Abstract
Bacillus subtilis synthesizes large amounts of the compatible solute proline as a cellular defense against high osmolarity to ensure a physiologically appropriate level of hydration of the cytoplasm and turgor. It also imports proline for this purpose via the osmotically inducible OpuE transport system. Unexpectedly, an opuE mutant was at a strong growth disadvantage in high-salinity minimal media lacking proline. Appreciable amounts of proline were detected in the culture supernatant of the opuE mutant strain, and they rose concomitantly with increases in the external salinity. We found that the intracellular proline pool of severely salinity-stressed cells of the opuE mutant was considerably lower than that of its opuE+ parent strain. This loss of proline into the medium and the resulting decrease in the intracellular proline content provide a rational explanation for the observed salt-sensitive growth phenotype of cells lacking OpuE. None of the known MscL- and MscS-type mechanosensitive channels of B. subtilis participated in the release of proline under permanently imposed high-salinity growth conditions. The data reported here show that the OpuE transporter not only possesses the previously reported role for the scavenging of exogenously provided proline as an osmoprotectant but also functions as a physiologically highly important recapturing device for proline that is synthesized de novo and subsequently released by salt-stressed B. subtilis cells. The wider implications of our findings for the retention of compatible solutes by osmotically challenged microorganisms and the roles of uptake systems for compatible solutes are considered.
INTRODUCTION
In its soil ecosystem, Bacillus subtilis is subjected to myriads of changing environmental conditions and restrictions in the supply of nutrients. These challenges require dynamic adjustments in gene transcription and metabolic networks to ensure survival, growth, and persistence of B. subtilis in this taxing habitat (12, 41). A key parameter affecting the integrity and well-being of microbial cells is the osmotic condition of the surroundings (8, 9, 64). Changes in this parameter trigger profound alterations in the transcriptional profile of B. subtilis cells in response to both sudden and sustained increases in salinity (21, 41, 49). Fluctuations in the osmotic conditions of the upper layers of the soil result from rainfall and desiccation, and they inevitably trigger water fluxes in or out of the cell (9, 64). B. subtilis counteracts these osmotically instigated water passages through its semipermeable cytoplasmic membrane by dynamically increasing or decreasing the osmotic potential of the cytoplasm (8).
Upon exposure to hypo-osmotic conditions, a reduction in water entry is achieved through the rapid and nonselective expulsion of water-attracting ions and organic solutes via the transient opening of mechanosensitive channels (6, 23). The gating of these safety valves (Fig. 1) prevents an undue rise in turgor (62) that might otherwise lead to bursting of the cell (24, 58). Conversely, under hyperosmotic conditions, the B. subtilis cell avoids dehydration of the cytoplasm and a reduction in turgor to physiologically unsustainable values through the accumulation of a selected group of water-attracting ions (primarily potassium) (26, 62) and organic compounds known as compatible solutes (8, 31). Proline is the only compatible solute that B. subtilis can synthesize de novo (35, 61), and its osmotically regulated production (Fig. 1) is critical for an effective cellular adjustment to sustained high-osmolarity surroundings (10).
Fig 1.
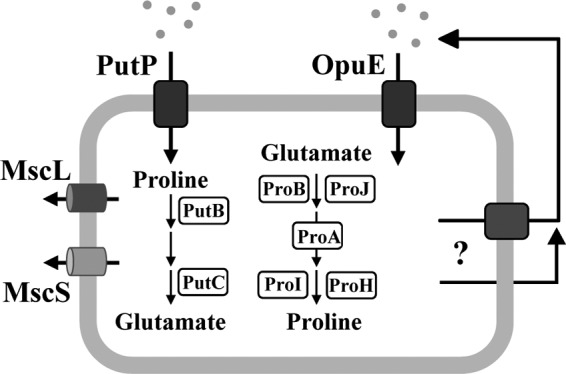
Synthesis, uptake, release, and consumption of proline in B. subtilis. Synthesis of proline for anabolic purposes is carried out by the ProB, ProA, and ProI enzymes; their structural genes (proBA, proI) are induced by proline starvation via a proline-sensing T-box regulatory mechanism (11). The ProJ, ProA, and ProH enzymes carry out synthesis of proline as an osmoprotectant; expression of the proHJ operon is induced by increases in the external salinity but that of the proA gene is not induced in response to this stimulus (10). Uptake of proline as an osmoprotectant is mediated by the OpuE transporter (56), whose structural gene is induced in cells grown at high salinity (47). Use of proline as a carbon or nitrogen source is catalyzed via import through the OpuE-related PutP transporter and subsequent degradation to glutamate via the PutB and PutC enzymes; expression of the putBCP operon is induced by the presence of proline in the growth medium (39). The excretion of proline is depicted either as a carrier/transporter-mediated process or through passive passage across the cytoplasmic membrane. MscL and MscS are mechanosensitive channels whose transient gating protects osmotically down-shocked cells from lysis (24, 58).
B. subtilis also makes extensive use of preformed compatible solutes (e.g., glycine betaine) to achieve enhanced osmotic stress tolerance. It scavenges these compounds from environmental sources through a set of five osmotically inducible uptake systems, the Opu family of transporters (8). One of the compatible solutes acquired by B. subtilis from exogenous sources (25) is proline, and the OpuE transporter is specifically used for its import as an osmoprotectant (Fig. 1) (56). Possible sources of proline in the soil ecosystem are root exudates (54) and decaying or osmotically down-shocked prokaryotic and eukaryotic cells (59). The role of OpuE as a proline uptake system involved in conferring osmotic stress tolerance is also reflected by the pattern of opuE expression, which is strongly induced in response to both an osmotic up-shock and to sustained high osmolarity (56). Transcription of the opuE gene is driven from osmotically controlled SigA- and SigB-type promoters (47), and the signal input and transcriptional activity of the latter type of promoter connect the functioning of OpuE to the SigB-controlled general stress regulon of B. subtilis (40).
The OpuE transporter, a member of the solute symporter family (29), has been considered to serve exclusively in the acquisition of exogenously provided proline from scarce environmental sources (8, 56). The following features make OpuE well suited for this task: (i) the osmotic induction of opuE expression, (ii) the high affinity of OpuE for its substrate proline (Km, about 20 μM) and its substantial transport capacity (Vmax, about 250 nmol min−1 mg protein−1) under osmotic stress conditions, and (iii) the resistance of OpuE transport activity to the inhibitory effects of high salinity, an environmental condition that impairs the working of the OpuE-related PutP transporter used by B. subtilis for the uptake of proline as a nutrient (Fig. 1) (39, 47, 56).
Here we focus on a new physiological role of OpuE in salinity-challenged cells cultivated in the absence of proline. We discovered that B. subtilis cells grown under these conditions engage in a cycle of de novo proline synthesis, release through an unknown mechanism, and subsequent recovery via the OpuE transporter (Fig. 1). Our finding that an opuE mutant cannot cope efficiently with high-osmolarity growth conditions underscores the role of OpuE as a physiologically important transporter system for the recapturing of proline synthesized as an osmoprotectant by the B. subtilis cell.
MATERIALS AND METHODS
Chemicals.
The antibiotics ampicillin, tetracycline, kanamycin and spectinomycin, the ninhydrin reagent, the chromogenic substrate 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (X-Gal) for the detection of β-galactosidase activity in Escherichia coli cells, isopropyl-beta-d-thiogalactopyranoside (IPTG), and para-nitrophenyl-α-d-glucopyranoside (α-PNPG), the chromogenic substrate for the TreA enzyme [phospho-α-(1,1)-glucosidase], were purchased from Sigma-Aldrich (Steinheim, Germany). Radiolabeled l-[U-14C]proline (40 mCi mmol−1) was purchased from DuPont NEN Research Products (Bad Homburg, Germany).
Growth media and cultivation conditions.
E. coli and B. subtilis strains were routinely maintained on Luria-Bertani agar plates. The B. subtilis strains were cultivated in Spizizen's minimal medium (SMM) with 0.5% (wt/vol) glucose as the carbon source and l-tryptophan (20 mg liter−1) and l-phenylalanine (18 mg liter−1) to satisfy the auxotrophic growth requirements of strain JH642 (trpC2 pheA1) and its derivatives (Table 1). A solution of trace elements was added to SMM (22). All B. subtilis cultures were inoculated from exponentially growing precultures in prewarmed minimal media to optical densities at 578 nm (OD578) of 0.1, and the cultures were subsequently propagated at 37°C in a shaking water bath set to 220 rpm. The B. subtilis cells were grown in 20-ml and 75-ml culture volumes in 100-ml and 500-ml Erlenmeyer flasks, respectively. The antibiotics tetracycline (10 μg ml−1), spectinomycin (100 μg ml−1), and kanamycin (5 μg ml−1) were used for the selection of gene disruption mutations in B. subtilis after DNA transformation with chromosomal DNA of previously constructed B. subtilis mutant strains (Table 1) and derivatives of the B. subtilis strain BLOB9 (Table 1) carrying the nonreplicative plasmid pBLOB15.2 (opuE+ sapB+) as a chromosomal insertion were selected on agar plates containing spectinomycin (Table 1).
Table 1.
B. subtilis strains used in this study
| Straina | Relevant genotype | Source or reference |
|---|---|---|
| JH642 | trpC2 pheA1 | BGSC 1A96b |
| BLOB9 | Δ(opuE::tet)1 | 56 |
| BLOB26c | Δ(opuE::tet)1 × (pBLOB15.2 opuE+ sapB+)1 | This study |
| SMB11 | Δ(putP::spc)1 | 39 |
| SMB12 | Δ(putP::spc)1 Δ(opuE::tet)1 | 39 |
| SMB80 | Δ(mscL::spc) Δ(ykuT::cat) Δ(yhdY::ery) Δ(yfkC::tet) | 24 |
| TMB99 | Δ(mscL::spc) Δ(ykuT::cat) Δ(yhdY::ery) Δ(yfkC::kan) | This study |
| TMB105 | Δ(opuE::tet)1 Δ(mscL::spc) Δ(ykuT::cat) Δ(yhdY::ery) Δ(yfkC::kan) | This study |
| BLOB20 | sapB::neo | This study |
| TMB140 | Δ(sapB::spc)2 | This study |
| TMB139 | Δ[(opuE-sapB)::spc]1 | This study |
| JSB36d | [amyE::(ΦproH153bp-treA)3 cat] (treA::neo)1 | 10 |
| TMB119 | Δ(opuE::tet)1 [amyE::(ΦproH153bp-treA)3 cat] (treA::neo)1 | This study |
| TRB2 | [amyE::(ΦopuE1091bp-treA) cat] (treA::neo)1 | 47 |
| TRB12 | Δ(opuE::tet)1 [amyE::(ΦopuE1091bp-treA) cat] (treA::neo)1 | F. Spiegelhalter |
All strains are derivatives of the B. subtilis wild-type strain JH642 (obtained from J. Hoch, Scripps Research Institute, CA) and therefore also carry, in addition to the genetic markers indicated, the trpC2 pheA1 mutations.
Bacillus Genetic Stock Center, Columbus, OH.
This strain carries a nonreplicative plasmid (Spcr) with copies of the intact opuE and sapB genes that was crossed into the B. subtilis chromosome in the vicinity of the Δ(opuE::tet)1 locus by selecting for spectinomycin resistance. The intact copy of the opuE gene is expressed from its natural promoter.
The designation [amyE::(ΦproH-treA)3 cat] indicates that the proH-treA reporter gene fusion is stably integrated via a double-recombination event into the chromosomal amyE gene of B. subtilis as a single copy, thereby rendering the fusion strain defective in the extracellular AmyE α-amylase. The reporter gene fusion construct is linked to a chloramphenicol resistance gene (cat).
Bacterial strains.
All B. subtilis strains used in this study are derivatives of strain JH642 (BGSC 1A96; a kind gift of J. Hoch, Scripps Research Institute, CA) and are listed in Table 1. JH642 is a member of the domesticated 168 lineage of B. subtilis laboratory strains (48) and served as the wild-type strain for our studies. The proline auxotrophic (proC46::Tn5) derivative of the wild-type Escherichia coli strain MG1655 (46) was kindly provided by G. Sezonov (Institut Pasteur, Paris, France), and the proline auxotroph E. coli strain RC711 (proA23 lac-28 his-51 trp-30) (CGSC 3456) was obtained from the E. coli Genetic Stock Center (Yale University, New Haven, CT).
Construction of B. subtilis mutant strains.
The Δ(yfkC::tet)1 mutation carried by strain SMB80 (Table 1) was changed to an Δ(yfkC::kan)1 allele to allow the introduction of the Δ(opuE::tet)1 mutation into the previously described mechanosensitive channel quadruple mutant strain SMB80 (24). This was done by long-flanking region PCR where an 895-bp upstream DNA fragment and a 1,010-bp downstream DNA fragment of the yfkC gene were joined with a 1,385-bp PCR fragment carrying a kan resistance gene derived from plasmid pDG783 (20). The resulting 3,290-bp fusion product was then transformed into strain TMB101 carrying gene disruptions in mscL, ykuT, and yhdY (Table 1) by selecting for kanamycin-resistant transformants, yielding strain TMB99 (Table 1). Subsequently, strain TMB99 was transformed with chromosomal DNA of strain BLOB9 [Δ(opuE::tet)1] (Table 1) and the transfer of the Δ(opuE::tet)1 allele was selected for by plating on LB agar plates containing tetracycline; one of the transformants was strain TMB105 (Table 1). Strain BLOB26 was constructed by transforming strain BLOB9 (Table 1) with circular DNA of the opuE+ sapB+ plasmid pBLOB15.2 (Spcr) and subsequently selecting for spectinomycin-resistant transformants. Since plasmid pBLOB15.2 does not carry an origin of replication that is functional in B. subtilis, the transformation of the Δ(opuE::tet)1 mutant strain BLOB9 (Table 1) with circular DNA of pBLOB15.2 results in a Campbell-type single-crossover integration of this plasmid into the B. subtilis chromosome in the vicinity of the opuE region (Table 1). The B. subtilis strain JSB36 carries a proH-treA reporter gene fusion inserted as a single copy into the chromosomal amyE locus (10). The Δ(opuE::tet)1 mutation was introduced into strain JSB36 by DNA transformation with chromosomal DNA of strain BLOB9, yielding strain TMB119 (Table 1). Strain TRB2 carries an opuE-treA reporter gene fusion inserted as a single copy into the chromosomal amyE locus and possesses an intact opuE gene (47); strain TRB12 is a derivative of TRB2 and was constructed by transforming strain TRB2 with chromosomal DNA of strain BLOB9 [Δ(opuE::tet)1] and subsequent selection for tetracycline-resistant colonies (Table 1). Strain BLOB20 (Table 1) was constructed by transforming strain JH642 with chromosomal DNA of the B. subtilis strain SL6216 (sapB::neo) (Table 1) (60) and selecting for the sapB::neo gene disruption mutation on agar plates containing kanamycin. Additional sapB disruption mutations were constructed using PCR-generated fusion constructs harboring 5′- and 3′-flanking regions of the corresponding genomic regions interrupted by a spectinomycin-resistant cassette which was derived from plasmid pDG1726 (20). These DNA constructs were introduced into the chromosome of the B. subtilis wild type JH642 by homologous recombination and subsequent selection for spectinomycin-resistant colonies. We generated strain TMB140 [Δ(sapB::spc)2] where an internal 627-bp fragment within the sapB gene was replaced by the spectinomycin-resistant cassette and the opuE sapB double mutant strain TMB139 {Δ[(opuE-sapB)::spc]1} carrying a 2,194-bp deletion of the opuE-sapB region replaced by a spectinomycin resistance cassette.
Plasmid construction.
Plasmid pBLOB15.2 was constructed by cleaving the opuE+ sapB+ plasmid pORT3 (56) at a SmaI site located upstream of the opuE gene and by inserting a spectinomycin resistance cassette derived from plasmid pIC156 (50).
TreA reporter enzyme activity assays.
The expression of proH-treA and opuE-treA operon gene fusions was monitored by assaying the TreA [phospho-α-(1,1)-glucosidase] enzyme activity of cells using the chromogenic substrate para-nitrophenyl-α-d-glucopyranoside (18, 47). TreA enzyme activity is expressed as U (mg protein−1) according to the definition used for the quantification of β-galactosidase (38). Protein concentrations of the samples were estimated from the optical density of the cell culture of the reporter strains (38).
Bioassays to assess the release of proline from B. subtilis cells.
To assess the excretion of proline from B. subtilis strains, we developed two bioassays that monitor the cross-feeding of proline auxotrophic E. coli strains by B. subtilis. For the first assay (“blue halo”), we grew the proC46::Tn5 proline auxotrophic derivative of the E. coli strain MG1655 in minimal medium A (MMA) (38) containing 0.4 M NaCl with 0.5% glucose as the carbon source and 10 mM proline to an OD578 of about 1 to 1.5. The culture was then washed three times with proline-free MMA containing 0.4 M NaCl, and 100 μl of the cell suspension was then mixed with 8 ml of prewarmed MMA top agar (0.7% agar) containing 0.5% glucose, 0.8 M NaCl, 1 mM IPTG, and 100 μl of an X-Gal stock solution (20 mg ml−1 dissolved in dimethylformamide). The top agar also contained l-tryptophan (20 mg liter−1) and l-phenylalanine (18 mg liter−1) to satisfy the auxotrophic growth requirements of the B. subtilis strain JH642 (trpC2 pheA1) and its various mutant derivatives (Table 1). This mixture was poured onto an MMA agar plate containing the same ingredients and an NaCl concentration of 0.8 M. Onto this lawn, 5 μl of various B. subtilis cultures grown in SMM to an OD578 of 1 to 1.5 was spotted; the plates were then incubated for 3 to 4 days at 37°C. The development of an intensive blue halo around the spotted B. subtilis cells was indicative of the release of proline from the tested B. subtilis strain and was caused by the growth of the lac+ proC46::Tn5 proline auxotrophic derivative of the E. coli strain MG1655 on the background lawn and the induction of the lac operon by IPTG.
For the second proline release bioassay, we plated 0.2 ml of an exponentially growing culture of the proline auxotrophic E. coli strain RC711 (proA23) onto an MMA agar plate containing 0.8 M NaCl. Onto this lawn, we then replica plated the B. subtilis wild-type strain JH642 and its mutant derivatives; the agar plates were incubated for 3 to 4 days at 37°C. Proline excretion was manifested by growth of the E. coli indicator strain RC711 around individual B. subtilis colonies. To satisfy the growth requirements of the B. subtilis strains for Trp and Phe and that of the E. coli strain RC711 for Trp and His, the MMA agar plates contained l-tryptophan (20 mg liter−1), l-phenylalanine (18 mg liter−1), and l-histidine (50 mg liter−1).
Quantification of proline.
The intracellular proline content of B. subtilis strains and the amount of proline released by these strains into the growth medium were quantified by a colorimetric assay developed by Bates et al. (2) that detects proline as a colored proline-ninhydrin complex that can be quantified by measuring the absorption of the solution at 480 nm. For this assay, B. subtilis cells were grown in SMM under various osmotic conditions until mid-exponential growth phase. For the measurement of the intracellular proline content, cells from 8 ml culture were harvested by centrifugation, extracted, and analyzed according to the procedure detailed by Bates et al. (2). The same procedure was also used to quantify the amount of proline released by B. subtilis cells into the growth medium. Proline concentrations were determined by establishing a standard curve with l-proline. Intracellular proline concentrations were calculated using a volume for a B. subtilis cell of 0.67 μl per 1 OD578 unit of cell culture (S. Moses, E. P. Bakker, and E. Bremer, unpublished data). The colorimetric proline assay was also used to quantify the amount of proline released by B. subtilis cells into the growth medium. To compare the amounts of extracellular proline between different strains and growth conditions, we normalized the values according to an OD578 of 1.0. All values for the intracellular and extracellular proline concentrations were analyzed from two subsamples of each culture. The standard error of the measured proline concentration was below 5%.
Transport assays with radiolabeled proline.
Uptake assays with radiolabeled proline were conducted as described previously (27, 56). To analyze the process of proline uptake and excretion, we precultured the B. subtilis wild-type strain JH642 in SMM containing 0.8 M NaCl and then used this preculture to inoculate a fresh culture (in SMM containing 0.8 M NaCl) to an optical density of 0.25 and allowed the cells to grow for about 4 h at 37°C until the culture reached an OD578 of 1. Two aliquots (2 ml each) of the culture were then transferred to 2-ml reaction vials and further incubated in a Thermomixer set to 37°C with vigorous shaking. The cells were then fed with 1 mM proline spiked with 0.63 μM radiolabeled [14C]proline. The accumulation of proline by the cells was followed over time by filtering 0.2-ml samples of each culture onto a cellulose filter (0.45 μm; Schleicher & Schuell, Dassel, Germany). The filters were washed with 20 ml of the proline-free cultivation medium, and the amount of [14C]proline retained by the cells collected on the cellulose filter was determined in a liquid scintillation counter (Coulter liquid scintillation analyzer 1900CA). We found that the intracellular [14C]proline content of the cells typically saturated after 10 min of incubation (see Fig. 6). After this time, one culture was exposed to a 100-fold excess of unlabeled proline and a second control culture was exposed to a 100-fold excess of unlabeled glycine. The intracellular [14C]proline pool of the cells from these two cultures was then monitored by collecting at certain time intervals 0.1-ml aliquots of cell culture and subsequently determining their [14C]proline content in a liquid scintillation counter.
Fig 6.
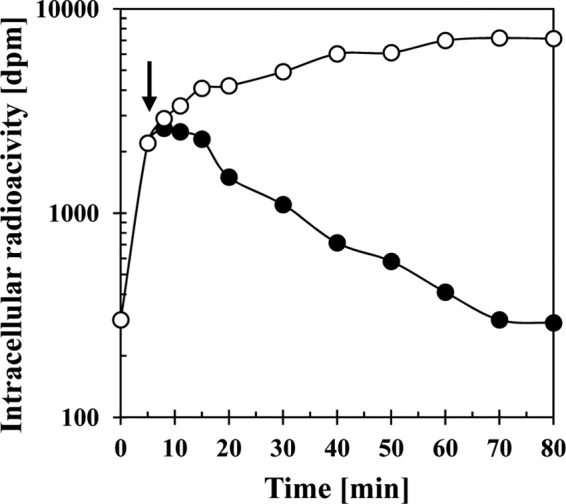
Efflux of [14C]proline from high-salinity-stressed B. subtilis cells in response to an exogenous supply of an excess proline. A culture of the wild-type strain JH642 was grown in SMM with 0.8 M NaCl to mid-exponential growth phase (OD578 of about 1) and l-proline that was spiked with 0.63 μM [14C]proline was added to a final concentration of 1 mM. After a 10-min incubation of the cultures (indicated by an arrow), 100 mM proline was added to one culture (●) and 100 mM glycine was added to the second culture (○), and both cell suspensions were then further incubated with vigorous shaking at 37°C. Samples were withdrawn at various time intervals, the cells were collected by filtration onto cellulose filters, and the intracellular [14C]proline content of the cells was determined by scintillation counting. This experiment was repeated three times, and the data given represent a typical result.
RESULTS
A defect of the OpuE proline transporter impairs growth at high salinity.
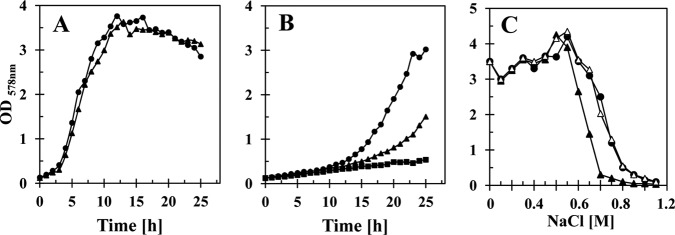
The B. subtilis opuE mutant strain BLOB9 [Δ(opuE::tet)1] grows like its isogenic opuE+ parent strain JH642 in a minimal medium (SMM) with glucose as the carbon source (Fig. 2A), indicating that the Δ(opuE::tet)1 gene disruption mutation does not confer indiscriminant negative effects on cell growth. However, the behavior of the opuE mutant strain changed significantly when we assessed its growth properties under high-osmolarity growth conditions (SMM containing 1.2 M NaCl). As expected from previously reported experiments (4), growth of the wild-type strain JH642 slowed considerably in the high-salinity medium in comparison with cells cultivated in SMM alone (Fig. 2B). Strain JSB8 [Δ(proHJ::tet)1], which lacks the ability to synthesize proline as an osmoprotectant (10) but is not a proline auxotroph (3, 11), showed the expected salt-sensitive growth phenotype (Fig. 2B). Surprisingly, growth of the opuE mutant strain BLOB9 was also strongly impaired in the high-salinity medium (Fig. 2B) and exhibited growth characteristics positioned between those of the wild-type strain JH642 and the proHJ mutant strain JSB8 (Fig. 2B). Impaired growth of BLOB9 became apparent when the salinity of the growth medium exceeded about 0.6 M NaCl (Fig. 2C). The salt-sensitive growth phenotype of BLOB9 was fully rescued when we reinserted, via homologous recombination, an intact copy of the opuE gene carried by a nonreplicating plasmid into the chromosome of strain BLOB9 [Δ(opuE::tet)1] in the vicinity of the opuE locus (strain BLOB26) (Fig. 2C). Taken together, these observations indicate that the loss of OpuE was responsible for the increased osmotic sensitivity of strain BLOB9.
Fig 2.
Effects of high salinity on the growth of the opuE mutant strain BLOB9 and its parent strain JH642. Cells of strain JH642 (opuE+) (●) and BLOB9 [Δ(opuE::tet)1] (▲) were grown at 37°C in SMM (A) or SMM with 1.2 M NaCl (B). Growth of strain JSB8 [Δ(proHJ::tet)1] (■) was also monitored at a high salinity for competitive purposes. (C) Cultures of strain JH642 (opuE+) (●), BLOB9 [Δ(opuE::tet)1] (▲), and BLOB26 [Δ(opuE::tet)1 × pBLOB15.2 (opuE+ sapB+)] (△) were cultivated in SMM with various salinities for 16 h at 37°C, and the optical densities of the cultures were then determined. The data shown represent a typical set of growth experiments.
The opuE mutant strain releases proline.
The salt-sensitive growth phenotype of the opuE mutant BLOB9 was unexpected in view of the fact that the tested growth conditions did not involve a medium where strain BLOB9 had to rely on the uptake of exogenously provided proline to cope with high salinity (56). However, it is known that different microorganisms release newly synthesized compatible solutes into the growth medium when they lack uptake systems to recapture these compounds (15, 19, 36, 37, 44, 51). We therefore considered the possibility that the increased osmotic sensitivity of the opuE mutant (Fig. 2B) was caused by release of proline into the growth medium that the salt-stressed cells then could not retrieve due to the missing OpuE transporter.
To test for the release of proline from the B. subtilis opuE mutant strain BLOB9, we developed two bioassays that relied on the cross-feeding of proline auxotrophic E. coli strains by B. subtilis. In the first of these bioassays, release of proline was manifested and visualized by the growth of a lac+ proline auxotrophic (proC46::Tn5) E. coli strain as a blue halo forming around B. subtilis cells spotted onto a lawn of the indicator cells in the presence of the LacZ indicator dye X-Gal; B. subtilis does not possess a strong β-galactosidase activity (43). As assessed by this bioassay, the Δ(opuE::tet)1 mutant BLOB9 released proline, whereas neither the opuE wild-type strain JH642 nor strain BLOB26, the above-described derivative of BLOB9 carrying an extra copy of the intact opuE gene, was surrounded by a blue halo, indicative of proline excretion (Fig. 3A).
Fig 3.
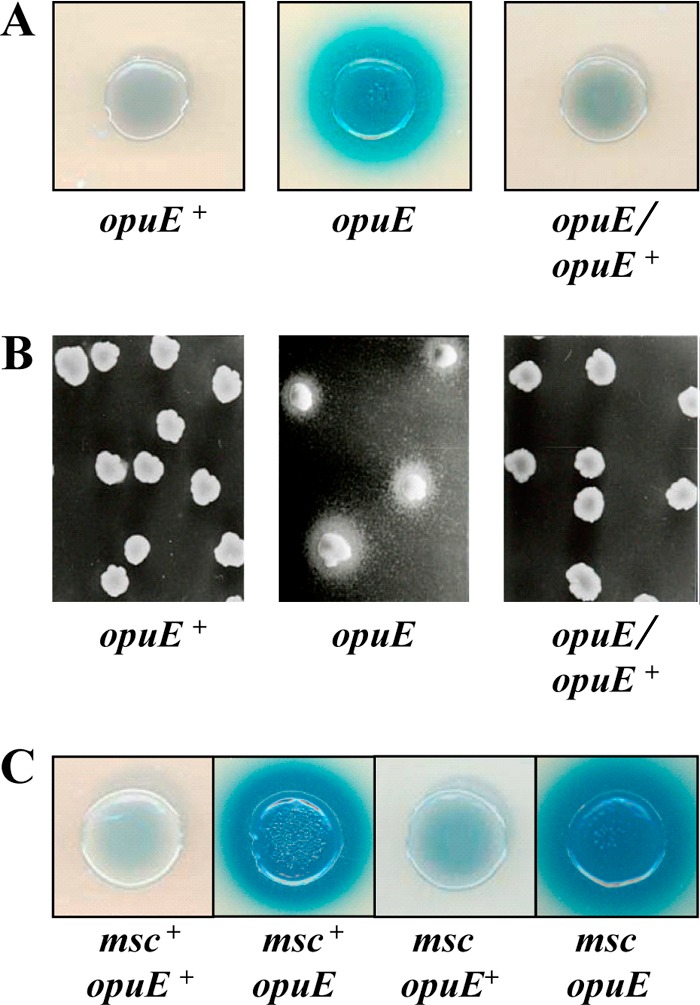
Detection of proline released from B. subtilis cells by bioassays. (A) Five-microliter aliquots of JH642 (opuE+), BLOB9 [Δ(opuE::tet)1], and BLOB26 [Δ(opuE::tet)1 × pBLOB15.2 (opuE+ sapB+)] were spotted onto a lawn of an E. coli lacIZYA+ proline auxotrophic derivative (proC46::Tn5) of strain MG1655 seeded in top agar. The MMA plates and top agar contained 0.8 M NaCl to osmotically stress B. subtilis and thereby trigger osmoadaptive proline synthesis; IPTG and X-Gal were included in the plates to induce the lac operon and to visualize β-galactosidase activity of this indicator strain. Cross-feeding of the E. coli Pro− auxotroph by proline released from the B. subtilis strain BLOB9 [Δ(opuE::tet)1] is visible as a blue halo around the spotted B. subtilis cells. (B) The E. coli proline auxotrophic strain RC711 (proA23) was plated onto a lawn of MMA plates containing 0.8 M NaCl, and B. subtilis colonies were replica plated onto this cell lawn. Cross-feeding of strain RC711 by proline released by the opuE mutant strain BLOB9 is evident from growth of E. coli cells around individual B. subtilis colonies. (C) Blue halo bioassay with strains JH642 (opuE+), BLOB9 [Δ(opuE::tet)1] and the mechanosensitive (msc) channel quadruple mutants SMB80 (opuE+) and TMB105 [Δ(opuE::tet)1], and the E. coli proC46::Tn5 proline auxotrophic indicator strain.
Release of proline by the Δ(opuE::tet)1 mutant strain BLOB9 was also evident from the second employed bioassay. In this assay, we replica plated colonies of the B. subtilis strains JH642, BLOB9, and BLOB26 onto a lawn of the proline auxotrophic E. coli strain RC711 (proA23) and observed growth of the E. coli cells only around colonies of the opuE mutant BLOB9 (Fig. 3B). Hence, both bioassays pointed toward the release of proline from salt-stressed cells of a B. subtilis opuE mutant strain.
Release of proline is not caused by polar effects of the Δ(opuE::tet)1 mutation on the expression of the sapB gene.
The opuE gene (56) in the B. subtilis genome is followed by sapB, a predicted integral membrane protein whose physiological function is not well defined (60). The 106-bp intragenic region between opuE and sapB contains an intrinsic transcriptional terminator sequence, but a minor part of the transcript initiating at the opuE promoters extends beyond this terminator and thus represents an mRNA species with a full-length opuE-sapB region (47). Furthermore, one of the promoters driving sapB expression resides within the opuE-coding region (41, 60). In view of the fact that SapB is a predicted integral membrane protein, we considered the possibility that the observed release of proline could be caused by polar effects exerted by the Δ(opuE::tet)1 mutation on the transcription of the sapB gene rather than by the loss of the OpuE transporter itself. To test whether proline release was triggered by loss of SapB, we measured the proline content of the supernatant of an isogenic set of strains with either opuE or sapB gene disruption and of a strain carrying a chromosomal deletion of the entire opuE-sapB region. The data summarized in Table 2 conclusively show that the release of proline into the growth medium is a consequence of the loss of the OpuE transporter and not of the SapB protein.
Table 2.
Proline content in the culture supernatants of cells grown either in SMM without additional NaCl or in the presence of 1 M additional NaCla
| Strain | Relevant genotype | Extracellular proline concn (μM) |
|
|---|---|---|---|
| Without NaCl | With 1 M NaCl | ||
| JH642 | Wild type | 6 ± 1 | 7 ± 1 |
| BLOB9 | Δ(opuE::tet)1 | 5 ± 1 | 406 ± 15 |
| BLOB20 | sapB::neo | 6 ± 1 | 13 ± 2 |
| TMB140 | Δ(sapB::spc)2 | 6 ± 1 | 7 ± 1 |
| TMB139 | Δ[(opuE-sapB)::spc]1 | 4 ± 1 | 340 ± 74 |
Cultures of the indicated strains were cultivated in SMM in the absence or in the presence of 1 M NaCl. When the cultures reached mid-exponential growth phase (OD578 of 1.5), the cells were removed from the growth medium by centrifugation and the proline contents of the supernatants were measured by a colorimetric assay (2). The proline values given were normalized for each culture corresponding to an OD578 of 1. The given proline concentrations represent measurements from three independently grown cultures and in each case included two technical replicates of the proline assay.
The data documented in Table 2 also provide a quantitative measure of the amounts of proline released by B. subtilis strains lacking OpuE. The proline content of the culture supernatants of strains BLOB9 [Δ(opuE::tet)1] and TMB139 [Δ(opuE-sapB::spc)1] cultivated to mid-exponential growth phase (OD578 of 1.5) in SMM containing 1 M NaCl was about 0.4 mM, whereas the proline content of the culture supernatants of the opuE+ strains BLOB20 [(sapB::neo)] and TMB140 [Δ(sapB::spc)1] was barely measurable (about 6 μM) (Table 2). Hence, the quantitative measurement of the proline content of the growth media of both opuE+ and opuE B. subtilis strains validates the qualitative assessment of their ability to release proline by the above-described bioassays (Fig. 3A and B).
A graded increase in the external salinity leads to a proportional increase in the level of the released proline.
The proline content of B. subtilis cells cultured under sustained high-salinity conditions increases linearly in response to increases in the external salinity once the external salinity of SMM is raised above 0.2 M (10). This leads to an increase in the proline pool from about 10 mM in cells cultivated in SMM to about 400 mM in cells cultivated in SMM containing 1 M NaCl (39). To assess the degree of proline excretion as a function of the external salinity and to investigate the influence of this process on the intracellular proline pool, we grew strains JH642 (opuE+) and BLOB9 [Δ(opuE::tet)1] in SMM with increasing salt concentrations to mid-exponential growth phase (OD578 of 1.5) and then measured the proline content of the cells and the amount of proline present in the growth medium. The intracellular proline content of the wild-type strain JH642 rose proportionally in response to the increase in the external salinity (Fig. 4A) in a pattern resembling that reported recently by Brill et al. (10) and reached a level of about 338 mM in cells grown in SMM containing 1 M NaCl; there was practically no proline detectable (about 10 μM) in the culture supernatants of these cells (Fig. 4B). The proline content of the cells of the opuE mutant strain BLOB9 also rose in response to increases in the external salinity and reached under these growth conditions a level of about 270 mM (Fig. 4A). However, the internal proline pool of the opuE mutant was substantially lower than that of the wild-type cells once the NaCl concentration in the medium exceeded about 0.5 M (Fig. 4A). As a matter of fact, the steady-state levels of the proline pools of the opuE+ and opuE strains differed by about 70 mM in cells cultivated in SMM containing 1 M NaCl (Fig. 4A). Hence, the proline content of the opuE mutant strain BLOB9 was about 20% lower than that of its opuE+ parent strain JH642. In contrast to the wild type, the proline content of the culture supernatant of the opuE mutant cells rose steadily in proportion to the increase in the external salinity and reached a level of 353 μM in the culture grown for 14 h up to an OD578 of about 1.5 in SMM containing 1 M NaCl (Fig. 4B). This corresponds to a level of proline in the growth medium of 43 mg liter−1.
Fig 4.
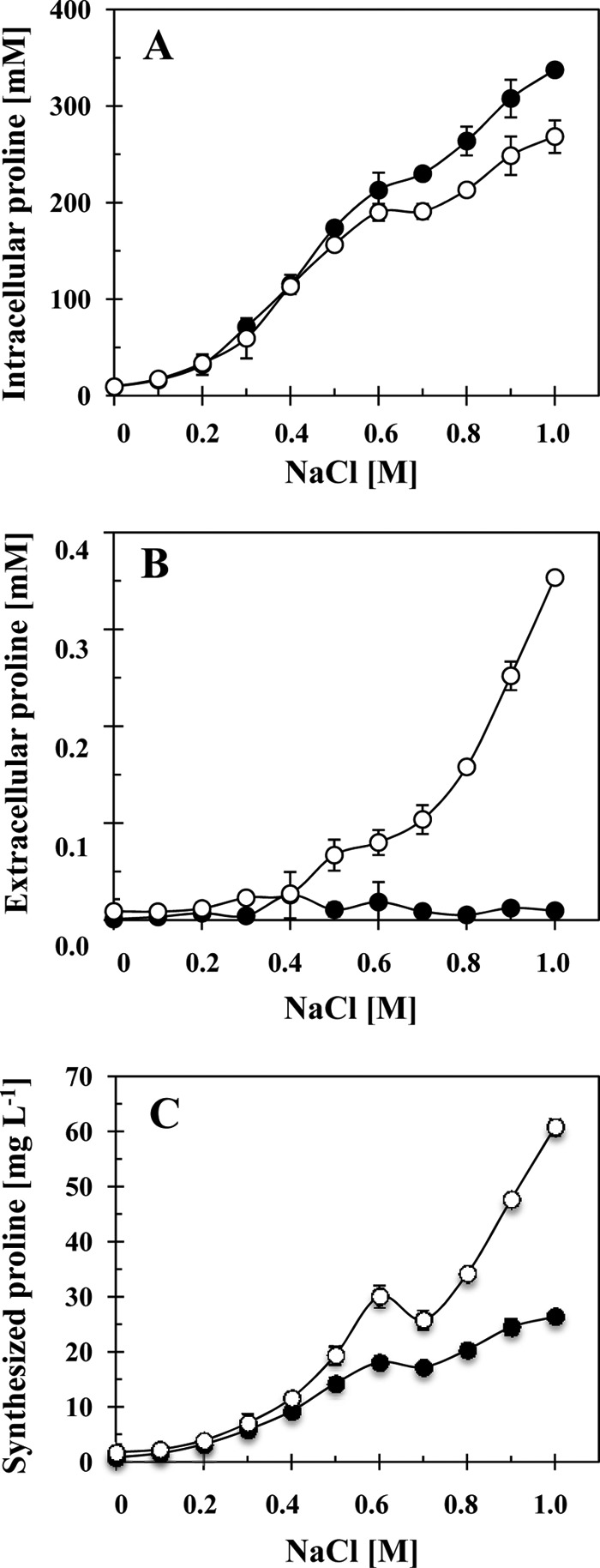
Intracellular and extracellular proline content of high-salinity grown B. subtilis cells. Cultures of the B. subtilis strains JH642 (opuE+) (●) and BLOB9 [Δ(opuE::tet)1] (○) were grown in SMM with various salinities. (A) Cells were harvested in mid-exponential growth phase (OD578 of about 1.5), and their proline content was determined by the colorimetric assay described by Bates et al. (2). (B) The same proline assay was also used to determine the proline content of the supernatants of these cultures. (C) The amounts of intracellular and extracellular proline were summed up for culture of strains JH642 and BLOB9 and are given as synthesized proline; we normalized these values according to an OD578 of 1.0. The error bars given are standard deviations of four separately grown B. subtilis cultures (n = 4).
We found that the expression, as assessed with a proH-treA reporter fusion, of the gene cluster (proHJ) encoding the central enzymes for osmoadaptive proline biosynthesis (10) was similar in the wild type and its opuE mutant derivative (Table 3). We also observed that the expression of an opuE-treA reporter fusion was not increased in an opuE mutant background (Table 3).
Table 3.
Influence of an opuE mutation on the transcriptional activity of the proH and the opuE promotersa
| Strain | treA fusion | opuEb | TreA enzyme activity (U/mg protein) |
||
|---|---|---|---|---|---|
| Without NaCl | With 0.5 M NaCl | With 1.0 M NaCl | |||
| JSB36 | proH-treA | + | 23 ± 1 | 46 ± 3 | 122 ± 5 |
| TMB119 | proH-treA | − | 24 ± 1 | 41 ± 8 | 109 ± 16 |
| TRB2 | opuE-treA | + | 25 ± 4 | 84 ± 5 | 190 ± 65 |
| TRB12 | opuE-treA | − | 17 ± 1 | 77 ± 12 | 213 ± 17 |
Cells carrying chromosomal copies of the indicated proH-treA and opuE-treA operon reporter fusions were cultivated in either SMM, SMM with 0.5 M NaCl, or SMM with 1.0 M NaCl to mid-exponential growth phase (OD578 of 1.5) and were then harvested for TreA reporter enzyme activity assays. Each TreA activity measurement was carried out with three independently grown cultures and in each case included two technical replicates of the TreA enzyme assay.
Strains carrying an opuE mutation (indicated by a minus) all harbored the Δ(opuE::tet)1 allele.
To test if the opuE mutant strain compensates for the loss of proline into the medium by an increased proline production, we calculated the amount of totally synthesized proline normalized to an OD578 of 1 of the cell culture. As shown in Fig. 4C, the salt-dependent proline production of the opuE mutant strain BLOB9 rose to a substantially higher level than that of the wild-type strain. At high salinity (SMM with 1.0 M NaCl), the total amount of proline (intracellular plus extracellular) produced by BLOB9 was 2.3-fold higher (61 mg liter−1) than the amount of proline produced by the wild-type strain (26 mg liter−1) (Fig. 4C). This finding implies, in view of the proHJ transcriptional data documented in Table 3, alterations in the kinetics of the proline biosynthetic enzymes in the opuE mutant strain BLOB9.
MscL- and MscS-type mechanosensitive channels do not participate in the release of proline under sustained high-salinity growth conditions.
We considered the possibility that the observed release of proline from cells continuously exposed to a high-salinity environment (Fig. 4B) was mediated by the gating of mechanosensitive channels (6, 23) operating in B. subtilis (24, 58).
B. subtilis possesses one channel protein of high conductance (MscL) and three different channel proteins of low conductance (MscS): YhdY, YfkC, and YkuT. Direct evidence for channel-forming activity in vivo is available only for the MscL and YkuT proteins since mutants lacking these proteins do not survive a severe osmotic down-shift (24, 58). In addition, the cellular levels of the MscL- and MscS-type mechanosensitive channel-forming proteins of B. subtilis vary with growth phase (57), suggesting that some of these mechanosensitive channel proteins might play not-yet-understood roles during certain periods of the growth cycle of individual B. subtilis cells.
To test the involvement of any of the known Msc-type proteins of B. subtilis in the release of proline, we compared the mechanosensitive quadruple mutant strain SMB80 [Δ(mscL::spc)1 Δ(yhdY::erm)1 Δ(yfkC::tet)1 Δ(ykuT::cat)1] and its corresponding Δ(opuE::tet) mutant derivative, strain TMB105. Release of proline was not prevented or impaired in the opuE mutant strain TMB105 or in the opuE+ strain SMB80 lacking the known MscL- and MscS-type channel proteins as assessed by the bioassay (Fig. 3C) and as quantified by a colorimetric proline assay (Fig. 5) (2). Hence, none of the known mechanosensitive channel proteins of B. subtilis participate in the excretion of proline in continuously salt-stressed cells.
Fig 5.
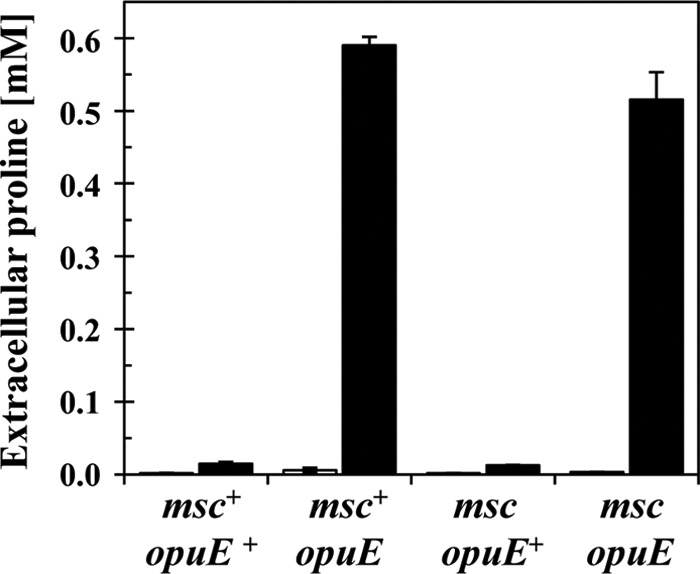
Release of proline into the growth medium is not mediated by MscL- and MscS-type channel proteins. Cells of strains JH642 (msc+ opuE+), BLOB9 (msc+ opuE), SMB80 (msc opuE+), and TMB105 (msc opuE) were grown to mid-exponential growth phase (OD578 of about 2) in either SMM (gray bars) or SMM containing 1 M NaCl (black bars). The proline content of the supernatants was determined by the colorimetric assay described by Bates et al. (2). The error bars given are standard deviations of two separately grown B. subtilis cultures (n = 2). msc mutant strains carry gene disruptions in the mscL, ykuT, yhdY, and yfkC loci, thereby interrupting the single MscL-type channel-forming protein and the three MscS-type channel-forming proteins from B. subtilis (24, 58). opuE mutant strains carry the Δ(opuE::tet)1 allele (56).
Release of proline is not caused by a reversal of the transport direction by the PutP proline importer.
B. subtilis can use proline as sole carbon and nitrogen source and imports proline for these purposes via the OpuE-related PutP transporter into the cell (39). Hence, it seemed possible that the high intracellular concentrations of proline amassed under high-salinity growth conditions by B. subtilis (10) would trigger a reversal of PutP from a proline import system to a proline export system. We tested this scenario by measuring the proline content of supernatants of strains SMB11 [Δ(putP::spc)1] and SMB12 [Δ(putP::spc)1 Δ(opuE::tet)1] cultured in SMM containing 1.2 M NaCl, growth conditions that lead to very high intracellular proline concentrations via de novo synthesis (10) (Fig. 4A). Both the wild-type strain JH642 and its Δ(putP::spc)1 derivative, strain SMB11, released hardly any proline (about 25 μM), whereas a substantial concentration of proline (about 576 μM) was found in the culture supernatants of strain SMB12 [Δ(putP::spc)1 Δ(opuE::tet)1], also lacking OpuE. Consequently, the PutP proline importer does not function as an export system under conditions where the intracellular proline concentration is very high.
Excess of external proline promotes efflux of preaccumulated proline.
The detection of substantial amounts of proline in the supernatants of cells lacking the OpuE transporter (Fig. 4B) suggests that B. subtilis wild-type cells continuously excrete part of the newly synthesized proline and then recover it again via OpuE. If this is correct, then one should be able to experimentally detect proline efflux from opuE wild-type cells. To test for proline efflux, we conducted an experiment similar to that reported by Jebbar et al. (28) for the uptake and efflux of the compatible solute ectoine in osmotically challenged E. coli cells. Cells of the B. subtilis wild-type strain JH642 were grown in SMM containing 0.8 M NaCl until the culture reached mid-exponential growth phase. The cells were then fed with 1 mM proline spiked with 0.63 μM radiolabeled [14C]proline. Uptake of proline was allowed to proceed for 10 min; thereafter, one portion of the cells received a 100-fold excess of unlabeled proline, and as a control, a second portion of the cells was exposed to a 100-fold excess of unlabeled glycine. Cells that were challenged with an excess of glycine continued to take up [14C]proline until saturation was achieved (Fig. 6). In contrast, the addition of an excess amount of unlabeled proline to the cells promoted [14C]proline efflux to such an extent that essentially no radiolabeled [14C]proline remained in the B. subtilis cells after 60 min (Fig. 6).
DISCUSSION
The data reported here expand our understanding of the function of the osmotically regulated proline import system OpuE (47, 56) within the physiological context of the acclimatization process of the B. subtilis cell to high-osmolarity environments (8, 9). Proline is the only compatible solute that B. subtilis can synthesize de novo (35, 61), and the buildup of an osmo-stress-responsive intracellular proline pool is critical for the ability of the B. subtilis cell to cope with the challenges posed by high salinity (Fig. 1) (10). Our data show that OpuE serves as a recapturing system for proline released from continuously salt-stressed cells (Fig. 4) and that loss of OpuE transporter activity causes osmotic sensitivity (Fig. 2B). Consequently, the function of OpuE as a proline recycling system contributes significantly to the efficiency with which B. subtilis adjusts to sustained high-osmolarity surroundings.
The detection of proline in the culture supernatant of salt-stressed opuE mutants implies that the B. subtilis wild-type strain engages in a cycle of proline synthesis, release, and OpuE-mediated recovery (Fig. 1). Our pulse-chase experiment with externally provided radiolabeled proline (Fig. 6) provides experimental support for this notion. Impaired growth of the opuE mutant in high-salinity media (Fig. 2B and C) is thus the consequence of continued release of proline (Fig. 4A), and the lack of OpuE transport activity then leads to a substantial reduction in the steady-state level of the intracellular proline pool (Fig. 4B). The B. subtilis cell does not compensate for the loss of proline into the growth medium by strongly upregulating the expression of the proHJ gene cluster encoding the central enzymes for osmoadaptive proline biosynthesis (Table 3) (10). Mutants of the moderate halophile Halomonas elongata, which lacks the ectoine-specific TRAP transporter TeaABC (19), and a mutant of Synechocystis sp. strain PCC6803, with a defect in the glycosylglycerol-specific ABC transporter GgtBCD (37), lose ectoine and glycosylglycerol in the growth medium. They compensate for the loss of the newly synthesized compatible solutes by enhanced production of these compounds so that the intracellular concentrations of ectoine and glycosylglycerol in the mutant strains are similar to that of the parent strains possessing the TeaABC and GgtBCD transport systems (19, 37). In contrast, the intracellular proline concentration in the B. subtilis strain with a defect in OpuE is lower than that of its parent, but nevertheless, the overall amount of proline produced under salt stress by the opuE mutant is substantially higher than that of the wild type (Fig. 4C). Since we did not observe increased expression of the gene cluster encoding the enzymes central for osmoadaptive proline biosynthesis in B. subtilis (10) (Table 3) but found increased amounts of proline in the opuE mutant (Fig. 4C), our data imply altered kinetic activities of the proline biosynthetic enzymes when the B. subtilis cell continuously loses newly made proline to the surroundings. This issue deserves further study. Despite the observed overproduction of proline by the opuE mutant (Fig. 4C), it cannot maintain a sufficient intracellular level of proline (Fig. 4A) to adjust effectively to high-salinity growth conditions (Fig. 2B and C).
As already indicated above for the tea mutant of H. elongata (19) and the ggt mutant of Synechocystis sp. PCC6803 (37), the release of newly synthesized compatible solutes under continuous osmotic stress growth conditions and their recapture are not a unique property of B. subtilis. To the best of our knowledge, Styrvold and Strom first reported this phenomenon in osmotically stressed bacteria. These authors observed that high-salinity-challenged E. coli cells continuously excrete the only compatible solute they can synthesize de novo, the disaccharide trehalose, into the growth medium (51), which is then recovered as a carbon source after hydrolysis to glucose by a periplasmic osmotically inducible trehalase (5). Strom and coworkers also found that E. coli cells synthesizing glycine betaine from an exogenous supply of the precursor choline release part of the newly produced glycine betaine into the growth medium and then reaccumulate it via the osmotically inducible compatible solute uptake systems ProP and ProU (36). Such a phenomenon has also been observed in Actinopolyspora halophila, which can synthesize glycine betaine both via oxidation of choline and via methylation of glycine (42). Release of compatible solutes has also been detected in microorganisms that have been selected or engineered to either overproduce or synthesize compatible solutes that they do not make naturally (13, 15, 44, 45).
If these previous reports and the findings reported here on proline excretion and OpuE-mediated recovery are not simply spurious incidents, then uptake systems for compatible solutes will frequently have a dual physiological function in microorganisms as scavengers of osmoprotectants provided by other organisms and as recapturing devices for the compatible solute synthesized and released by a given bacterium. If this hypothesis is correct, then many bacteria are predicted to possess a retrieval system for the main compatible solute they produce, for reuse either as an osmoprotectant or as a nutrient. Release and recapture of compatible solutes might also be a mechanism for cooperativity in microbial communities and biofilms, as suggested by an analysis of the role of newly synthesized and then released glycine betaine from osmotically challenged Vibrio cholerae cells (30). However, the OpuE-mediated recovery of the released proline is so effective in B. subtilis, at least under laboratory conditions, that the growth of an E. coli proline auxotroph in the vicinity of the salt-stressed B. subtilis cells is prevented (Fig. 3A and B).
Our data raise the obvious question of how proline is released from the B. subtilis cell. Osmotically stressed microbial cells typically amass compatible solutes to exceedingly high intracellular concentrations (14, 31, 63), and this also holds true for the proline pool of B. subtilis (10, 39, 61). It reaches intracellular levels of around 400 mM (Fig. 4A) (39) in cells cultivated to mid-exponential growth phase in a minimal medium containing 1 M NaCl (Fig. 4A) and even temporarily exceeds this value in cells that are subjected to a sudden osmotic up-shock with 0.4 M NaCl (61). Hence, the proline gradient across the cytoplasmic membrane of B. subtilis might be so steep that a certain amount of proline simply leaks passively through the cytoplasmic membrane (44) (Fig. 1).
Alternatively, the release of proline might be an active, protein-catalyzed process (Fig. 1). Efflux carriers for selected amino acids are well known to exist in many microorganisms (16, 34), and they function in export of their substrates across the cytoplasmic membrane at much lower gradients (53) than are typically attained through the synthesis of compatible solutes in response to osmotic stress (14, 31, 63). Hence, B. subtilis might possess either a specific proline efflux system or an efflux system with broad substrate specificity for osmoprotectants. Biochemical evidence for the existence of a glycine betaine/proline efflux system in osmotically stressed cells of Salmonella enterica serovar Typhimurium has been provided through modification experiments of membrane proteins in intact cells with sulfhydryl reagents (33). Likewise, a kinetic analysis of the osmotically triggered glycine betaine fluxes in Lactobacillus plantarum also suggests the existence of a carrier-based efflux system for this compatible solute (17).
Mechanosensitive channels gate in response to increased tension in the cytoplasmic membrane that is caused by a buildup of turgor elicited by severe osmotic down-shifts (6, 23). It thus seemed possible to us that these channels might also transiently gate in B. subtilis cells cultivated under steady-state high-salinity growth conditions and thereby release proline. However, our data clearly rule out the involvement of any of the known MscL- and MscS-type mechanosensitive channels operating in B. subtilis (24, 58) in this process (Fig. 5). This finding does not imply that no mechanosensitive channels are involved in the release of proline, because these types of pressure-sensitive safety valves might exist in B. subtilis (1, 52) but have not yet been recognized at the molecular level. Our data rule out the remote possibility that the OpuE-related proline importer PutP, which is involved in the uptake of proline as a nutrient by B. subtilis (39), reverses its transport direction and exports proline when the intracellular concentration of this compatible solute is very high.
Although a cycle of synthesis, release, and recapture of proline (Fig. 1) at first seems wasteful in terms of the overall energetic balance sheet, it might provide B. subtilis with a flexible tool to fine-tune its turgor. Maintenance of turgor is generally regarded as critical for cell viability and growth (63). The turgor of B. subtilis has been estimated as 1.9 MPa (62), a value that approaches 10 times that of a standard car tire. Active water management is a central aspect of the cell's response to rapidly fluctuating or long-lasting changes in the osmotic conditions of the environment in order to set and maintain the magnitude of turgor to physiologically acceptable values (8, 9). However, turgor might vary in much more subtle ways in individual cells that are not subjected to rapid osmotic up- or down-shifts.
Temporary fluctuations in turgor might result from the transitory overaccumulation of newly synthesized compatible solutes or their import from exogenous sources. Indeed, in Corynebacterium glutamicum, part of the newly imported compatible solute glycine betaine is released again through the MscS-variant mechanosensitive channel MscSG. Börngen et al. have called this behavior “pump and leak” and have suggested a role for this process in the fine-tuning of the steady-state concentration of compatible solutes accumulated in response to hyperosmotic stress (7). Temporary fluctuations in turgor might also arise during the growth cycle of individual cells. Turgor is considered a key driving force for cell elongation (32), a process in which the cell continuously extends its peptidoglycan sacculus through synthesis (55) and concomitantly increases its volume before it eventually divides into two cells of equal volume and most likely of equal turgor. Hence, during elongation, the cell has to strike a fine balance between maintenance of turgor within physiologically acceptable boundaries and maintaining the mechanical stability of the peptidoglycan sacculus that resists the turgor pressure. Under osmotic stress conditions, B. subtilis might therefore engage, at times, in a temporary reduction or increase of its turgor-promoting proline pool through an active excretion and recapturing process. When the cell cannot recapture the released proline via the osmotically induced OpuE system, B. subtilis is at a distinct growth disadvantage in high-osmolarity surroundings.
If the release of proline from salt-stressed B. subtilis cells is indeed a protein-catalyzed process (Fig. 1), then the bioassays that we have developed (Fig. 3) might be useful to set up screening or selection procedures to genetically identify the involved transporters or channels.
ACKNOWLEDGMENTS
We thank P. J. Piggot, G. Sezonov, the E. coli Genetic Stock Center, and F. Spiegelhalter for generously providing bacterial strains. We are very grateful to J. Gade for her expert and dedicated technical assistance, and we greatly appreciate the help of V. Koogle in the language editing of our manuscript. We highly value our discussions with L. N. Csonka, R. Krämer, and J. M. Wood on the mechanisms by which proline might permeate through the cytoplasmic membrane.
Financial support for this study was provided by grants from the BMBF via the Bacell-SysMo2 consortium, the LOEWE program of the State of Hessen via the Centre for Synthetic Microbiology (SYNMIKRO; Marburg, Germany), and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print 8 June 2012
REFERENCES
- 1. Alcayaga C, Venegas R, Carrasco A, Wolff D. 1992. Ion channels from the Bacillus subtilis plasma membrane incorporated into planar lipid bilayers. FEBS Lett. 311:246–250 [DOI] [PubMed] [Google Scholar]
- 2. Bates LS, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207 [Google Scholar]
- 3. Belitsky BR, Brill J, Bremer E, Sonenshein AL. 2001. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 183:4389–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boch J, Kempf B, Bremer E. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boos W, Ehmann U, Bremer E, Middendorf A, Postma P. 1987. Trehalase of Escherichia coli. Mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions. J. Biol. Chem. 262:13212–13218 [PubMed] [Google Scholar]
- 6. Booth IR, Edwards MD, Black S, Schumann U, Miller S. 2007. Mechanosensitive channels in bacteria: signs of closure? Nature Rev. Microbiol. 5:431–440 [DOI] [PubMed] [Google Scholar]
- 7. Börngen K, et al. 2010. The properties and contribution of the Corynebacterium glutamicum MscS variant to fine-tuning of osmotic adaptation. Biochim. Biophys. Acta 1798:2141–2149 [DOI] [PubMed] [Google Scholar]
- 8. Bremer E. 2002. Adaptation to changing osmolarity, p 385–391 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 9. Bremer E, Krämer R. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes, p 79–97 In Storz G, Hengge-Aronis R. (ed), Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 10. Brill J, Hoffmann T, Bleisteiner M, Bremer E. 2011. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J. Bacteriol. 193:5335–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brill J, Hoffmann T, Putzer H, Bremer E. 2011. T-box-mediated control of the anabolic proline biosynthetic genes of Bacillus subtilis. Microbiology 157:977–987 [DOI] [PubMed] [Google Scholar]
- 12. Buescher JM, et al. 2012. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335:1099–1103 [DOI] [PubMed] [Google Scholar]
- 13. Carvalho AL, Cardoso FS, Bohn A, Neves AR, Santos H. 2011. Engineering trehalose synthesis in Lactococcus lactis for improved stress tolerance. Appl. Environ. Microbiol. 77:4189–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Csonka LN. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Csonka LN. 1988. Regulation of cytoplasmic proline levels in Salmonella typhimurium: effect of osmotic stress on synthesis, degradation, and cellular retention of proline. J. Bacteriol. 170:2374–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eggeling L, Sahm H. 2003. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 180:155–160 [DOI] [PubMed] [Google Scholar]
- 17. Glaasker E, Konings WN, Poolman B. 1996. Glycine betaine fluxes in Lactobacillus plantarum during osmostasis and hyper- and hypo-osmotic shock. J. Biol. Chem. 271:10060–10065 [DOI] [PubMed] [Google Scholar]
- 18. Gotsche S, Dahl MK. 1995. Purification and characterization of the phospho-alpha(1,1)glucosidase (TreA) of Bacillus subtilis 168. J. Bacteriol. 177:2721–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grammann K, Volke A, Kunte HJ. 2002. New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581T. J. Bacteriol. 184:3078–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61 [DOI] [PubMed] [Google Scholar]
- 21. Hahne H, et al. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192:870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harwood CR, Archibald AR. 1990. Growth, maintenance and general techniques, p 1–26 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus, vol 1 Wiley & Sons Ltd., Chichester, United Kingdom [Google Scholar]
- 23. Haswell ES, Phillips R, Rees DC. 2011. Mechanosensitive channels: what can they do and how do they do it? Structure 19:1356–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoffmann T, Boiangiu C, Moses S, Bremer E. 2008. Responses of Bacillus subtilis to hypotonic challenges: physiological contributions of mechanosensitive channels to cellular survival. Appl. Environ. Microbiol. 74:2454–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann T, Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 193:1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holtmann G, Bakker EP, Uozumi N, Bremer E. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185:1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holtmann G, Bremer E. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jebbar M, Talibart R, Gloux K, Bernard T, Blanco C. 1992. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J. Bacteriol. 174:5027–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jung H, Hilger D, Raba M. 2012. The Na/L-proline transporter PutP. Front. Biosci. 17:745–759 [DOI] [PubMed] [Google Scholar]
- 30. Kapfhammer D, Karatan E, Pflughoeft KJ, Watnick PI. 2005. Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities. Appl. Environ. Microbiol. 71:3840–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high osmolality environments. Arch. Microbiol. 170:319–330 [DOI] [PubMed] [Google Scholar]
- 32. Koch AL. 1983. The surface stress theory of microbial morphogenesis. Adv. Microb. Physiol. 24:301–366 [DOI] [PubMed] [Google Scholar]
- 33. Koo SP, Higgins CF, Booth IR. 1991. Regulation of compatible solute accumulation in Salmonella typhimurium: evidence for a glycine betaine efflux system. J. Gen. Microbiol. 137:2617–2625 [DOI] [PubMed] [Google Scholar]
- 34. Krämer R. 1994. Systems and mechanisms of amino acid uptake and excretion in prokaryotes. Arch. Microbiol. 162:1–13 [DOI] [PubMed] [Google Scholar]
- 35. Kuhlmann AU, Bremer E. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68:772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lamark T, Styrvold OB, Strom AR. 1992. Efflux of choline and glycine betaine from osmoregulating cells of Escherichia coli. FEMS Microbiol. Lett. 75:149–154 [DOI] [PubMed] [Google Scholar]
- 37. Mikkat S, Hagemann M. 2000. Molecular analysis of the ggtBCD gene cluster of Synechocystis sp. strain PCC6803 encoding subunits of an ABC transporter for osmoprotective compounds. Arch. Microbiol. 174:273–282 [DOI] [PubMed] [Google Scholar]
- 38. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 39. Moses S, et al. 2012. Proline utilization by Bacillus subtilis: uptake and catabolism. J. Bacteriol. 194:745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nannapaneni P, et al. 2012. Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification. Microbiology 158:696–707 [DOI] [PubMed] [Google Scholar]
- 41. Nicolas P, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106 [DOI] [PubMed] [Google Scholar]
- 42. NyyssölÄ A, Leisola M. 2001. Actinopolyspora halophila has two separate pathways for betaine synthesis. Arch. Microbiol. 176:294–300 [DOI] [PubMed] [Google Scholar]
- 43. Pozsgai ER, Blair KM, Kearns DB. 2012. Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis. Appl. Environ. Microbiol. 78:778–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rancourt DE, Stephenson JT, Vickell GA, Wood JM. 1984. Proline excretion by Escherichia coli K12. Biotechnol. Bioeng. 26:74–80 [DOI] [PubMed] [Google Scholar]
- 45. Schubert T, Maskow T, Benndorf D, Harms H, Breuer U. 2007. Continuous synthesis and excretion of the compatible solute ectoine by a transgenic, nonhalophilic bacterium. Appl. Environ. Microbiol. 73:3343–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spiegelhalter F, Bremer E. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis—contributions of the SigA and SigB-dependent stress-responsive promoters. Mol. Microbiol. 29:285–296 [DOI] [PubMed] [Google Scholar]
- 48. Srivatsan A, et al. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4:e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steil L, Hoffmann T, Budde I, Völker U, Bremer E. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Steinmetz M, Richter R. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis through in vivo recombination. Gene 142:79–83 [DOI] [PubMed] [Google Scholar]
- 51. Styrvold OB, Strom AR. 1991. Synthesis, accumulation, and excretion of trehalose in osmotically stressed Escherichia coli K-12 strains: influence of amber suppressors and function of the periplasmic trehalase. J. Bacteriol. 173:1187–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Szabó I, Petronilli V, Zoratti M. 1992. A patch-clamp study of Bacillus subtilis. Biochim. Biophys. Acta 1112:29–38 [DOI] [PubMed] [Google Scholar]
- 53. Trötschel C, Deutenberg D, Bathe B, Burkovski A, Krämer R. 2005. Characterization of methionine export in Corynebacterium glutamicum. J. Bacteriol. 187:3786–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vílchez S, Molina L, Ramos C, Ramos JL. 2000. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 182:91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vollmer W, Seligman SJ. 2010. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 18:59–66 [DOI] [PubMed] [Google Scholar]
- 56. von Blohn C, Kempf B, Kappes RM, Bremer E. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175–187 [DOI] [PubMed] [Google Scholar]
- 57. Wahome PG, Cowan AE, Setlow B, Setlow P. 2009. Levels and localization of mechanosensitive channel proteins in Bacillus subtilis. Arch. Microbiol. 191:403–414 [DOI] [PubMed] [Google Scholar]
- 58. Wahome PG, Setlow P. 2008. Growth, osmotic downshock resistance and differentiation of Bacillus subtilis strains lacking mechanosensitive channels. Arch. Microbiol. 189:49–58 [DOI] [PubMed] [Google Scholar]
- 59. Welsh DT. 2000. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24:263–290 [DOI] [PubMed] [Google Scholar]
- 60. Whalen MB, Piggot PJ. 1997. Gain-of-function mutation of sapB that affects formation of alkaline phosphatase by Bacillus subtilis in sporulation conditions. Microbiology 143:577–583 [DOI] [PubMed] [Google Scholar]
- 61. Whatmore AM, Chudek JA, Reed RH. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527–2535 [DOI] [PubMed] [Google Scholar]
- 62. Whatmore AM, Reed RH. 1990. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136:2521–2526 [DOI] [PubMed] [Google Scholar]
- 63. Wood JM. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63:230–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wood JM, et al. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437–460 [DOI] [PubMed] [Google Scholar]