Abstract
Resistance to Bacillus thuringiensis Cry1Ac toxin was characterized in a population of Helicoverpa zea larvae previously shown not to have an alteration in toxin binding as the primary resistance mechanism to this toxin. Cry1Ac-selected larvae (AR1) were resistant to protoxins and toxins of Cry1Ab, Cry1Ac, and the corresponding modified proteins lacking helix α-1 (Cry1AbMod and Cry1AcMod). When comparing brush border membrane vesicles (BBMVs) prepared from susceptible (LC) and AR1 larval midguts, there were only negligible differences in overall Cry1Ac toxin binding, though AR1 had 18% reversible binding, in contrast to LC, in which all binding was irreversible. However, no differences were detected in Cry1Ac-induced pore formation activity in BBMVs from both strains. Enzymatic activities of two putative Cry1Ac receptors (aminopeptidase N [APN] and alkaline phosphatase [ALP]) were significantly reduced (2-fold and 3-fold, respectively) in BBMVs from AR1 compared to LC larvae. These reductions corresponded to reduced protein levels in midgut luminal contents only in the case of ALP, with an almost 10-fold increase in specific ALP activity in midgut fluids from AR1 compared to LC larvae. Partially purified H. zea ALP bound Cry1Ac toxin in ligand blots and competed with Cry1Ac toxin for BBMV binding. Based on these results, we suggest the existence of at least one mechanism of resistance to Cry1A toxins in H. zea involving binding of Cry1Ac toxin to an ALP receptor in the larval midgut lumen of resistant larvae.
INTRODUCTION
Insecticidal proteins derived from the entomopathogenic bacterium Bacillus thuringiensis have been exploited in agriculture for many years as a leading alternative or complement to chemical pest control agents. However, it was with the introduction of cry genes into plants (Bt crops) such as cotton (1996) and corn (1997) that the intensive use of B. thuringiensis proteins spread worldwide (23). Due to their high specificity, B. thuringiensis-based insecticidal technologies are safe to the environment and nontarget fauna. The most important concern with the wide adoption of transgenic Bt crops is the evolution of resistance in target pest populations. To avoid evolution of resistance to Bt crops, several insect resistance management (IRM) strategies are utilized: refugia (either “structured” or “natural”), a high dose (typically used when only one insecticidal compound is expressed in the crop), and/or simultaneous expression of multiple insecticidal compounds with diverse modes of action (gene pyramiding) (29).
Transgenic Bt cotton expressing Cry1Ac was commercialized in 1996 as Bollgard in the United States (9). The cotton bollworm, Helicoverpa zea Boddie, is one of the primary target pests of Bt cotton in the United States along with tobacco budworm (Heliothis virescens, F.) and pink bollworm (Pectinophora gossypiella, Saunders). However, H. zea is at higher risk for development of resistance than either H. virescens or P. gossypiella because the former is less susceptible to Cry1Ac and is also exposed to a similar B. thuringiensis protein (Cry1Ab) in Bt maize (22, 48). An additional protein (Cry2Ab) was commercialized in 2003 pyramided with Cry1Ac in Bollgard II to help reduce the risk of resistance evolution (and increase efficacy) against all three target pests of cotton (9, 41). Although current EPA-mandated monitoring has yet to detect any changes in H. zea susceptibility in the cotton-growing regions of the United States (10, 30), it is important to know how insects such as H. zea develop resistance to Cry proteins so that methods to suppress resistance mechanisms can be developed.
Resistance to Cry1A toxins in lepidopteran pests can result from alterations in any of the steps in the intoxication process, including protoxin solubilization, toxin activation, binding to receptors on the midgut brush border membrane, and pore formation, leading to osmotic cell death and disruption of the midgut (reviewed in reference 43). While alterations in toxin processing have been reported in some Cry-resistant insects, in most cases of laboratory selection, resistance relates to reduced toxin binding to midgut receptors (11). Although there have been numerous attempts to select and characterize resistance to Cry1Ac in H. zea, the only stable Cry1Ac-resistant H. zea population that has been at least partially characterized was reported by Anilkumar et al. (1, 2). This population (AR) displayed greater than 100-fold resistance to Cry1Ac toxin, but no changes were detected regarding Cry1Ac and Cry1Aa binding to midgut receptors (1), a major mechanism of Cry protein resistance (11).
In the present work, we have further explored potential Cry1Ac toxin resistance mechanisms in a Cry1Ac toxin-selected H. zea population (AR1). We have characterized this population in terms of cross-resistance to related Cry1A and modified Cry1A (Cry1AMod) toxins and protoxins, Cry1Ac binding properties, Cry1Ac pore formation activity, and enzymatic activities of two putative Cry1Ac receptors: aminopeptidase N (APN) and alkaline phosphatase (ALP) (36). Our data suggest that alterations in toxin receptor concentrations in the midgut lumen and brush border membranes are associated with Cry1A toxin resistance in H. zea.
MATERIALS AND METHODS
Insects and selection.
A laboratory-susceptible colony of H. zea was established in September 2004 from a laboratory colony from Monsanto (Union City, TN). A resistant strain (AR) resulted from the continuous selection of the laboratory colony on an artificial diet containing up to 500 μg Cry1Ac toxin/g diet for 25 generations (1). As is common with H. zea colonies in general and resistant colonies specifically, AR was crossed with the Monsanto susceptible strain (Union City, TN) in 2007 and reselected with Cry1Ac toxin (500 μg Cry1Ac toxin/g diet), which resulted in a strain designated as AR1 (3). This process was repeated in October 2010 using 100 μg Cry1Ac toxin/g diet from another source (similar in toxicity to 500 μg Cry1Ac toxin/g diet observed previously) (reference 3 and W. J. Moar, unpublished data). In both cases, AR1 displayed at least a 100-fold level of resistance compared to susceptible insects (reference 3 and Moar, unpublished).
All larvae from both strains were reared until ca. 18 to 24 h after molting into 5th instar for biochemical analyses; susceptible larvae (LC) were reared exclusively on an untreated artificial diet, while Cry1Ac-resistant larvae (AR1) were reared on an artificial diet containing 500 μg Cry1Ac toxin/g diet (2009) or 100 μg Cry1Ac toxin/g diet (2010) until 3rd instar (1), when survivors were transferred to an untreated artificial diet until 5th instar as described above.
B. thuringiensis Cry protein purification and biotin-labeling.
The cry1Ac protoxin gene from B. thuringiensis subsp. kurstaki strain HD-1 was expressed in Escherichia coli. The Cry1Ac protoxin was activated with trypsin, purified, lyophilized as indicated elsewhere (1), and used for selection.
Solubilized Cry1AbMod and Cry1AcMod protoxin and toxin (42), together with the corresponding wild-type Cry1Ab and Cry1Ac protoxin and toxin, were kindly provided by M. Soberón and A. Bravo (Instituto de Biotecnología, Universidad Nacional Autónoma de México, Cuernavaca, México). These protoxins and toxins were produced and purified as described in references 32 and 42.
For ligand blot experiments, Cry1Ac was produced and purified from B. thuringiensis strain HD-73, obtained from the Bacillus Genetic Stock Center (Columbus, OH). Toxin was solubilized, activated, and purified as described elsewhere (35). The activated toxin sample was quantified using the method of Bradford (5) with bovine serum albumin (BSA) as the standard and stored at −80°C until used. This purified Cry1Ac toxin (1 mg) was biotinylated using a 1:30 molar ratio of EZ link N-hydroxysuccinimide (NHS)-LC-biotin (Pierce) following the manufacturer's instructions. After biotinylation, labeled toxin samples were extensively dialyzed in 20 mM Na2CO3, pH 9.8, 150 mM NaCl at 4°C. Labeled toxins were quantified as above before use for ligand blotting.
Bioassays.
Neonate LC and AR1 were bioassayed using diet incorporation as described in reference 1. At least 50 μg/g diet of wild-type and modified Cry1Ab and Cry1Ac toxin and protoxin were used for bioassays based on availability and previous results with wild-type toxin and protoxin against susceptible H. zea (1). Each bioassay was replicated twice (Cry1Ab proteins) or three times (Cry1Ac proteins). All treatments for a particular Cry protein (Cry1Ab or Cry1Ac) were bioassayed concurrently for each replicate. Mortality in untreated controls in all replicates averaged less than 5% (data not shown). Percent mortality for all replicates was averaged.
Midgut isolation and sample preparation.
Actively feeding fifth-instar H. zea larvae on untreated diet were dissected in cold MET buffer (250 mM mannitol, 5 mM EGTA, 17 mM Tris-HCl, pH 7.5), and the isolated midguts were frozen in liquid nitrogen and stored at −80°C. Brush border membrane vesicles (BBMVs) were prepared by the differential centrifugation method (51). Final BBMV pellets were resuspended in one-half MET buffer at pH 7.2, quantified as described for Cry1Ac toxin, and frozen in liquid nitrogen and stored at −80°C until required. For the pore formation assays, freshly prepared BBMVs were resuspended in a small volume of MET buffer at pH 7.2 to obtain a final concentration of around 5 mg/ml.
To isolate midgut luminal contents, actively feeding fifth-instar H. zea larvae were cut longitudinally to dissect the midgut, which was opened lengthwise to isolate the lumen contents, which were immediately frozen in liquid nitrogen or on dry ice and stored at −80°C. Samples were collected from individual Cry1Ac-resistant and susceptible H. zea larvae in 2009 and in 2010 (separated by 17 to 19 generations). Midgut content samples were homogenized with a pestle in a microcentrifuge tube containing 100 μl of phosphate-buffered saline (PBS; 8 mM Na2HPO4, 2 mM KH2PO4, 150 mM NaCl, pH 7.4), vigorously vortexed, and centrifuged at 4°C (10 min at 16,000 × g). Supernatants were used for enzymatic assay measurements.
Purification of midgut HzALP.
Neonate H. zea larvae (Benzon Research, Carlisle, PA) were reared on tobacco budworm artificial diet (Bio-Serv, Frenchtown, NJ) at 26°C, 65% relative humidity (RH), and a 14-h-light:10-h-dark (14L:10D) photoperiod. Midguts from fifth-instar larvae were dissected and BBMVs purified as described above. Purified BBMVs were quantified using the Quant-iT protein assay kit (Invitrogen, Carlsbad, CA) with BSA as the standard, and then BBMVs (59 mg) were solubilized in PBS containing 1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} and Complete EDTA-free protease inhibitors (Roche) for 2 h at 4°C. Unsolubilized debris was removed by centrifugation (21,130 × g for 30 min at 4°C), and the supernatant dialyzed against buffer A (20 mM Tris-HCl, pH 8.5, plus 1 mM MgCl2). Solubilized BBMV proteins were fractionated by anion-exchange fast-performance liquid chromatography (FPLC) using a Mono Q 5/50 GL column (GE Healthcare) and a gradient of buffer A containing 1 M NaCl. The presence of ALP in the eluted fractions was determined using activity assays as described elsewhere (20), and fractions with high activity (eluted between 0.2 and 0.3 M NaCl) were pooled. Pooled fractions were concentrated using iCON 9-kDa molecular mass cutoff (MWCO) concentrators (Thermo Scientific, Rockford, IL) and then further purified using size exclusion chromatography in a HiLoad 16/60 Superdex 200 prep-grade column (GE Healthcare). The sample was eluted with 20 mM Tris-HCl, pH 8.5, plus 1 mM MgCl2. Fractions containing ALP activity, as determined by activity assays, were pooled as partially purified H. zea alkaline phosphatase (HzALP).
The presence of an alkaline phosphatase in the purified HzALP sample was confirmed using in-gel activity assays following SDS-PAGE under reducing conditions as described elsewhere (21). To detect HzALP in Western blots and test Cry1Ac binding using ligand blotting, partially purified HzALP (2 μg) was heat denatured at 95°C for 5 min in sample buffer (24) and loaded on SDS-10% polyacrylamide gels. After electrophoresis, proteins were silver stained (18) or transferred to polyvinylidene difluoride (PVDF) filters overnight at 20 V and 4°C. Upon transfer, filters were blocked for 1 h at room temperature in PBS containing 0.1% Tween 20 and 3% BSA. For ligand blots, purified Cry1Ac biotinylated toxin was used as described elsewhere (19). Filters were incubated with biotinylated Cry1Ac toxin alone (0.5 μg/ml) or in combination with 500-fold unlabeled Cry1Ac toxin for 1 h in blocking buffer. For Western blotting, blocked filters were probed with a 1:2,000 dilution of antisera against membrane-bound alkaline phosphatase (ALP) from Anopheles gambiae (kindly provided by G. Hua and M. Adang, University of Georgia, Athens, GA) or a 1:1,000 dilution of antisera against 130-kDa aminopeptidase N (APN) from Heliothis virescens (kindly provided by S. Gill, University of California, Riverside, CA) for 1 h in blocking buffer. Biotinylated toxin and ALP/APN antisera were detected on blocked filters using streptavidin and anti-rabbit antisera, respectively, conjugated to horseradish peroxidase, and developed using enhanced chemiluminescence (SuperSignal West Pico; Pierce, Rockford, IL).
Binding assays with 125I-labeled Cry1Ac.
Cry1Ac was labeled by incubating 25 μg of toxin with 0.5 mCi of [125I]NaI (Perkin Elmer, Madrid, Spain) using chloramine-T as previously described (49). Two different batches of labeled toxin were used throughout the study. Specific activities obtained were 10 and 3 mCi/mg. For binding assays, BBMVs were centrifuged for 10 min at 16,000 × g and resuspended in binding buffer (8 mM Na2HPO4, 2 mM KH2PO4, 150 mM NaCl, pH 7.4, 0.1% BSA) before the assay.
To check for the presence of specific binding and to determine the optimal concentration of BBMVs to use in competition and dissociation experiments, increasing amounts of BBMVs were incubated with 0.04 nM 125I-Cry1Ac in a final volume of 0.1 ml of binding buffer for 1 h at 25°C. An excess of unlabeled Cry1Ac toxin (0.3 μM) was used to calculate the nonspecific binding. After incubation, samples were centrifuged at 16,000 × g for 10 min and the pellet was washed once with 500 μl of cold binding buffer. Radioactivity retained in the pellet was measured in an LKB model 1282 CompuGamma CS gamma counter (LKB Wallac Pharmacia, Turku, Finland). Specific binding was calculated by subtracting nonspecific binding from total binding.
Competition experiments were conducted by incubating 3 μg of BBMV protein with a fixed amount of 125I-Cry1Ac (0.04 nM) in a final volume of 0.1 ml of binding buffer for 1 h at 25°C in the presence of increasing amounts of unlabeled Cry1Ac. Binding reactions were stopped by centrifugation as described above. Radioactivity retained in the BBMV pellet was measured in a gamma counter. Dissociation constants (Kd) and concentration of binding sites (Rt) were estimated using the LIGAND software (31).
For the dissociation experiments, binding reaction mixtures containing BBMV protein (3 μg) and 125I-Cry1Ac (0.04 nM) proceeded for 1 h, and then a 25-fold excess of unlabeled toxin was added to the mixtures. Upon addition of unlabeled toxin, reactions were stopped by centrifugation at different intervals. Nonspecific binding was determined by incubating an aliquot in the presence of unlabeled toxin (0.4 μM) added at the beginning of the experiment.
To measure the inhibition of 125I-Cry1Ac binding to BBMVs from larvae of the susceptible strain by partially purified HzALP at different times, 125I-Cry1Ac (0.04 nM) was preincubated with 5 μg/ml of HzALP for 30 min, 1 h, and 2 h at room temperature prior to initiating binding assays with the addition of 4 μg BBMVs. Specific binding was estimated by subtracting nonspecific binding (determined by adding an excess of unlabeled toxin) from total binding. The reaction was stopped by centrifugation as described above. Radioactivity retained in the BBMV pellet was measured in a gamma counter.
In a second set of experiments to measure inhibition of Cry1Ac binding by partially purified HzALP, 4 μg of BBMV protein was incubated with a fixed amount of 125I-Cry1Ac (0.04 nM) in a final volume of 0.1 ml of binding buffer for 1 h at 25°C in the presence of increasing amounts of unlabeled Cry1Ac or partially purified HzALP.
BBMV pore formation assays.
BBMV permeability to K+ was measured by recording the fluorescence quenching of the voltage-sensitive cyanine dye 3,3′-dipropylthiodicarbocyanine iodide DiSC3 (5) (Molecular Probes, Società Italiana Chimici, Italy). The basis of this method consists of the difference in the segregation of the dye in membrane vesicles, and thus in the change of fluorescence, in response to changes in the electrochemical gradient across the BBMV due to the flux of cations (40). Experiments were performed as per reference 37, with minor modifications. BBMV pellets were resuspended in a syringe containing a small volume of MET buffer at pH 7.2 for a final concentration of ca. 5 mg/ml. BBMVs (29 μg of protein) were preincubated for 30 min at 25°C in polyacryl cuvettes containing 1 ml of MET buffer, pH 7.2, 6 μM DiSC3 (5), and either 100 nM Cry1Ac toxin or toxin buffer (20 mM Tris-HCl at pH 8.6, 150 mM CsCl) as the control. After preincubation, cuvettes containing suspensions were transferred to a spectrofluorimeter (Cary Eclipse, Varian), and DiSC3 (5) fluorescence was measured using excitation and emission wavelengths of 645 and 665 nm, respectively. The ionophore valinomycin (23 μM) was added in the positive control samples just before the introduction of the cuvettes into the instrument. Extravesicular increments of K+ concentrations were made by three successive additions of 2 M KCl to give final concentrations of 40, 80, and 120 mM. Pore formation measurements were replicated four times and the results averaged. Measurements were replicated with two independent BBMV preparations.
Enzymatic assays.
Aminopeptidase N (APN) (E.C. 3.4.11.2) and alkaline phosphatase (ALP) (E.C.3.1.3.1) activities in homogenate, BBMV, and midgut content samples were determined spectrophotometrically as described before (37) by measuring the release of ρ-nitroaniline from l-leucine-ρ-nitroanilide in 40 mM Tris-HCl at pH 7.5 and of ρ-nitrophenol from ρ-nitrophenylphosphate in 1 M Tris-HCl at pH 8, respectively.
RESULTS
Cross-resistance to Cry1Mod in AR1 larvae.
Cry1AMod toxins have been reported to reduce resistance levels to B. thuringiensis proteins in some insect species (42, 46). Bioassays with LC and AR1 showed substantial differences in susceptibility to Cry1AbMod and Cry1AcMod toxin and protoxin between the two strains, and these differences were similar to those observed for Cry1Ab and Cry1Ac toxin and protoxin (Table 1). Modified toxins and protoxins were always substantially less toxic than their wild-type counterparts. These results show that AR1 is cross-resistant to both Cry1AbMod and Cry1AcMod (toxins and protoxins), indicating that the modified proteins do not overcome resistance in the AR1 strain.
Table 1.
Toxicity of wild-type and modified Cry1Ab and Cry1Ac toxins and protoxins against LC and AR1 Helicoverpa zeac
| Protein (concn)a | % mortalityb |
|
|---|---|---|
| LC | AR1 | |
| Cry1Ab toxin (50) | 87.5 | 7.5 |
| Cry1AbMod toxin (50) | 18.9 | 4.5 |
| Cry1Ab protoxin (100) | 100 | 7.5 |
| Cry1AbMod protoxin (100) | 55 | 2.5 |
| Cry1Ac toxin (64) | 98 | 33 |
| Cry1AcMod toxin (64) | 32 | 6.3 |
| Cry1Ac protoxin (94) | 92 | 45 |
| Cry1AcMod protoxin (94) | 30 | 14.7 |
The highest concentration (μg protein/gram diet) tested is indicated in parentheses. Two replicates comprising 40 insects tested for all Cry1Ab treatments. Three replicates comprising 48 insects tested for all Cry1Ac treatments.
Mortality evaluated as dead insects plus insects failing to molt to 2nd instar after 7 days.
LC, susceptible; AR1, Cry1Ac resistant.
Binding of 125I-labeled Cry1Ac to BBMVs from susceptible (LC) and resistant (AR1) larvae.
Binding of 125I-labeled Cry1Ac was evaluated at increasing concentrations of BBMVs from both H. zea strains. Specific 125I-Cry1Ac binding was observed for BBMVs from LC and AR1, with just small quantitative differences. Specific binding was slightly higher in BBMVs from the LC (maximum 25%) compared to the specific binding in AR1 (maximum 19%) at all tested BBMV concentrations (Fig. 1).
Fig 1.
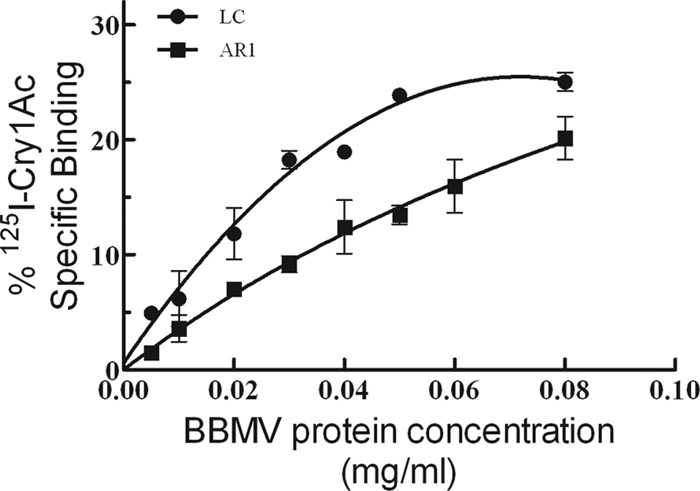
Specific binding of 125I-labeled Cry1Ac as a function of BBMV protein concentration from midguts of susceptible (LC) and Cry1Ac-resistant (AR1) H. zea larvae. Each data point is the mean of at least two replicates, and the bars indicate the standard error of the mean.
Binding competition experiments showed similar displacement curves of 125I-Cry1Ac binding to BBMVs from the susceptible (LC) and resistant (AR1) strains (Fig. 2). The quantitative binding parameters estimated from the curves are shown in Table 2. Only negligible differences were found in equilibrium dissociation constant (Kd) and binding site concentration (Rt) among strains.
Fig 2.
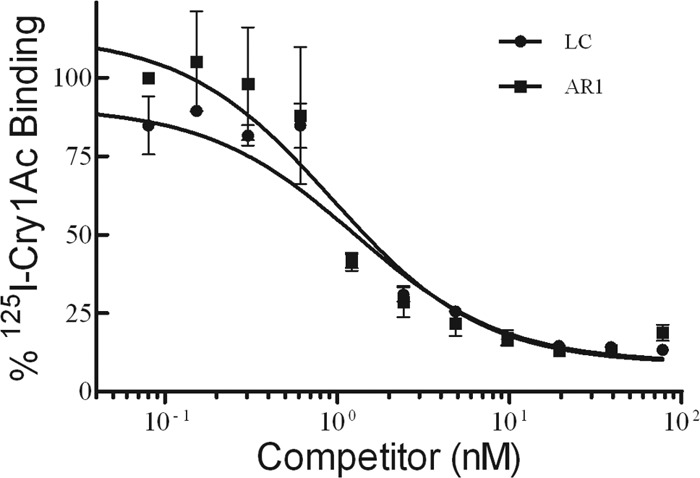
Binding of 125I-labeled Cry1Ac to BBMVs from susceptible (LC) and Cry1Ac-resistant (AR1) H. zea larvae at increasing concentrations of unlabeled toxin. Each data point is a mean of three replicates. The bars indicate the standard error of the mean.
Table 2.
Kd and Rt values calculated from the competition assays with BBMVs from LC and AR1 H. zea larvaec
Mean ± SEM.
Values are expressed in picomoles per milligram of BBMV protein.
LC, susceptible; AR1, Cry1Ac-resistant.
Dissociation kinetics of 125I-Cry1Ac binding to BBMVs.
Because we did not detect relevant differences in Cry1Ac binding affinity, we next examined the irreversibility of 125I-Cry1Ac binding to BBMVs from susceptible and resistant H. zea larvae by measuring the amount of bound toxin remaining over time after dilution with an excess of unlabeled homologous competitor. In these conditions, unlabeled toxin should displace the reversibly bound 125I-Cry1Ac but not the fraction bound irreversibly. After reaching equilibrium (1 h), the addition of excess unlabeled Cry1Ac to the reaction did not affect the amount of bound 125I-Cry1Ac to BBMVs from LC, suggesting that all detected binding was irreversible (Fig. 3). In contrast, in AR1 the reversible component of the binding accounted for about 18% of the total binding (Fig. 3).
Fig 3.
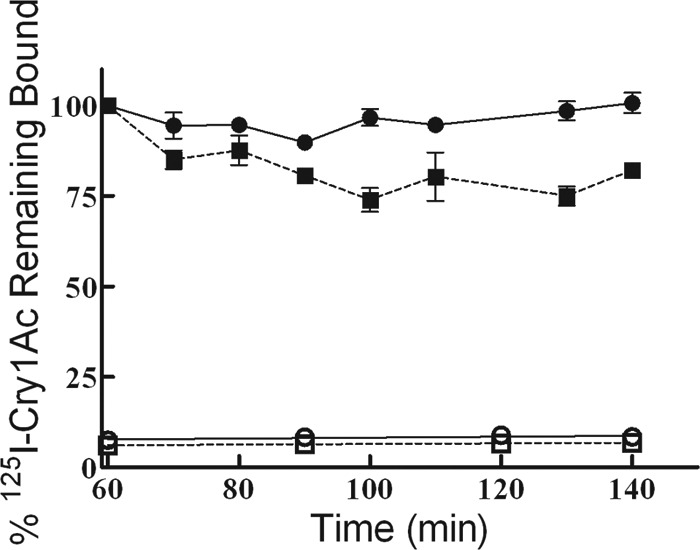
Dissociation kinetics of the bound 125I-labeled Cry1Ac to BBMVs from susceptible (circles, solid lines) and Cry1Ac-resistant (squares, broken lines) H. zea larvae. After 60 min of incubation, a 25-fold excess of unlabeled Cry1Ac was added to the reaction mixture, and the radioactivity in the pellet was measured at different times (filled symbols). The nonspecific binding (open symbols) was determined by adding an excess of unlabeled toxin at the beginning of the assay. Values represent the binding as a percentage of the total binding after 60 min of association binding. Each data point is a mean of four replicates, and the bars indicate the standard error of the mean.
Pore formation.
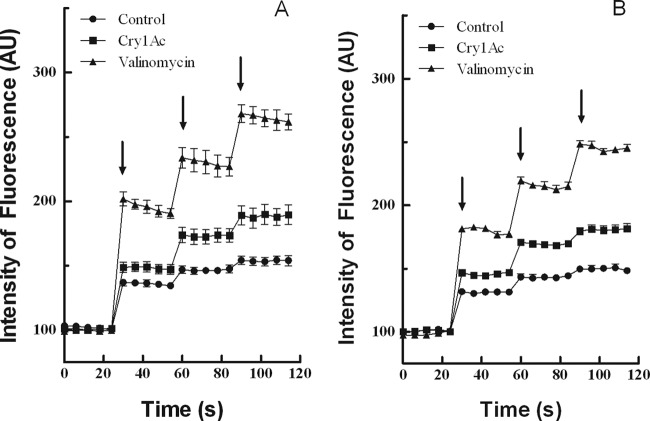
To determine whether detected differences in 125I-Cry1Ac irreversible binding resulted in altered Cry1Ac toxin pore formation, we monitored toxin-induced permeation of BBMVs as described previously (37). As expected due to the intrinsic BBMV permeability to K+, fluorescence intensity increased in control samples upon the addition of KCl (Fig. 4). As a positive control for permeation, we used the K+ ionophore valinomycin, which gave a maximal increase in fluorescence intensity corresponding to the free influx of K+ (Fig. 4). When BBMVs from LC and AR1 larvae were preincubated with 100 nM Cry1Ac toxin, a substantial increase in fluorescence intensity compared to negative controls was observed (Fig. 4). However, no differences were detected at the level of BBMV permeabilization by Cry1Ac when comparing vesicles from LC and AR1.
Fig 4.
Effect of Cry1Ac toxin on K+ permeability of BBMVs from the midgut of LC (susceptible) (A) and AR1 (Cry1Ac-resistant) (B) H. zea larvae. BBMVs, preloaded with MET buffer at pH 7.2, were preincubated at room temperature with the buffer in which the toxin was dissolved (control and valinomycin samples) or with 100 nM Cry1Ac in the cuvette with 1 ml of the preloading buffer supplemented with 6 μM DiSC3 (5). After preincubation, the cuvettes with the suspensions were transferred to the spectrophotometer, and the recording of the fluorescence started. The ionophore valinomycin was added to the cuvettes of positive controls at the moment of the introduction in the instrument. KCl was added at the times indicated by the arrows to obtain extravesicular final concentrations of 40, 80, and 120 mM. Each trace represents the mean ± SEM of at least four replicates. Each experiment was repeated for two independent BBMV preparations. AU, arbitrary fluorescence units.
Comparison of APN and ALP activities in midgut.
We quantified APN- and ALP-specific activity in midgut homogenate, brush border membrane, and midgut lumen contents obtained from LC and AR1 5th-instar larvae to compare levels of these putative receptors. In midgut homogenates, APN activity was only slightly higher in AR1, whereas ALP activity in AR1 was approximately 50% reduced compared to that for LC (Table 3). This reduction in ALP activity in midgut homogenates corresponded with reduced ALP levels (about 3-fold) in BBMVs from larvae of the AR1 strain, while APN levels were reduced about 2-fold in these vesicles compared to samples from LC larvae (Table 3). Most notably, when testing enzymatic activities in midgut luminal contents, ALP specific activity was significantly increased (almost 10-fold) in midgut contents from AR1 compared to LC larvae (Student's t test, P < 0.001), while APN activity was not significantly different (Table 3).
Table 3.
APN and ALP activity in homogenates, BBMV, and midgut contents from LC and AR1 H. zea larvaea
| Strain | APN |
ALP |
||||
|---|---|---|---|---|---|---|
| H | BBMV | MC | H | BBMV | MC | |
| LC | 867 ± 26 (12) | 10,523 ± 384 (11) | 975 ± 173 (33) | 157 ± 4 (10) | 1,701 ± 65 (11) | 127 ± 17 (27) |
| AR1 | 1,134 ± 59 (8)* | 5,278 ± 83 (11)* | 1,121 ± 87 (47) | 83 ± 14 (8)* | 518 ± 35 (17)* | 1,194 ± 110 (60)* |
LC, susceptible; AR1, Cry1Ac resistant. H, homogenates; MC, midgut contents. Activities are expressed in mU/mg of proteins and the values reported are the mean ± SEM, with the number of repetitions in parenthesis. Student's t test versus LC: *, P < 0.001.
Role of HzALP in Cry1Ac binding and resistance.
The detected increased levels of ALP in luminal midgut contents from AR1 larvae suggested a role for solubilized ALP as a potential binding site for Cry1Ac, possibly preventing or reducing interactions between Cry1Ac and the brush border membrane. To test this hypothesis, we partially purified HzALP from susceptible H. zea BBMVs and determined its Cry1Ac toxin binding capabilities. Immunoblots detected a band of the predicted size for HzALP (∼68 kDa) in our partially purified sample (Fig. 5B), which was the only protein band in the partially purified HzALP sample that displayed ALP activity in activity assays (data not shown). As shown in Fig. 5A, the partially purified HzALP sample also contained protein bands of approximately 110 kDa and 120 kDa. Because ALP and APN are usually reported to copurify (38), we predicted that these higher-molecular-size proteins were APN. Immunoblots with antisera to the 130-kDa APN of H. virescens (33) recognized the 110-kDa and 120-kDa bands (Fig. 5C) as well as other smaller protein bands that were weakly detected by silver staining. In ligand blots with biotinylated Cry1Ac, we observed that most of the toxin bound to the ALP band (Fig. 5D), although binding was also observed with the 110-kDa APN band. Competition of Cry1Ac binding using a 500-fold excess of unlabeled Cry1Ac eliminated toxin binding to ALP, while binding to the APN band was almost unaffected (Fig. 5E). These data supported Cry1Ac binding to ALP and APN proteins in our partially purified HzALP sample, although only the ALP protein bound toxin specifically.
Fig 5.
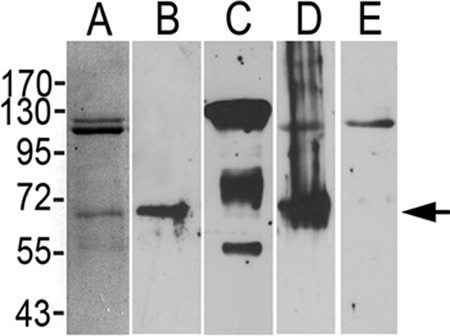
Characterization of partially purified H. zea ALP. Partially purified HzALP samples (2 μg to 5 μg) were silver stained for total protein (A) or transferred to PVDF filters and probed with antisera against ALP of A. gambiae (B), antisera against 130-kDa APN from H. virescens (C), biotinylated Cry1Ac alone (D), or biotinylated Cry1Ac and a 500-fold excess of unlabeled Cry1Ac (E). Arrow indicates the position of the HzALP protein band for reference.
To test the effect of soluble HzALP on the amount of Cry1Ac toxin binding to BBMVs, we performed in vitro binding assays with 125I-Cry1Ac and partially purified HzALP. When 125I-Cry1Ac was preincubated for different times with 5 μg/ml partially purified HzALP prior to the addition of BBMVs to the binding reaction, we observed a significant reduction (up to 17.3% at 2 h, Student's t test, P < 0.001) in Cry1Ac binding to BBMVs compared to controls in the absence of HzALP (Fig. 6). However, when testing binding of 125I-Cry1Ac to BBMVs as a function of increasing concentrations of partially purified HzALP (without any previous preincubation) or unlabeled Cry1Ac (2.5 ng/ml to 5 μg/ml), we detected no inhibition of 125I-Cry1Ac binding when using HzALP as the competitor (Fig. 7).
Fig 6.
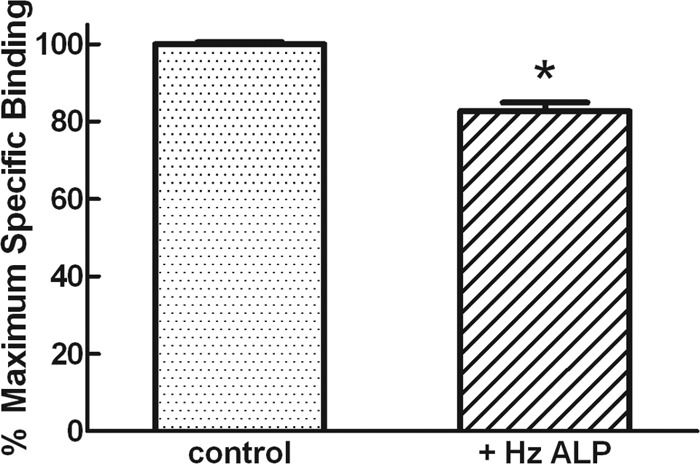
Specific binding of 125I-labeled Cry1Ac to BBMVs from LC (susceptible) H. zea in the absence or in the presence of 5 μg/ml of HzALP. 125I-labeled Cry1Ac was incubated for 2 h with HzALP prior to initiating the binding experiment. Each point bar is the mean of at least 10 determinations, and the bars show standard error of the mean. Student's t test versus control: *, P < 0.001.
Fig 7.
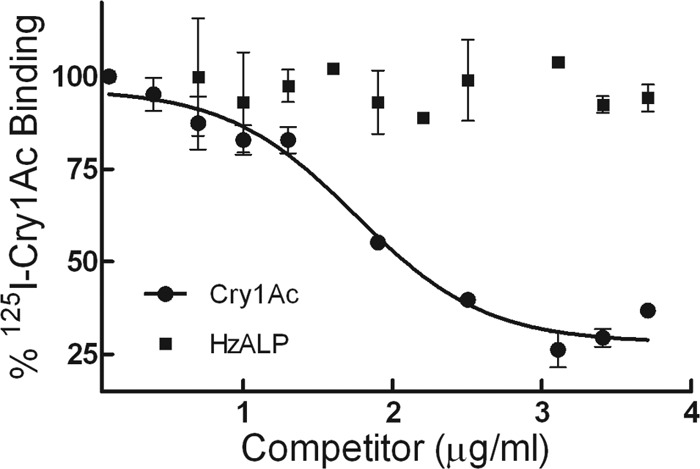
Binding of 125I-labeled Cry1Ac to BBMVs from LC (susceptible) H. zea at increasing concentrations of unlabeled toxin (Cry1Ac) and partially purified alkaline phosphatase (HzALP). Each data point is a mean of three replicates. The bars indicate the standard error of the mean.
DISCUSSION
Altered binding of Cry1 toxins to their midgut receptors is the most commonly described mechanism conferring the highest levels of resistance, and it is the only described mechanism for populations that evolved resistance to B. thuringiensis Cry proteins in field or semifield conditions (8, 11, 12, 39, 45, 50, 52). Alternative resistance mechanisms described to date include alterations in protoxin activation (34), mutations in ABC transporter proteins (14), toxin degradation (13), faster replacement of midgut epithelial cells (28), elevated immune response (16), and toxin sequestration by esterases (15) or lipophorin (27).
To characterize Cry1Ac resistance in our H. zea colony, we first investigated the binding of Cry1Ac in BBMVs from susceptible and resistant insects. Although Anilkumar et al. (1) reported no binding differences between the susceptible and the Cry1Ac-resistant AR strain of H. zea, subsequent backcrossing of this resistant colony to a laboratory-susceptible colony that had received an influx of wild individuals warranted a repeat of the binding experiments. Specific binding and associated parameters (Rt and Kd) obtained in this study were similar to those obtained by Anilkumar et al. (1) and differed only marginally between LC and AR1. Therefore, we conclude (and as reported previously with the AR resistant strain [1]) that the slight observed differences in overall binding between LC and AR1 cannot explain the >100-fold level of resistance observed in AR1. Additionally, these results suggest that the addition of wild H. zea into the laboratory colony had little impact, if any, on the resistance binding phenotype of the selected colony.
As expected from previous reports of cross-resistance to Cry1Ab in AR larvae (1), AR1 larvae were cross-resistant to Cry1Ab. Most notably, AR1 larvae were also cross-resistant to Cry1AbMod and Cry1AcMod protoxins and toxins (42). This is the first report documenting resistance to both Cry1AMod toxins and protoxins. Cry1AMod protoxins have been shown to reduce Cry1A resistance in eight of nine insect strains tested, involving six lepidopteran species (46). Contrary to a previous hypothesis (42), this reduced resistance by Cry1AMod toxins was independent of mutations in the gene coding for the cadherin receptor (46). However, a shared feature of all insect strains for which Cry1AbMod and Cry1AcMod could overcome resistance is that they have moderately to severely reduced binding of at least one Cry1A toxin (46). More recently, a mutation in an ABC transporter protein has been linked to Cry1Ac resistance in strains of three insect species with severely reduced Cry1Ac binding (4, 14). Interestingly, Cry1A resistance was significantly reduced in two of these three species using Cry1AbMod or Cry1AcMod protoxins (42, 46). However, this was not the case with either Cry1AbMod or Cry1AcMod toxin or protoxin against AR1, which is consistent with previous reports that a major alteration in binding is probably not the predominant mechanism of resistance in this species. Cross-resistance to Cry1AMod toxins and protoxins, along with the lack of binding alteration, suggests that the mechanism of Cry1Ac resistance in AR1 is different from those conferring resistance in lepidopteran species susceptible to Cry1AMod toxins.
High levels of cross-resistance in Cry1Ac toxin-resistant H. zea do not extend to protoxin forms of Cry1Ac (MVP II and purified protoxin) (reference 1 and Moar, unpublished). Although results in Table 1 suggest relatively high levels of cross-resistance to protoxin (especially Cry1Ab), the use of a single relatively low concentration of protein (to document susceptibility to Cry1AMod proteins) typically does not provide the accuracy of a dose-response curve. Anilkumar et al. (1) suggested that this observation may be indicative of a differential activation of protoxin in the resistant insect midgut or that the C-terminal end of the full-length protein contained in the protoxin may protect the active toxin from the degradative action of midgut proteases, resulting in a higher yield of the fully active toxin. Results presented here, in which AR1 does not lose resistance in the presence of modified toxin or protoxin (these toxins were produced by deleting part of the N-terminal portion of the protein [42] where there is relatively little cleavage during initial proteolysis [6]), is consistent with the hypothesis that the C-terminal end of the protoxin may provide protection from midgut proteases found in Cry1Ac-resistant H. zea (1).
Dissociation kinetics of Cry1Ac binding showed that, in contrast to the complete irreversibility of binding in LC BBMVs, binding to AR1 BBMVs is characterized by a partial (18%) reversibility. This might be due to a reduction in second-step receptors (such as membrane-bound APN and ALP) responsible for the insertion of the oligomeric protein into the brush border membrane (7). We thus analyzed pore formation activity of Cry1Ac in both strains, which reflect the capacity of the irreversibly bound toxin to permeabilize BBMVs to potassium ions and, indirectly, the capacity of BBMVs to elicit oligomerization of the toxin. Despite the difference in irreversible binding between LC and AR1 BBMVs, our results showed similar levels of Cry1Ac permeabilization with BBMVs from both strains, indicating no major effect on the oligomerization and subsequent membrane permeabilization process, at least in vitro.
Mutations or altered expression of APN or ALP has previously been associated with Cry1 resistance (17, 20, 53). More recently, reduced expression of ALP has been proposed as a B. thuringiensis resistance biomarker in Lepidoptera (21), although the specific role of reduced ALP expression in resistance has not been described in detail. Interestingly, we found a significant decrease in APN and ALP activity in BBMVs from AR1 compared to LC. This result is in agreement with the detected reduction in irreversible binding, as both APN and ALP are considered to be responsible for localizing Cry1A toxin oligomers to lipid rafts (54) and the subsequent toxin insertion into the membrane (irreversible binding). Reduced ALP levels have previously been detected in BBMVs from Cry1A-resistant H. virescens larvae (20, 21). Interestingly, we also detected a 10-fold-increased ALP activity in the midgut lumen of AR1 compared to LC larvae, while APN activity in these samples was similar. The fact that we observed similar results of ALP and APN enzyme activity in two populations of Cry1Ac-resistant H. zea that were separated by 17 to 19 generations, including a backcross with a laboratory colony and subsequent selection with a different Cry1Ac toxin source, suggests that these differences in enzyme activity are strongly associated with B. thuringiensis resistance. These observations led us to hypothesize a possible relationship between the increased presence of ALP in the lumen and Cry1Ac resistance.
In particular, because the depletion of ALP activity in the brush border of resistant larvae is reflected in its increase in the midgut lumen, it is reasonable to assume the presence of a receptor-shedding mechanism from the brush border of resistant larvae. Receptor shedding is an already described phenomenon consequent to Cry1 exposure for Lymantria dispar (47) and Spodoptera exigua (15) larvae. After intoxication of L. dispar larvae with different Cry1 and Cry2 toxins, a massive shedding of glycosylphosphatidylinositol (GPI)-anchored APN and ALP into the midgut lumen was observed. Also S. exigua larvae exposed to Cry1Ca showed a high increase of APN activity in the midgut lumen (17). The role of the observed receptor shedding in response to Cry1 exposure has not been clarified yet, although Los et al. (25) recently showed RAB-11-dependent expulsion of microvilli from the apical side of intestinal epithelial cells in Cry5B-intoxicated nematode Caenorhabditis elegans. Cleavage of APN from the midgut brush border membrane after toxin binding by an endogenous phospholipase C was speculated as a potential resistance mechanism in Heliothis virescens (26). In our case, the shedding of ALP is probably not related to Cry1Ac exposure but rather to Cry1Ac resistance, because exposure to Cry1Ac toxin was stopped after 7 days (3rd instar) and experiments were performed in 5th-instar larvae (ca. an additional 5 days); molting at the end of the 3rd and 4th instar should have removed most/all Cry toxin from the gut. Furthermore, the observation that ALP levels, but not APN, are highly increased in the lumen suggests a selective mechanism targeting ALP shedding rather than the membrane expulsion mechanism observed in C. elegans (25). Moreover, the higher differences in ALP levels between susceptible and resistant larvae detected for midgut fluids compared to BBMVs may suggest the increased synthesis and secretion of a soluble form of ALP from the midgut cells in resistant larvae. While reductions in ALP but not APN levels in BBMVs have been reported in diverse resistant strains (21), the potential increase in ALP levels in the midgut lumen of these insects was not tested. Our data add an important piece to the puzzle on ALP involvement in Cry resistance and open a new perspective on markers to be used in the field to monitor resistance. Further studies on other resistant colonies would be pivotal to clarify whether ALP increase in the midgut lumen of resistant insects could be used as marker of resistance as ALP expression on the brush border.
Once in the lumen of resistant larvae, ALP might interact with Cry1Ac to reduce toxin binding to the brush border membrane, as has also been suggested for esterases and Cry1Ac resistance in Helicoverpa armigera (15). Our ligand blotting results support that Cry1Ac binds specifically to ALP in H. zea BBMVs, while binding to a copurifying APN protein seemed nonspecific. In agreement with this observation, preincubation of Cry1Ac toxin with HzALP resulted in reduced Cry1Ac binding to BBMVs, although the reduction was only moderate. However, complete inhibition of Cry1Ac toxin binding to the BBMVs by ALP would not be expected, considering that Cry1Ac binding to the BBMVs is not exclusively mediated by ALP and that the affinity of the toxin for this receptor is much lower than for the cadherin primary receptor (7). The low-affinity interaction with ALP in the midgut lumen could account for a sufficient hindrance in the binding of Cry1Ac to its receptors on the microvillar membrane, preventing Cry1Ac crossing through the peritrophic membrane. A reduced amount of binding could be critical for resistance because it may allow gut-healing mechanisms to recover from the weak damage produced by the toxin that was able to bind (28, 44). Additionally, because ALP does bind specifically to Cry1Ac toxin and not protoxin, and AR1 is resistant to toxin with little cross-resistance to protoxin, ALP might also play a role in changing the three-dimensional profile of the Cry1Ac toxin, exposing additional protease cleavage sites not found in susceptible insects. Based on these observations, we propose one potential hypothesis that ALP in the midgut lumen contributes to Cry1Ac resistance by binding to Cry1Ac toxin, thus reducing the amount of Cry1Ac toxin available to interact with the surface of the midgut cells as well as changing the three-dimensional conformation of the Cry1Ac toxin allowing additional proteolysis to occur. Further studies are under way to investigate alterations in proteolytic degradation of Cry1Ac toxin in AR1.
The characterization of Cry1Ac resistance reported herein and elsewhere suggests that resistance is complex and possibly polygenic, in which the final result of preventing binding of the toxin to the midgut epithelial membrane is not accomplished by a major alteration of the overall binding affinity to the membrane binding sites. Instead, the increase in ALP within the midgut lumen is most likely a mechanism contributing to resistance to Cry1Ac in the AR1 strain of H. zea and would represent a novel resistance mechanism that should be considered in alternative models.
ACKNOWLEDGMENTS
We thank N. Adams, Monsanto Co., Union City, TN, for help in rearing H. zea; and S. Penn and J. Fridley, Monsanto Co., St. Louis, MO, for technical support. We thank M. Soberón and A. Bravo (Universidad Nacional Autonoma de Mexico, Cuernavaca, Mexico) for providing us wild-type and modified Cry1Ab and Cry1Ac trypsin-activated and full-length proteins and R. de Maagd (Plant Research International, Wageningen, The Netherlands) for the E. coli Cry1Ac-producing clone.
This research was partially supported by the Biotechnology Risk Assessment Program competitive grant no. 2008-39211-19577 from the USDA National Institute of Food and Agriculture (NIFA). Support for this project at the University of Valencia was provided by the Spanish Ministry of Science and Innovation (AGL2006-11914 and AGL2009-13340-C02-01), by the Generalitat Valenciana (GVARVIV2007-090 and ACOMP/2009/313), and by European FEDER funds. S.C. was supported by a Marie Curie grant (contract no. PIEF-GA-2008-219993) from the EU.
Footnotes
Published ahead of print 8 June 2012
REFERENCES
- 1. Anilkumar KJ, et al. 2008. Production and characterization of Bacillus thuringiensis Cry1Ac-resistant cotton bollworm Helicoverpa zea (Boddie). Appl. Environ. Microbiol. 74:462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anilkumar KJ, Pusztai-Carey M, Moar WJ. 2008. Fitness costs associated with Cry1Ac-resistant Helicoverpa zea (Lepidoptera: Noctuidae): a factor countering selection resistance to Bt cotton? J. Econ. Entomol. 101:1421–1431 [DOI] [PubMed] [Google Scholar]
- 3. Anilkumar KJ, et al. 2009. Synergistic interactions between Cry1Ac and natural cotton defenses limit survival of Cry1Ac-resistant Helicoverpa zea (Lepidoptera: Noctuidae) on Bt cotton. J. Chem. Ecol. 35:785–795 [DOI] [PubMed] [Google Scholar]
- 4. Baxter WS, et al. 2011. Parallel evolution of Bt toxin resistance in Lepidoptera. Genetics 189:675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. J. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6. Bravo A, Sanchez J, Kouskoura T, Crickmore N. 2002. N-terminal activation is an essential early step in the mechanism of action of the Bacillus thuringiensis Cry1Ac toxin. J. Biol. Chem. 277:23985–23987 [DOI] [PubMed] [Google Scholar]
- 7. Bravo A, et al. 2004. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim. Biophys. Acta 1667:38–46 [DOI] [PubMed] [Google Scholar]
- 8. Caccia S, et al. 2010. Binding site alteration is responsible for field-isolated resistance to Bacillus thuringiensis Cry2A insecticidal proteins in two Helicoverpa species. PLoS One 5:e9975 doi:10.1371/journal.pone.0009975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chitkowski RL, Turnipseed SG, Sullivan MJ, Bridges WC. 2003. Field and laboratory evaluations of transgenic cottons expressing one or two Bacillus thuringiensis var. kurstaki Berliner proteins for management of noctuid (Lepidoptera) pests. J. Econ. Entomol. 96:755–762 [DOI] [PubMed] [Google Scholar]
- 10. Dennehy TJ, et al. 2011. 2010-Season update on monitoring of resistance to Bt cotton in key lepidopteran pests in the U.S.A., p 1061–1062 Proc. Beltwide Cotton Conf. 2011 National Cotton Council of America, Memphis, TN [Google Scholar]
- 11. Ferré J, Van Rie J. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47:501–533 [DOI] [PubMed] [Google Scholar]
- 12. Ferré J, Van Rie J, MacIntosh SC. 2008. Insecticidal genetically modified crops and insect resistance management (IRM), p 41–85 In Romeis J, Shelton AM, Kennedy GG. (ed). Integration of insect resistant genetically modified crops within IPM programs. Springer, New York, NY [Google Scholar]
- 13. Forcada C, Alcácer E, Garcerá MD, Martínez R. 1996. Differences in the midgut proteolytic activity of two Heliothis virescens strains, one susceptible and one resistant to Bacillus thuringiensis toxins. Arch. Insect Biochem. Physiol. 31:257–272 [DOI] [PubMed] [Google Scholar]
- 14. Gahan LJ, Pauchet Y, Vogel H, Heckel DG. 2010. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6:e1001248 doi:10.1371/journal.pgen.1001248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gunning RV, Dang HT, Kemp FC, Nicholson IC, Moores GD. 2005. New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl. Environ. Microbiol. 71:2558–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hernández-Martínez P, et al. 2010. Constitutive activation of the midgut response to Bacillus thuringiensis in Bt-resistant Spodoptera exigua. PLoS One 5:e12795 doi:10.1371/journal.pone.0012795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrero S, Gechev T, Bakker PL, Moar WJ, de Maagd RA. 2005. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four aminopeptidase N genes. BMC Genomics 6:96 doi:10.1186/1471-2164-6-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heukeshoven J, Dernick R. 1985. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6:103–112 [Google Scholar]
- 19. Jurat-Fuentes JL, Adang MJ. 2001. Importance of Cry1 delta-endotoxin domain II loops for binding specificity in Heliothis virescens (L.). Appl. Environ. Microbiol. 67:323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jurat-Fuentes JL, Adang MJ. 2004. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 271:3127–3135 [DOI] [PubMed] [Google Scholar]
- 21. Jurat-Fuentes JL, et al. 2011. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS One 6:e17606 doi:10.1371/journal.pone.0017606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karim S, Riazuddin S, Gould F, Dean DH. 2000. Determination of receptor binding properties of Bacillus thuringiensis delta-endotoxins to cotton bollworm (Helicoverpa zea) and pink bollworm (Pectinophora gossypiella) midgut brush border membrane vesicles. Pestic. Biochem. Physiol. 67:198–216 [Google Scholar]
- 23. Kumar S, Chandra A, Pandey KC. 2008. Bacillus thuringiensis (Bt) transgenic crop: an environment friendly insect-pest management strategy. J. Environ. Biol. 29:641–653 [PubMed] [Google Scholar]
- 24. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 25. Los FCO, et al. 2011. RAB-5 and RAB-11 dependent vesicle-trafficking pathways are required for plasma membrane repair after attack by bacterial pore-forming toxin. Cell Host Microbe 9:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo K, et al. 1997. The Heliothis virescens 170 kDa aminopeptidase functions as “receptor A” by mediating specific Bacillus thuringiensis Cry1A delta-endotoxin binding and pore formation. Insect Biochem. Mol. Biol. 27:735–743 [DOI] [PubMed] [Google Scholar]
- 27. Ma G, et al. 2005. Is the mature endotoxin Cry1Ac from Bacillus thuringiensis inactivated by a coagulation reaction in the gut lumen of resistant Helicoverpa armigera larvae? Insect Biochem. Mol. Biol. 35:729–739 [DOI] [PubMed] [Google Scholar]
- 28. Martínez-Ramírez AC, Gould F, Ferré J. 1999. Histopathological effects and growth reduction in a susceptible and a resistant strain of Heliothis virescens (Lepidoptera: Noctuidae) caused by sublethal doses of pure Cry1A crystal proteins from Bacillus thuringiensis. Biocontrol. Sci. Technol. 9:239–246 [Google Scholar]
- 29. Moar WJ, Anilkumar KJ. 2007. Plant science: the power of the pyramid. Science 318:1561–1562 [DOI] [PubMed] [Google Scholar]
- 30. Moar WJ, Dennehy T, Anilkumar K, Head G. 2010. Bt resistance in Helicoverpa zea (Boddie): from biology to monitoring. Southwest. Entomol. 35:395–398 [Google Scholar]
- 31. Munson P, Rodbard D. 1980. LIGAND: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 107:220–239 [DOI] [PubMed] [Google Scholar]
- 32. Muñoz-Garay C, et al. 2009. Characterization of the mechanism of action of the genetically modified Cry1AbMod toxin that is active against Cry1Ab-resistant insects. Biochim. Biophys. Acta 1788:2229–2237 [DOI] [PubMed] [Google Scholar]
- 33. Oltean DI, Pullikuth AK, Lee HK, Gill SS. 1999. Partial purification and characterization of Bacillus thuringiensis Cry1A toxin receptor A from Heliothis virescens and cloning of the corresponding cDNA. Appl. Environ. Microbiol. 65:4760–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oppert B, Kramer KJ, Johnson DE, MacIntosh SC, McGaughey WH. 1994. Altered protoxin activation by midgut enzymes from a Bacillus thuringiensis resistant strain of Plodia interpunctella. Biochem. Biophys. Res. Commun. 198:940–947 [DOI] [PubMed] [Google Scholar]
- 35. Perera OP, Willis JD, Adang MJ, Jurat-Fuentes JL. 2009. Cloning and characterization of the Cry1Ac-binding alkaline phosphatase (HvALP) from Heliothis virescens. Insect Biochem. Mol. Biol. 39:294–302 [DOI] [PubMed] [Google Scholar]
- 36. Pigott CR, Ellar DJ. 2007. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71:255–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodrigo-Simón A, Caccia S, Ferré J. 2008. Bacillus thuringiensis Cry1Ac toxin-binding and pore-forming activity in brush border membrane vesicles prepared from anterior and posterior midgut regions of lepidopteran larvae. Appl. Environ. Microbiol. 74:1710–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sangadala S, Walters FS, English LH, Adang MJ. 1994. A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA (c) toxin binding and 86Rb(+)-K+ efflux in vitro. J. Biol. Chem. 269:10088–10092 [PubMed] [Google Scholar]
- 39. Sayyed AH, Haward R, Herrero S, Ferré J, Wright DJ. 2000. Genetic and biochemical approach for characterization of resistance to Bacillus thuringiensis toxin Cry1Ac in a field population of the diamondback moth, Plutella xylostella. Appl. Environ. Microbiol. 66:1509–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sims PJ, Waggoner AS, Wang C-H, Hoffman JF. 1974. Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry 13:3315–3330 [DOI] [PubMed] [Google Scholar]
- 41. Sivasupramaniam S, et al. 2008. Toxicity and characterization of cotton expressing Bacillus thuringiensis Cry1Ac and Cry2Ab2 proteins for control of lepidopteran pests. J. Econ. Entomol. 101:546–554 [DOI] [PubMed] [Google Scholar]
- 42. Soberón M, et al. 2007. Engineering modified Bt toxins to counter insect resistance. Science 318:1640–1642 [DOI] [PubMed] [Google Scholar]
- 43. Soberón M, Gill SS, Bravo A. 2009. Signaling versus punching hole: how do Bacillus thuringiensis toxins kill insect midgut cells? Cell. Mol. Life Sci. 66:1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spies AG, Spence KD. 1985. Effect of sublethal Bacillus thuringiensis crystal endotoxin treatment on the larval midgut of a moth, Manduca: SEM study. Tissue Cell 17:379–394 [DOI] [PubMed] [Google Scholar]
- 45. Tabashnik BE. 1994. Evolution of resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 39:47–79 [Google Scholar]
- 46. Tabashnik BE, et al. 2011. Efficacy of genetically modified Bt toxins against insects with different genetic mechanisms of resistance. Nat. Biotechnol. 29:1128–1131 [DOI] [PubMed] [Google Scholar]
- 47. Valaitis AP. 2008. Bacillus thuringiensis pore-forming toxins trigger massive shedding of GPI-anchored aminopeptidase N from gypsy moth midgut epithelial cells. Insect Biochem. Mol. Biol. 38:611–618 [DOI] [PubMed] [Google Scholar]
- 48. Van Frankenhuyzen K. 2009. Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertbr. Pathol. 101:1–16 [DOI] [PubMed] [Google Scholar]
- 49. Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. 1989. Specificity of Bacillus thuringiensis δ-endotoxins: importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur. J. Biochem. 186:239–247 [DOI] [PubMed] [Google Scholar]
- 50. Wang P, et al. 2007. Mechanism of resistance to Bacillus thuringiensis toxin Cry1Ac in a greenhouse population of the cabbage looper, Trichoplusia ni. Appl. Environ. Microbiol. 73:1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wolfersberger MG, et al. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 86A:301–308 [DOI] [PubMed] [Google Scholar]
- 52. Wright DJ, Iqbal M, Granero F, Ferré J. 1997. A change in a single midgut receptor in the diamondback moth (Plutella xylostella) is only in part responsible for the field resistance to Bacillus thuringiensis subsp. kurstaki and B. thuringiensis subsp. aizawai. Appl. Environ. Microbiol. 63:1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang S, et al. 2009. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 39:421–429 [DOI] [PubMed] [Google Scholar]
- 54. Zhuang M, et al. 2002. Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation. J. Biol. Chem. 19:13863–13872 [DOI] [PubMed] [Google Scholar]