Abstract
The temporal dynamics of planktonic protists in river water have received limited attention despite their ecological significance and recent studies linking phagotrophic protists to the persistence of human-pathogenic bacteria. Using molecular-based techniques targeting the 18S rRNA gene, we studied the seasonal diversity of planktonic protists in Southwestern Alberta rivers (Oldman River Basin) over a 1-year period. Nonmetric multidimensional scaling analysis of terminal restriction fragment length polymorphism (T-RFLP) data revealed distinct shifts in protistan community profiles that corresponded to season rather than geographical location. Community structures were examined by using clone library analysis; HaeIII restriction profiles of 18S rRNA gene amplicons were used to remove prevalent solanaceous plant clones prior to sequencing. Sanger sequencing of the V1-to-V3 region of the 18S rRNA gene libraries from spring, summer, fall, and winter supported the T-RFLP results and showed marked seasonal differences in the protistan community structure. The spring library was dominated by Chloroplastidae (29.8%), Centrohelida (28.1%), and Alveolata (25.5%), while the summer and fall libraries contained primarily fungal clones (83.0% and 88.0%, respectively). Alveolata (35.6%), Euglenozoa (24.4%), Chloroplastida (15.6%), and Fungi (15.6%) dominated the winter library. These data demonstrate that planktonic protists, including protozoa, are abundant in river water in Southwestern Alberta and that conspicuous seasonal shifts occur in the community structure.
INTRODUCTION
Protists play an integral role in aquatic ecosystems. Protists are responsible for primary production, mobilizing trace nutrients, and controlling bacterial populations, yet they have not been extensively studied. They form a complex group of organisms spanning all eukaryotic kingdoms and vary substantially in size, shape, and motility. These characteristics were once relied upon exclusively by taxonomists for classification; however, the complexity of protistan morphology demands years of study and experience for accurate taxonomic placement. Furthermore, morphological-based identification is time-consuming and not always accurate. Recent studies have shown that organisms that were once considered to be of single morphospecies have highly variable 18S rRNA gene sequences and represent many distinct species (39). Conversely, protists that possess distinctly different morphologies (i.e., considered to be different morphospecies) have been shown to possess identical 18S rRNA gene sequences (44). As a result, protistologists now rely on molecular-based methods, such as terminal restriction fragment (T-RF) length polymorphism (T-RFLP), denaturing gradient gel electrophoresis (DGGE), and 18S rRNA gene libraries, to study these polyphyletic organisms.
The majority of protistan studies conducted to date have focused on protists in oceanic ecosystems (15, 21–23, 43, 49), as they produce upwards of half of the world's oxygen and are the basis of aquatic food webs. Freshwater systems (41) and water distribution systems (70) are attracting more attention as of late, due in part to experimental evidence linking freshwater protists with the protection of human pathogens (40). Phagotrophic protists (i.e., protozoans), such as Acanthamoeba spp., have been shown to play a role in the persistence of human pathogens, including Campylobacter jejuni (8, 67, 68), Legionella pneumophila (3, 40), Mycobacterium avium subsp. paratuberculosis (31), and Vibrio cholerae (1, 63), and thus may play a role in the transmission of infectious bacterial cells to humans. This presents a significant risk for public health, as Acanthamoeba cells are highly resistant to UV irradiation and oxidative treatments, and they may survive water treatment processes (40). To date, limited research has examined protists in freshwater ecosystems (14, 18, 41, 42, 48, 59, 65). Given the potential importance of protists in freshwater ecosystems to public health, it is of the utmost importance that we achieve a better understanding of planktonic protistan diversity.
According to the Public Health Agency of Canada's Notifiable Diseases On-Line website (http://dsol-smed.phac-aspc.gc.ca/dsol-smed/ndis/index-eng.php) and Alberta Health's 2004 Notifiable Diseases Report (http://www.health.alberta.ca/documents/Notifiable-Diseases-Report-2004.pdf), Southwestern Alberta has substantially higher rates of campylobacteriosis than the Canadian and Provincial averages (36); however, reasons for the high rates of campylobacteriosis in this region remain enigmatic. The waterborne transmission of C. jejuni has been suggested to be an important factor in the epidemiology of sporadic campylobacteriosis. Recent laboratory studies which showed the prolonged persistence of C. jejuni in the presence of freshwater protozoans suggested a possible link to waterborne transmission (8, 68). Consequently, there is a need for studies to investigate the diversity of protists in freshwater systems.
We hypothesized that rich protistan communities exist in river water in Southwestern Alberta and that community structures will differ among sampling sites and between seasons. The following objectives were constructed to test this hypothesis: (i) to longitudinally study freshwater protistan diversity in river water in Southwestern Alberta (Oldman River Basin) over a 1-year period and (ii) to identify protists in this region which may be contributing to the environmental persistence of human pathogens.
MATERIALS AND METHODS
Ethidium monoazide validation. (i) Culture of protists.
Acanthamoeba polyphaga, Tetrahymena pyriformis, Chlamydomonas moewusii, and Euglena gracilis were selected for ethidium monoazide (EMA) validation. A. polyphaga was cultured in peptone-yeast-glucose (PYG) growth medium at 30°C and at 100 rpm for 1 week and then for an additional week in fresh growth medium. T. pyriformis was grown in PPG growth medium (Culture Collection of Algae and Protozoa, Oban, Scotland) for 48 h at 28°C. C. moewusii and E. gracilis were grown for 1 week in Ward's basic culture medium (Wards Scientific, Rochester, NY) and a modified salts growth medium (61), with exposure to a southeast-facing window. Cells were washed once by centrifugation at 1,000 × g for 10 min, followed by the resuspension of the cells in 1× phosphate-buffered saline (PBS). Cell densities were estimated by using a hemocytometer, and viability was verified with trypan blue staining. Cultures were adjusted to a final concentration of 5 × 104 cells ml−1 and were divided into four 200-μl samples in 2-ml tubes for each protist.
(ii) EMA treatment.
For each protist, two arbitrarily selected samples were incubated at 100°C for 20 min (heat treated), and the remaining two samples were maintained at 4°C for 20 min (non-heat treated). EMA is a photoreactive cross-linker that binds irreversibly to free DNA, thereby inhibiting PCR amplification (60), and EMA (3 μl; Molecular Probes) was added to one heat-treated sample and one non-heat-treated sample (final concentration of 100 μg ml−1). Following the addition of EMA, samples were placed on ice in the dark for 5 min, and tubes (with lids open) were then exposed to light emitted from a 500-W halogen light bulb for two 1-min intervals; the light source was situated 10 cm from the top of the tubes, and samples were vortexed between light exposures. All samples were then stored at −20°C until subsequent processing.
(iii) DNA extraction and PCR amplification.
DNA was extracted by using the DNeasy blood and tissue kit (Qiagen Inc., Mississauga, Ontario, Canada) according to the manufacturer's instructions, with the exception that the final elution volume was reduced to 50 μl. PCR was performed by using 5 μl of extracted DNA. PCR primers Euk1A (5′-CTGGTTGATCCTGCCAG-3′) and Euk516R (5′-ACCAGACTTGCCCTCC-3′) were used; these primers amplify variable regions V1 to V3 of the 18S rRNA gene, which correspond to positions 4 to 563 of the Saccharomyces cerevisiae (GenBank accession number AY251630) 18S rRNA gene. PCR conditions were described previously by Diez et al. (20). PCR products were resolved by capillary electrophoresis using the MED250 protocol with a QIAxcel capillary system (Qiagen Inc.).
River water sample collection and processing.
Southwestern Alberta is a semiarid ecosystem dominated by a short-grass grassland ecosystem. The region contains the headwaters of the Oldman River and its tributaries, which constitute the primary watershed of the region. A total of five river sites in the Oldman watershed were sampled on a monthly basis over a 1-year period (April 2008 to May 2009). Three of the five sites were along the Oldman River from west to east (49°44′35.15″N, 113°22′27.60″W; 49°43′5.10″N, 112°51′56.97″W; and 49°51′24.41″N, 112°37′27.10″W) and subjected to a gradation of nutrient inputs. The remaining two river sites were tributaries of the Oldman River: Willow Creek (49°45′15.32″N, 113°24′23.72″W) and the Little Bow River (49°54′5.22″N, 112°30′24.19″W) (Fig. 1). Latitude and longitude coordinates of the sampling sites were determined by using Google Earth (version 6.1.0.5001; Google Inc. [http://earth.google.com/]). Samples were collected at 4-week intervals, from 7 May 2008 to 13 May 2009. At each site, samples of approximately 800 to 900 ml were obtained by using 1-liter Nalgene bottles attached to the end of a sampling pole; water was collected at a depth of ∼20 to 30 cm in the flowing portion of the river. Samples were stored on ice and processed within 6 h of collection. Samples (250 ml) were filtered through a 0.45-μm GF/F prefilter (Whatman, Florham Park, NJ), followed by a 0.2-μm Iso-Grid final filter (Neogen Corp., Lansing, MI) under a vacuum using a six-place filtration manifold (Advantec MFS Inc., Dublin, CA) fitted with an Iso-Grid filtration unit (Neogen Corp.). The two filters per sample were combined in a 50-ml Falcon tube with 20 ml PBS (0.1 M; pH 7.2) and mixed vigorously to detach cells. Filters were subsequently removed, and the suspension was then centrifuged at 14,900 × g for 10 min at 4°C. The top 17 ml of the supernatant was discarded, and pellets were resuspended in the remaining 3 ml of PBS. Two-hundred-microliter aliquots were dispensed into each of two 2-ml tubes, and EMA was added to samples as described above. Following EMA treatment, samples were stored at −20°C until subsequent processing.
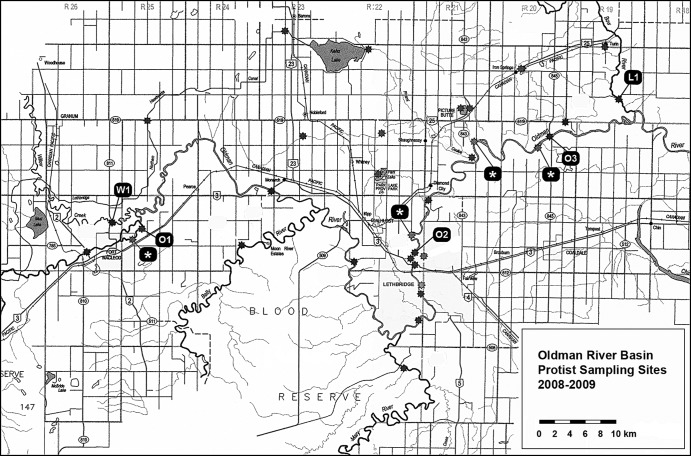
Fig 1.
Map of Southwestern Alberta showing river sampling sites along the Oldman River (O1, O2, and O3), Willow Creek (W1), and Little Bow River (L1). Wastewater outflows for the Town of Fort MacLeod, City of Lethbridge, Town of Picture Butte, and Town of Coaldale are marked with asterisks from left to right. (Source: Alberta Environment and Sustainable Resource Development.)
DNA extraction and quantification.
DNA was extracted from samples by using the QIAamp DNA stool minikit (Qiagen Inc., Mississauga, Ontario, Canada) according to the manufacturer's protocol, with the exception that the entire lysate volume was carried forward following InhibitEX treatment, and subsequent reagent concentrations were adjusted accordingly to compensate for the increased volume. DNA extractions were quantified by using a Nanodrop ND-3300 fluorimeter (Thermo Scientific) with Hoechst double-stranded DNA (dsDNA) labeling (Life Technologies, Carlsbad, CA), using calf thymus dsDNA as a quantification standard.
PCR conditions for T-RFLP community profiling.
PCR was performed by using 5 ng of DNA or 5 μl for samples in which the DNA concentration was less than 1 ng μl−1. The PCR primers used were 6-carboxyfluorescein (FAM)-labeled Euk1A and Euk516R. To repair single-stranded fragments following PCR, which may result in pseudo-T-RFs (24, 25), samples were incubated with Klenow polymerase (New England BioLabs, Ipswich, MA) according to the manufacturer's protocol. After Klenow treatment, samples were digested with HaeIII and HhaI (Invitrogen) according to the manufacturer's specifications prior to ethanol precipitation and separation by capillary electrophoresis on an ABI-3130 genetic analyzer in two independent runs (Applied Biosystems, Foster City, CA).
Analysis of T-RFLP community profiles.
T-RFLP data quality was manually inspected with Genemapper (Applied Biosystems) and was subsequently imported into T-REX (19). T-REX was used to standardize the total peak height across samples (i.e., to compensate for small differences in the amounts of digested DNA resolved per sample) and to filter “noise” according to a method described previously by Abdo et al. (2); 3 standard deviations were used as the threshold for discriminating between noise and true peaks. Profiles were aligned by using a clustering threshold of 0.5 (66). Peaks not present in replicate T-RFLP profiles were removed from subsequent analyses. Nontransposed (T-RFs as columns and samples as rows) and transposed (samples as rows and T-RFs as columns) presence-absence data matrices were exported from T-REX for the generation of a four-way Venn diagram using a custom Excel macro (Microsoft Inc., Redmond, WA) and for use with BioNumerics (Applied Maths, St-Martin-Latem, Belgium), respectively. BioNumerics was used to generate a distance matrix using the Bray-Curtis distance measure, which formed the basis for the group significance test (17), and three-dimensional nonmetric multidimensional scaling (NMS) analysis was conducted by using the MDS procedure in SAS (SAS Institute Inc., Cary, NC). NMS is an ordination technique which arranges individual data points (in this case, T-RFLP profiles) in three-dimensional space based on similarity or dissimilarity; data points which cluster together are more alike than those further apart.
Emulsion PCR and clone library construction.
Sampling periods throughout the year were categorized as spring (March 21 to June 20), summer (June 21 to September 21), fall (September 22 to December 21), and winter (December 22 to March 20) based on the dates for equinoxes (March 20 and September 21) and solstices (June 20 and December 21) according to the U.S. Naval Observatory (http://www.usno.navy.mil/USNO/astronomical-applications/data-services/earth-seasons).
Community DNAs from representative sampling times for the spring (late April), summer (late August), fall (mid-October), and winter (late December), based on the clustering of T-RFLP profiles with NMS, were pooled and amplified with primers Euk1A and Euk516r in an oil-in-water emulsion, according to the EMBL-90 protocol described previously by Williams et al. (72). Emulsion PCR was performed to reduce PCR biases, such as the bias toward 1:1 product ratios due to heteroduplex formation and an underrepresentation of sequences which have mismatches between primer and template sequences (13, 56). Following amplification, the emulsion was broken by two extractions with diethyl ether, followed by one extraction with ethyl acetate and two additional extractions with diethyl ether.
PCR products were purified by using the Qiagen PCR purification kit (Qiagen) and subsequently ligated into the pGEM-T Easy vector (Promega) at a 3:1 vector-to-insert ratio. The chemical transformation of Escherichia coli JM109 cells (Promega) was performed according to the manufacturer's protocol. Blue-white screening was used to differentiate colonies with inserts from those without; white colonies were picked into 96-well plates containing LB medium supplemented with 4% glycerol and 100 μg ml−1 ampicillin by using a Qpix robot (Genetix, San Jose, CA) prior to screening. Approximately 500 clones from each library were prescreened by HaeIII restriction digestion of M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) PCR-amplified products. Digested fragments were sized by using the QIAxcel capillary system. Clones matching the restriction profile of prevalent solanaceous plants (∼80 to 90% of the clones) were excluded from further analyses.
Clone library sequencing and analysis.
Sequencing was performed by a single pass with primer M13F on an ABI-3130 genetic analyzer (Applied Biosystems). Sequences were imported into Geneious v5.1 (Biomatters Ltd., Auckland, New Zealand), trimmed manually, and screened for chimeras by using Bellerophon (34). Putative chimeras were then examined on an individual basis with Pintail (7), with S. cerevisiae (GenBank accession number AY251630) used as the reference. Sequences were aligned by the SINA alignment service using the SILVA SSU, release 106, database (57), imported into Geneious, and manually curated. Alignments were then exported, and operational taxonomic units (OTUs) were determined by using the Mothur software package (64), with a 1% sequence divergence cutoff (9, 70). Sequences were classified according to the new higher-level classification of eukaryotes (5).
The degree to which microbial communities differed based on 18S rRNA gene sequence was assessed by using the Unifrac P-test (33). All 198 protistan 18S rRNA gene sequences from river water, along with Giardia lamblia (ATCC 50803) as the outgroup, were aligned within the SILVA database and imported into Mothur to remove common gaps prior to tree building in Geneious. A rooted phylogenetic tree was constructed from the trimmed alignment by using the neighbor-joining method and the Tamura-Nei distance metric. The Newick-formatted tree was then analyzed by using Unifrac (33). Chao1 and ACE species richness estimates were performed by using Mothur (64).
Nucleotide sequence accession numbers.
The 18S rRNA gene sequence data were deposited in the GenBank database under accession numbers JX068881 to JX069077.
RESULTS
Ethidium monoazide validation.
A substantial reduction in PCR amplicon band intensity (ranging from a weak product to no detectable product) was observed for samples heated at 100°C for 20 min and exposed to EMA for all protists tested (Fig. 2). Unheated samples treated with EMA yielded PCR products slightly less intense than the no-EMA samples, indicating a minimal penetration of live cells.
Fig 2.

QIAxcel capillary electrophoresis of 18S rRNA gene amplicons from Acanthamoeba polyphaga (lanes 1 to 4), Tetrahymena pyriformis (lanes 5 to 8), Chlamydomonas moewusii (lanes 9 to 12), and Euglena gracilis (lanes 13 to 16). EMA-treated samples are presented in odd-numbered lanes, and non-EMA-treated samples are presented in even-numbered lanes. The first two lanes for each protist are cells that were not heat treated, whereas the second two lanes per protist are cells that were heat treated. The markers on the left side of the image correspond to fragment sizes (base pairs).
T-RFLP community profiles.
T-RFLP profiles targeting the 18S rRNA gene were determined for 70 river water samples from the Oldman watershed of Southwestern Alberta. Considerable variability in the number of T-RFs was observed among samples, and the numbers of T-RFs ranged from 17 to 112 (Fig. 3). The total number of unique T-RFs detected throughout the study was 331. The highest numbers of T-RFs for any given season were detected during the spring (253 T-RFs) and winter (236 T-RFs) (Fig. 4). Protistan diversity was reduced in the summer and fall, and 127 and 107 T-RFs were observed, respectively. The T-RFLP fingerprints had high heterogeneity, with a beta diversity value of 6.59, as determined with T-REX. NMS analysis of T-RFLP profiles revealed a pattern of seasonal relatedness independent of sample location; the three-dimensional plot exhibited a stress value of 14 (Fig. 5). The group significance test supported the clustering of samples into distinct groups by season (P ≤ 0.05); however, there was greater variation in the spring-summer, spring-winter, and fall-summer comparisons (Table 1). The numbers of T-RFs exclusive to the spring, summer, fall, and winter sampling periods were 59, 8, 6, and 46, respectively (Fig. 4). A total of 59 T-RFs were common to all four seasons.
Fig 3.
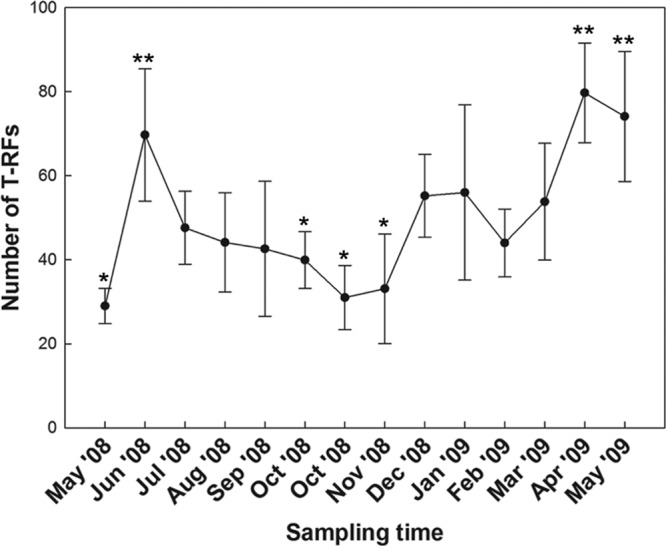
Number of terminal restriction fragments over a 1-year sampling period (May 2008 to May 2009). Vertical bars associated with means represent the standard deviations across the five sample sites. Means denoted with single asterisks differ (P ≤ 0.05) from means indicated by double asterisks.
Fig 4.
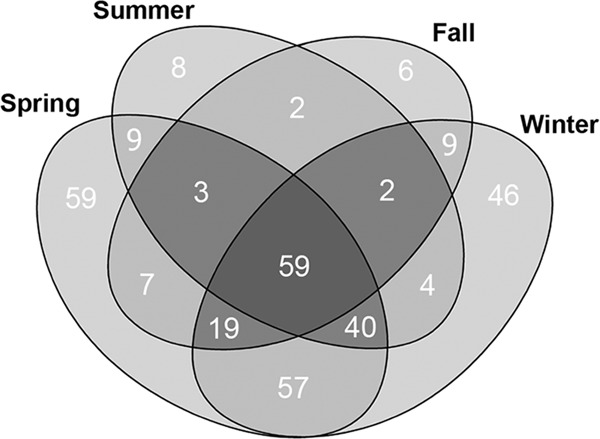
Four-way Venn diagram of unique terminal restriction fragments detected in river water by season.
Fig 5.
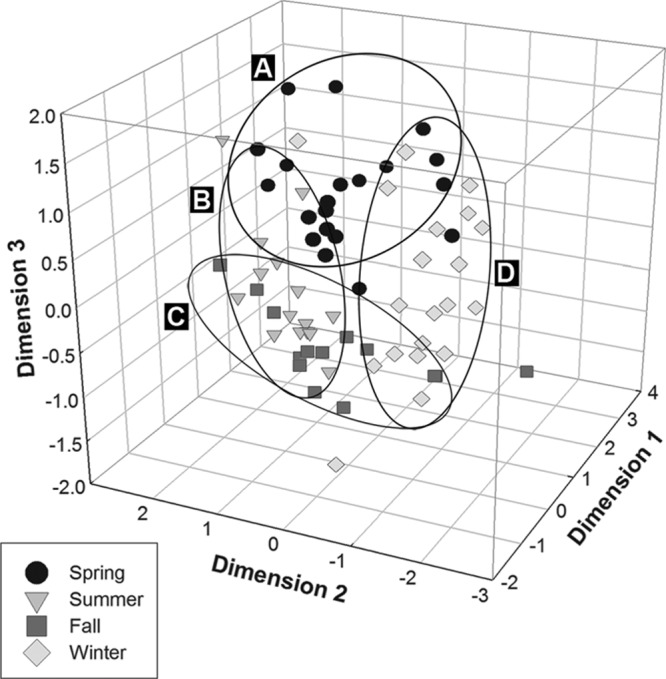
Nonmetric multidimensional scaling plots of T-RFLP community profiles from river water (stress value of 14), with ellipses A to D encompassing 90% of the respective markers by season. Markers that appear close together are more similar than distant markers.
Table 1.
Pairwise significance matrix of protist T-RFLP community profiles of seasonally partitioned data
| Season |
P valuea |
|||
|---|---|---|---|---|
| Spring | Summer | Fall | Winter | |
| Spring | 0.391 | <0.001* | 0.131 | |
| Summer | <0.001* | <0.001* | <0.001* | |
| Fall | 0.017* | 0.125 | <0.001* | |
| Winter | <0.001* | <0.001* | <0.001* | |
Values indicated with an asterisk are significantly different (P ≤ 0.05).
Clone libraries.
Eighty-five OTUs from 198 18S rRNA gene sequences were observed for river water. The spring library was dominated by centrohelid, stramenopile, and Chloroplastida clones (∼29, 20, and 20%, respectively) (Fig. 6). Summer and fall libraries were dominated by Saccharomycetes, primarily Candida spp. (Table 2), which accounted for 83% and 88% of the clones, respectively. Alveolata (33%), Euglenozoa (23%), Chloroplastida (14%), and fungal (14%) clones were most common in the winter library. ACE predicted 94, 29, 131, and 79 OTUs for the spring, summer, fall, and winter libraries, respectively, while Chao1 predicted 87, 15, 97, and 37, respectively (Table 2). The Unifrac P-test confirmed differences between seasonal clone libraries (P < 0.001).
Fig 6.
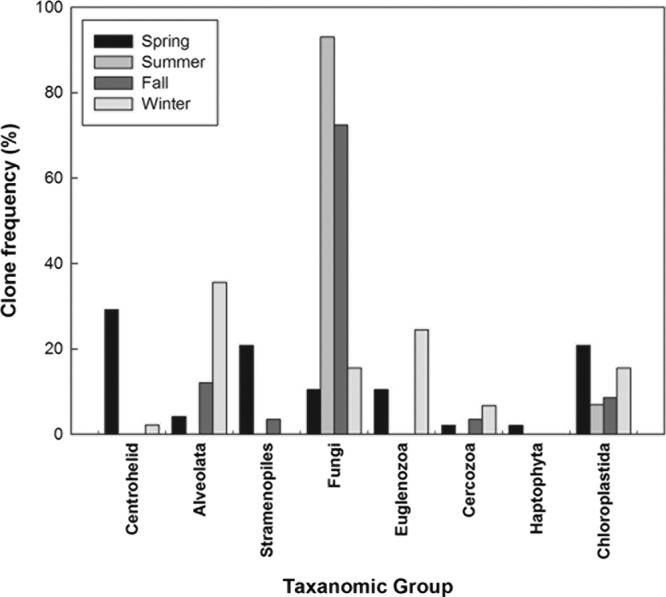
Distribution of 18S rRNA gene sequences (percent) at the first-rank taxonomic level (level 5) by season.
Table 2.
Summary of 18S rRNA gene libraries, including ACE and Chao1 nonparametric richness estimates
| Library | No. of clones | No. of OTUs | No. of predicted OTUs |
|||||
|---|---|---|---|---|---|---|---|---|
| ACE |
Chao1 |
|||||||
| Mean | Lower | Upper | Mean | Lower | Upper | |||
| Spring | 61 | 29 | 94 | 51 | 217 | 87 | 48 | 208 |
| Summer | 46 | 8 | 29 | 10 | 274 | 15 | 9 | 50 |
| Fall | 58 | 32 | 131 | 65 | 332 | 97 | 55 | 219 |
| Winter | 47 | 21 | 79 | 52 | 130 | 37 | 25 | 79 |
DISCUSSION
The temporal dynamics of the protistan community structure associated with freshwater systems, particularly rivers, have yet to benefit from the explosion of molecular-based investigations into microbial ecology and biogeography. Such studies have focused mostly on Bacteria and Archaea, using 16S rRNA gene sequences as a target and, to a lesser extent, oceanic picoeukaryotes (i.e., protists ≤0.2 μm in size) targeting 18S rRNA gene sequences. Using T-RFLP and clone library analyses, we observed conspicuous temporal shifts in protistan community structures in Southwestern Alberta rivers.
NMS analysis of T-RFLP profiles revealed a high level of similarity between sampling sites over time, as samples clustered according to time (season) rather than location. This occurred despite the fact that sampling sites were located in areas subject to differing natural (i.e., organic matter or an embankment) and anthropogenic (i.e., upstream/downstream from wastewater treatment plant effluent, proximity to highways, agricultural runoff, and urban versus rural sites) factors. Anthropogenic factors, in particular, have been shown to have pronounced effects on microbial planktonic communities in freshwater systems (29). The stress value of the ordination was 14, which indicates that the ordination is satisfactory and is a good representation of the T-RFLP data (46).
There are currently two predominant models of protistan biogeography: the moderate endemicity model (26, 27) and the ubiquity model (69). The ubiquity model proposes that protists are cosmopolites and can be found anywhere in which their niche requirements are met, whereas the moderate endemicity model postulates that while the majority of protists are cosmopolites, a significant amount (roughly one-third) exhibit a limited biogeographical range (27). Despite the high variance in T-RF richness between sampling sites at any given time, the collective seasonality of T-RFLP community profiles suggests that a stable core group of protists exists in river water in Southwestern Alberta. As such, our results for protists in river water are consistent with the moderate endemicity model. Similarly, Nolte et al. (48) previously observed seasonality in the distribution of protists in Lake Fuschlsee (Salzhammergut, Austria); they observed that changes in community structure were not merely quantitative but also qualitative, as taxa observed in 1 month would drop below the detection threshold the following month.
T-RFLP targeting the 18S rRNA gene proved to be instrumental in providing a snapshot of protist communities and assisted in the selection of sites for Sanger sequencing in the current study. Due to the costs associated with the sequencing of clone libraries, it was not logistically possible to process all 70 libraries in this manner. In previous studies using T-RFLP to characterize bacterial communities (i.e., targeting the 16S rRNA gene), T-RFs were identified via in silico restriction digests of sequences from known databases (4). However, recent evidence suggests that this is not a robust analysis method due to the differential migration of T-RFs with variable pyrimidine contents and inaccurate sizing resulting from the differential migration of the LIZ-labeled size standard and FAM-labeled PCR products (38). Furthermore, multiple taxa may occupy a single T-RF. As these biases are consistent between samples and replicate runs, T-RFLP remains a valid method for community profiling. Given that the identification of T-RFs based on in silico analyses of sequence databases is not reliable, we selected sites for Sanger sequencing based on the similarities and differences in T-RFLP profiles rather than specific protistan assemblages inferred from T-RFs.
The study of protists in freshwater systems, especially rivers, poses the added challenge that terrestrial and aquatic plant matter and microeukaryotes are present in high numbers, and there are no PCR primers capable of excluding them while retaining the ability to amplify a broad array of protists. We observed that all 18S rRNA gene libraries generated in the current study were dominated by clones later identified as a solanaceous plant, perhaps that of a common agricultural weed in the study area (10). As plants have been shown to have between 500 and 40,000 copies of the 18S rRNA gene per diploid cell (73), a single contaminating cell could severely hinder the detection of protists. Our regimen for sample processing may have benefitted from a prefiltration step; however, it is uncertain if this would have impacted the recovery of large or attached protists.
In our study, we used EMA to prevent the PCR amplification of nonviable protists. EMA has been used extensively in studies involving prokaryotes (60); however, to our knowledge, this is the first study regarding the ecology of protists to use EMA for the differentiation of live from dead cells. Previous uses of EMA involving eukaryotic cells included distinguishing viable from dead cells using flow cytometry (50) and removing contaminating eukaryotic DNA from yeast extracts (58). EMA has been documented to penetrate some bacterial species with intact cell membranes (52), which could result in an underestimation of diversity; however, prior to this study, it was unclear if this occurs with protists. Our results indicate that although some penetration of live cells did occur, EMA is suitable for reducing DNA from dead cells in the study of protistan diversity. In some ecosystems, such as human fecal samples, a large proportion of microbial cells are not viable, and community analyses in the absence of EMA may result in a dramatic overestimation of community richness and even inaccurate conclusions, for example, concluding no change in the community composition of all organisms when there may me a marked shift in viable organisms (51).
There was a significant disparity between the T-RFLP and clone library analyses in measured community richness. For example, 331 unique T-RFs were observed, compared to only 89 OTUs. Furthermore, richness estimates based on Chao1 and ACE suggest that additional taxa are present. This could be due to the use of Sanger sequencing, which often fails to fully illuminate the rare biosphere due to throughput constraints. In contrast, alternatives such as second-generation sequencing platforms (i.e., 454 pyrosequencing) are prone to sequencing errors, which result in an overestimation of the rare biosphere (28). Sanger libraries offer unmatched accuracy, and at the time of this study, Sanger read lengths more than doubled those of 454 pyrosequencing (45); the limited pyrosequencing read lengths restrict the taxonomic assignment of protists to the second rank (6). To compensate for the relatively small number of clones that we could logistically sequence, we prescreened clones based on HaeIII restriction profiles to avoid the sequencing of clones belonging to solanaceous plants. This allowed us to screen nearly 2,000 clones. A similar method was utilized previously by Diez et al. (21) to study the genetic diversity of protists in oceanic water, except that the restriction digest profiles were used to group clones to maximize OTU discovery via Sanger sequencing libraries.
Heterophrys spp., the dominant centrohelids detected in the spring (14 clones) and winter (1 clone) libraries, are a group of phagotrophic protists (16) and thus may affect the persistence of enteric bacteria in rivers. The spring is a particularly interesting time for studies regarding the fate of enteric bacteria due to the large amount of agricultural runoff (i.e., animal fecal matter), which has been associated with increases in levels of coliform bacteria (30) and spikes in enteric disease (53). Phagotrophic protists such as Acanthamoeba spp., Hartmanella spp., Naegleria spp., and Tetrahymena spp. have been linked with the environmental persistence of a wide array of pathogenic bacteria (12). Legionella pneumophila, for example, escapes the phagosome into the cytoplasm and is able to replicate within the protozoan host (47). While the protists mentioned above were not detected in the present study, this may be due to the common association of amoeboid protists with biofilms (35). Nonetheless, studies targeting Heterophrys sp. in relation to the persistence of enteric pathogens in river water are warranted.
We observed that centrohelids and Chloroplastidae dominated river water during the spring, representing 28.1% and 29.8% of clones, respectively. Dramatic declines in both were noted for the summer and fall libraries, which were dominated by fungi, specifically Candida spp., a genus containing the ubiquitous human pathogens Candida albicans and C. tropicalis (11). Kopylov and Kosolapov (42) also reported a reduction in numbers of heterotrophic protists in early summer in the Ob river of West Siberia, which corresponded with increased levels of heterotrophic bacteria. Photosynthetic protists (i.e., Chloroplastidae) face increased competition from cyanobacteria, which bloom during the increased temperatures that accompany the summer months as they enter their temperature optima (37). Cyanobacteria are also smaller, thus providing advantages over the larger Chloroplastidae, such as a lower sinking rate (71) and more efficient nutrient acquisition due to the lower diffusion boundary layer, which is a result of having a higher surface-area-to-volume ratio (55). Salmanian et al. (62) suggested previously that Candida spp. play a role in the environmental persistence of Helicobacter pylori, which was once considered part of the genus Campylobacter and is associated with gastric ulcers in humans (54). Those researchers found bacterium-like bodies (BLBs) present in vacuoles of Candida spp. isolated from foods that were confirmed to be viable through live/dead staining. Furthermore, Candida spp. containing BLBs were PCR positive for the H. pylori ureAB genes in 9 out of 15 cases (32).
In conclusion, we observed through the use of T-RFLP and clone library analyses that protists were common in river water in Southwestern Alberta, and seasonal succession trends were consistent with the moderate endemicity model. The majority of T-RFs were found in the spring and winter, which were dominated by Alveolata, Centrohelida, and Chloroplastidae (spring) and Alveolata, Euglenozoa, Chloroplastida, and fungi (winter). Summer and fall were dominated by saccharomycetous fungi. Our study also illustrates some of the complications that researchers face in studying protists using the 18S rRNA gene as a target.
ACKNOWLEDGMENTS
We thank Ashley Moore and Anthony Russell, University of Lethbridge, for providing the strain of E. gracilis.
This work was supported in part by a Peer Review Sustainable Agriculture Environmental Systems (SAGES) grant from Agriculture and Agri-Food Canada to G.D.I. and by a graduate student assistantship from the University of Lethbridge to M.C.T.
Footnotes
Published ahead of print 8 June 2012
REFERENCES
- 1. Abd H, Saeed A, Weintraub A, Sandstrom G. 2009. Vibrio cholerae O139 requires neither capsule nor LPS O side chain to grow inside Acanthamoeba castellanii. J. Med. Microbiol. 58:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdo Z, et al. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929–938 [DOI] [PubMed] [Google Scholar]
- 3. Abu Kwaik Y, Gao L-Y, Stone BJ, Venkataraman C, Harb OS. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adiba S, Nizak C, van Baalen M, Denamur E, Depaulis F. 2010. From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882 doi:10.1371/journal.pone.0011882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adl SM, et al. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399–451 [DOI] [PubMed] [Google Scholar]
- 6. Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372 doi:10.1371/journal.pone.0006372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Axelsson-Olsson D, Waldenstrom J, Broman T, Olsen B, Holmberg M. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl. Environ. Microbiol. 71:987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bailly J, et al. 2007. Soil eukaryotic functional diversity, a metatranscriptomic approach. ISME J. 1:632–642 [DOI] [PubMed] [Google Scholar]
- 10. Blackshaw RE. 1991. Hairy nightshade (Solanum sarrachoides) interference in dry beans (Phaseolus vulgaris). Weed Sci. 39:48–53 [Google Scholar]
- 11. Brinkman NE, et al. 2003. Evaluation of a rapid, quantitative real-time PCR method for enumeration of pathogenic Candida cells in water. Appl. Environ. Microbiol. 69:1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown MRW, Barker J. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7:46–50 [DOI] [PubMed] [Google Scholar]
- 13. Bru D, Martin-Laurent F, Philippot L. 2008. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl. Environ. Microbiol. 74:1660–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carrias JF, Amblard C, Bourdier G. 1996. Protistan bacterivory in an oligomesotrophic lake: importance of attached ciliates and flagellates. Microb. Ecol. 31:249–268 [DOI] [PubMed] [Google Scholar]
- 15. Casamayor EO, et al. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338–348 [DOI] [PubMed] [Google Scholar]
- 16. Cavalier-Smith T, von der Heyden S. 2007. Molecular phylogeny, scale evolution and taxonomy of centrohelid heliozoa. Mol. Phylogenet. Evol. 44:1186–1203 [DOI] [PubMed] [Google Scholar]
- 17. Costa E, Puhl NJ, Selinger LB, Inglis GD. 2009. Characterization of mucosa-associated bacterial communities of the mouse intestine by terminal restriction fragment length polymorphism: utility of sampling strategies and methods to reduce single-stranded DNA artifacts. J. Microbiol. Methods 78:175–180 [DOI] [PubMed] [Google Scholar]
- 18. Creer S. 2010. Second-generation sequencing derived insights into the temporal biodiversity dynamics of freshwater protists. Mol. Ecol. 19:2829–2831 [DOI] [PubMed] [Google Scholar]
- 19. Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH. 2009. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10:171 doi:10.1186/1471-2105-10-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diez B, Pedros-Alio C, Marsh TL, Massana R. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diez B, Pedros-Alio C, Massana R. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edgcomb V, et al. 2011. Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness. ISME J. 5:1344–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgcomb VP, Kysela DT, Teske A, de Vera Gomez A, Sogin ML. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. U. S. A. 99:7658–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Egert M, Friedrich MW. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Egert M, Friedrich MW. 2005. Post-amplification Klenow fragment treatment alleviates PCR bias caused by partially single-stranded amplicons. J. Microbiol. Methods 61:69–75 [DOI] [PubMed] [Google Scholar]
- 26. Foissner W. 1999. Protist diversity: estimates of the near-imponderable. Protist 150:363–368 [DOI] [PubMed] [Google Scholar]
- 27. Foissner W. 2008. Protist diversity and distribution: some basic considerations. Biodivers. Conserv. 17:235–242 [Google Scholar]
- 28. Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. 2012. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J. 6:554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frost JA. 2001. Current epidemiological issues in human campylobacteriosis. J. Appl. Microbiol. 90:85S–95S doi:10.1046/j.1365-2672.2001.01357.x [DOI] [PubMed] [Google Scholar]
- 30. Gannon JJ, Busse MK. 1989. E. coli and enterococci levels in urban stormwater, river water and chlorinated treatment plant effluent. Water Res. 23:1167–1176 [Google Scholar]
- 31. Gardner TJ, et al. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin. Infect. Dis. 53:26–32 [DOI] [PubMed] [Google Scholar]
- 32. González JM, Iriberri J, Egea L, Barcina I. 1990. Differential rates of digestion of bacteria by freshwater and marine phagotrophic protozoa. Appl. Environ. Microbiol. 56:1851–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 35. Huws SA, McBain AJ, Gilbert P. 2005. Protozoan grazing and its impact upon population dynamics in biofilm communities. J. Appl. Microbiol. 98:238–244 [DOI] [PubMed] [Google Scholar]
- 36. Inglis GD, Boras VF, Houde A. 2011. Enteric campylobacteria and RNA viruses associated with healthy and diarrheic humans in the Chinook Health Region of Southwestern Alberta, Canada. J. Clin. Microbiol. 49:209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jöhnk KD, et al. 2008. Summer heatwaves promote blooms of harmful cyanobacteria. Glob. Change Biol. 14:495–512 [Google Scholar]
- 38. Kaplan CW, Kitts CL. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54:121–125 [DOI] [PubMed] [Google Scholar]
- 39. Kim E, Wilcox L, Graham L, Graham J. 2004. Genetically distinct populations of the dinoflagellate Peridinium limbatum in neighboring northern Wisconsin lakes. Microb. Ecol. 48:521–527 [DOI] [PubMed] [Google Scholar]
- 40. King CH, Shotts EB, Jr, Wooley RE, Porter KG. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kiss ÁK, Ács E, Kiss KT, Török JK. 2009. Structure and seasonal dynamics of the protozoan community (heterotrophic flagellates, ciliates, amoeboid protozoa) in the plankton of a large river (River Danube, Hungary). Eur. J. Protistol. 45:121–138 [DOI] [PubMed] [Google Scholar]
- 42. Kopylov A, Kosolapov D. 2011. The structure of the planktic microbial community in the lower reaches of the Ob River near Salekhard. Contemp. Probl. Ecol. 4:1–7 [Google Scholar]
- 43. Li L-Y, et al. 2011. Diversity and distribution of planktonic protists in the northern South China Sea. J. Plankton Res. 33:445–456 [Google Scholar]
- 44. Logares R, et al. 2007. Phenotypically different microalgal morphospecies with identical ribosomal DNA: a case of rapid adaptive evolution? Microb. Ecol. 53:549–561 [DOI] [PubMed] [Google Scholar]
- 45. Mardis ER. 2008. The impact of next-generation sequencing technology on genetics. Trends Genet. 24:133–141 [DOI] [PubMed] [Google Scholar]
- 46. McCune B, Grace JB. 2003. Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR [Google Scholar]
- 47. Molmeret M, Bitar DM, Han L, Kwaik YA. 2004. Disruption of the phagosomal membrane and egress of Legionella pneumophila into the cytoplasm during the last stages of intracellular infection of macrophages and Acanthamoeba polyphaga. Infect. Immun. 72:4040–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nolte V, et al. 2010. Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol. Ecol. 19:2908–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Not F, del Campo J, Balagué V, de Vargas C, Massana R. 2009. New insights into the diversity of marine picoeukaryotes. PLoS One 4:e7143 doi:10.1371/journal.pone.0007143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Brien MC, Bolton WE. 1995. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytometry 19:243–255 [DOI] [PubMed] [Google Scholar]
- 51. Olsen SJ, et al. 2001. An outbreak of Campylobacter jejuni infections associated with food handler contamination: the use of pulsed-field gel electrophoresis. J. Infect. Dis. 183:164–167 [DOI] [PubMed] [Google Scholar]
- 52. Pearson AD, et al. 2000. Continuous source outbreak of campylobacteriosis traced to chicken. J. Food Prot. 63:309–314 [DOI] [PubMed] [Google Scholar]
- 53. Pebody RG, Ryan MJ, Wall PG. 1997. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7:R33–R37 [PubMed] [Google Scholar]
- 54. Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537–546 [DOI] [PubMed] [Google Scholar]
- 55. Ploug H, Stolte W, Epping EHG, Jorgensen BB. 1999. Diffusive boundary layers, photosynthesis, and respiration of the colony-forming plankton algae, Phaeocystis sp. Limnol. Oceanogr. 44:1949–1958 [Google Scholar]
- 56. Polz MF, Cavanaugh CM. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rawsthorne H, Phister TG. 2009. The presence of Saccharomyces cerevisiae DNA in various media used to propagate yeasts and its removal by ethidium monoazide. Lett. Appl. Microbiol. 49:652–654 [DOI] [PubMed] [Google Scholar]
- 59. Ribblett SG, Palmer MA, Coats DW. 2005. The importance of bacterivorous protists in the decomposition of stream leaf litter. Freshw. Biol. 50:516–526 [Google Scholar]
- 60. Rudi K, Moen B, Dromtorp SM, Holck AL. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Russell AG, Schnare MN, Gray MW. 2004. Pseudouridine-guide RNAs and other Cbf5p-associated RNAs in Euglena gracilis. RNA 10:1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Salmanian A-H, et al. 2012. Foodborne yeasts serve as reservoirs of Helicobacter pylori. J. Food Saf. 32:152–160 [Google Scholar]
- 63. Sandström G, Saeed A, Abd H. 2010. Acanthamoeba polyphaga is a possible host for Vibrio cholerae in aquatic environments. Exp. Parasitol. 126:65–68 [DOI] [PubMed] [Google Scholar]
- 64. Schloss PD, et al. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shi X, Liu X, Liu G, Sun Z, Xu H. 2012. An approach to analyzing spatial patterns of protozoan communities for assessing water quality in the Hangzhou section of Jing-Hang Grand Canal in China. Environ. Sci. Pollut. Res. Int. 19:739–747 [DOI] [PubMed] [Google Scholar]
- 66. Smith CJ, et al. 2005. T-Align, a Web-based tool for comparison of multiple terminal restriction fragment length polymorphism profiles. FEMS Microbiol. Ecol. 54:375–380 [DOI] [PubMed] [Google Scholar]
- 67. Snelling W, et al. 2008. Colonization of broilers by Campylobacter jejuni internalized within Acanthamoeba castellanii. Arch. Microbiol. 189:175–179 [DOI] [PubMed] [Google Scholar]
- 68. Snelling WJ, McKenna JP, Lecky DM, Dooley JS. 2005. Survival of Campylobacter jejuni in waterborne protozoa. Appl. Environ. Microbiol. 71:5560–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tauxe RV, Hargrett-Bean N, Patton CM, Wachsmuth IK. 1988. Campylobacter isolates in the United States, 1982-1986. MMWR CDC Surveill. Summ. 37:1–13 [PubMed] [Google Scholar]
- 70. Valster RM, Wullings BA, Bakker G, Smidt H, van der Kooij D. 2009. Free-living protozoa in two unchlorinated drinking water supplies, identified by phylogenic analysis of 18S rRNA gene sequences. Appl. Environ. Microbiol. 75:4736–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Walsby AE, Holland DP. 2006. Sinking velocities of phytoplankton measured on a stable density gradient by laser scanning. J. R. Soc. Interface 3:429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Williams R, et al. 2006. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 3:545–550 [DOI] [PubMed] [Google Scholar]
- 73. Zia S, et al. 2003. Health problems following Campylobacter jejuni enteritis in a Lancashire population. Rheumatology 42:1083–1088 [DOI] [PubMed] [Google Scholar]