Health and Human Services, Washington, DC [Google Scholar]
- 9. Fernández-No IC, et al. 2011. Detection and quantification of spoilage and pathogenic Bacillus cereus, Bacillus subtilis and Bacillus licheniformis by real-time PCR. Food Microbiol. 28:605–610 [DOI] [PubMed] [Google Scholar]
- 10. Francis KP, Mayr R, von Stetten F, Stewart GS, Scherer S. 1998. Discrimination of psychrotrophic and mesophilic strains of the Bacillus cereus group by PCR targeting of major cold shock protein genes. Appl. Environ. Microbiol. 64:3525–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franks JF, Yousef AE. 2004. Tests for groups of microorganisms, p 227–248 In Wehr HM, Frank JF. (ed), Standard methods for the examination of dairy products, 17th ed American Public Health Association, Washington, DC [Google Scholar]
- 12. Fromm H, Boor K. 2004. Characterization of pasteurized fluid milk shelf-life attributes. J. Food Sci. 69:M207–M214 doi:10.1111/j.1365-2621.2004.tb09889.x [Google Scholar]
- 13. Gracias KS, McKillip JL. 2011. Triplex PCR-based detection of enterotoxigenic Bacillus cereus ATCC 14579 in nonfat dry milk. J. Basic Microbiol. 51:147–152 [DOI] [PubMed] [Google Scholar]
- 14. Gram L, et al. 2002. Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 78:79–97 [DOI] [PubMed] [Google Scholar]
- 15. Guinebretiere MH, et al. 2001. Identification of bacteria in pasteurized zucchini purées stored at different temperatures and comparison with those found in other pasteurized vegetable purées. Appl. Environ. Microbiol. 67:4520–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gustavsson J, Cederberg C, Sonesson U. 2011. Global food losses and food waste. FAO report. FAO, Rome, Italy [Google Scholar]
- 17. Haakensen M, Ziola B. 2008. Identification of novel horA-harbouring bacteria capable of spoiling beer. Can. J. Microbiol. 54:321–325 [DOI] [PubMed] [Google Scholar]
- 18. Hayes W, White CH, Drake MA. 2002. Sensory aroma characteristics of milk spoilage by Pseudomonas species. J. Food Sci. 67:448–454 [Google Scholar]
- 19. Huck JR, Sonnen M, Boor KJ. 2008. Tracking heat-resistant, cold-thriving fluid milk spoilage bacteria from farm to packaged product. J. Dairy Sci. 91:1218–1228 [DOI] [PubMed] [Google Scholar]
- 20. Huck JR, Woodcock NH, Ralyea RD, Boor KJ. 2007. Molecular subtyping and characterization of psychrotolerant endospore-forming bacteria in two New York State fluid milk processing systems. J. Food Prot. 70:2354–2364 [DOI] [PubMed] [Google Scholar]
- 21. Ivy RA, et al. 2012. Identification and characterization of psychrotolerant sporeformers associated with fluid milk production and processing. Appl. Environ. Microbiol. 78:1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jang JH, et al. 2011. Detection of Alicyclobacillus species in fruit juice using a random genomic DNA microarray chip. J. Food Prot. 74:933–938 [DOI] [PubMed] [Google Scholar]
- 23. Kantor LS, Lipton K, Manchester A. 1997. Estimating and addressing America's food losses. Food Rev. 20:2–12 [Google Scholar]
- 24. Luo H, Yousef AE, Wang HH. 2004. A real-time polymerase chain reaction-based method for rapid and specific detection of spoilage Alicyclobacillus spp. in apple juice. Lett. Appl. Microbiol. 39:376–382 [DOI] [PubMed] [Google Scholar]
- 25. Martin NH, et al. 2011. Results from raw milk microbiological tests do not predict the shelf-life performance of commercially pasteurized fluid milk. J. Dairy Sci. 94:1211–1222 [DOI] [PubMed] [Google Scholar]
- 26. Martínez J, Simon V, Gonzalez B, Conget P. 2010. A real-time PCR-based strategy for the detection of Paenibacillus larvae vegetative cells and spores to improve the diagnosis and the screening of American foulbrood. Lett. Appl. Microbiol. 50:603–610 [DOI] [PubMed] [Google Scholar]
- 27. Martínez-Blanch JF, Sánchez G, Garay E, Aznar R. 2009. Development of a real-time PCR assay for detection and quantification of enterotoxigenic members of Bacillus cereus group in food samples. Int. J. Food Microbiol. 135:15–21 [DOI] [PubMed] [Google Scholar]
- 28. Mayr R, Eppert I, Scherer S. 1999. Incidence and identification of psychrotrophic (7 degrees C-tolerant) Bacillus spp. in German HTST pasteurized milk. Milchwissenschaft 54:26–30 [Google Scholar]
- 29. Naghmouchi K, et al. 2011. Paenibacillus polymyxa JB05-01-1 and its perspectives for food conservation and medical applications. Arch. Microbiol. 193:169–177 [DOI] [PubMed] [Google Scholar]
- 30. Pirttijärvi TS, Graeffe TH, Salkinoja-Salonen MS. 1996. Bacterial contaminants in liquid packaging boards: assessment of potential for food spoilage. J. Appl. Bacteriol. 81:445–458 [DOI] [PubMed] [Google Scholar]
- 31. Piuri M, Sanchez-Rivas C, Ruzal SM. 1998. A novel antimicrobial activity of a Paenibacillus polymyxa strain isolated from regional fermented sausages. Lett. Appl. Microbiol. 27:9–13 [DOI] [PubMed] [Google Scholar]
- 32. Postollec F, et al. 2010. A multiparametric PCR-based tool for fast detection and identification of spore-forming bacteria in food. Int. J. Food Microbiol. 142:78–88 [DOI] [PubMed] [Google Scholar]
- 33. Ralyea RD, Wiedmann M. 1998. Bacterial tracking in a dairy production system using phenotypic and ribotyping methods. J. Food Prot. 61:1336–1340 [DOI] [PubMed] [Google Scholar]
- 34. Ranieri ML, Boor KJ. 2009. Short communication: bacterial ecology of high-temperature, short-time pasteurized milk processed in the United States. J. Dairy Sci. 92:4833–4840 [DOI] [PubMed] [Google Scholar]
- 35. Ranieri ML, Boor KJ. 2010. Tracking and eliminating sporeformers in dairy systems. Aust. J. Dairy Technol. 65:74–80 [Google Scholar]
- 36. Ranieri ML, Huck JR, Sonnen M, Barbano DM, Boor KJ. 2009. High temperature, short time pasteurization temperatures inversely affect bacterial numbers during refrigerated storage of pasteurized fluid milk. J. Dairy Sci. 92:4823–4832 [DOI] [PubMed] [Google Scholar]
- 37. Sakai M, Ezaki S, Suzuki N, Kurane R. 2005. Isolation and characterization of a novel polychlorinated biphenyl-degrading bacterium, Paenibacillus sp. KBC101. Appl. Microbiol. Biotechnol. 68:111–116 [DOI] [PubMed] [Google Scholar]
- 38. Scheldeman P, Goossens K. 2004. Paenibacillus lactis sp. nov., isolated from raw and heat-treated milk. Int. J. Syst. Evol. Microbiol. 54:885–891 [DOI] [PubMed] [Google Scholar]
- 39. Schmidt VSJ, Kaufmann V, Kulozik U, Scherer S, Wenning M. 2011. Microbial biodiversity, quality and shelf life of microfiltered and pasteurized extended shelf life (ESL) milk from Germany, Austria and Switzerland. Int. J. Food Microbiol. 154:1–9 [DOI] [PubMed] [Google Scholar]
- 40. Shaheen M, Li J, Ross AC, Vederas JC, Jensen SE. 2011. Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem. Biol. 18:1640–1648 [DOI] [PubMed] [Google Scholar]
- 41. Stulova I, et al. 2010. Microbiological quality of raw milk produced in Estonia. Lett. Appl. Microbiol. 51:683–690 [DOI] [PubMed] [Google Scholar]
- 42. te Giffel MC, Wagendorp A, Herrewegh A, Driehuis F. 2002. Bacterial spores in silage and raw milk. Antonie Van Leeuwenhoek 81:625–630 [DOI] [PubMed] [Google Scholar]
- 43. Wehrle E, Didier A, Moravek M, Dietrich R, Märtlbauer E. 2010. Detection of Bacillus cereus with enteropathogenic potential by multiplex real-time PCR based on SYBR green I. Mol. Cell. Probes 24:124–130 [DOI] [PubMed] [Google Scholar]
- 44. Yegorenkova IV, Tregubova KV, Matora LY, Burygin GL, Ignatov VV. 2011. Biofilm formation by Paenibacillus polymyxa strains differing in the production and rheological properties of their exopolysaccharides. Curr. Microbiol. 62:1554–1559 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.