Abstract
Free-living diazotrophs are diverse and ubiquitous in soil, contributing the nitrogen pool in natural ecosystems. The isolation of nitrogen-fixing microorganisms has relied on semisolid nitrogen-free medium enrichment, followed by multiple subculturing steps. These procedures limit the diversity of recovered isolates. In the current study, we investigated three different isolation strategies for free-living diazotrophs using a soil sample from the Amazon forest. The methods were (i) direct plating on solid nitrogen-free medium under a 2% O2 concentration, (ii) enrichment in semisolid nitrogen-free medium before plating on solid nitrogen-free medium under 2% O2, and (iii) enrichment followed by subculturing in the semisolid nitrogen-free medium before plating on nitrogen containing medium under a 21% O2 concentration. A total of 794 isolates were differentiated by their genomic fingerprinting patterns, and strains with unique profiles were identified on the basis of sequencing of their 16S rRNA gene. Isolates belonged to four bacterial phyla: Proteobacteria, Firmicutes, Actinobacteria, and Bacteriodetes. The novel strategy of combining a solid N-free medium and hypoxic conditions showed an increase of 62.6% in the diversity of diazotrophs in comparison to that obtained by the conventional semisolid medium-based methods. All isolates grew on the nitrogen-free medium under a 2% O2 concentration, 78% of them showed the presence of the nifH gene, and 39% tested positive for acetylene reduction activity. Our results suggest that direct plating of soil dilutions on nitrogen-free solid medium under a 2% O2 concentration is a useful strategy for the isolation of the diverse diazotrophic communities.
INTRODUCTION
Biological nitrogen fixation is considered one of the most important natural processes on Earth, second to photosynthesis. The distinctive characteristic of the diazotrophs, i.e., ability to fix nitrogen, provides them with a selective growth advantage over other microorganisms under N-limited conditions. Most nitrogenases, a class of enzymes responsible for the fixation of atmospheric N2 gas, are irreversibly inactivated in the presence of high O2 concentrations (41, 43). Hence, maintenance of a continuously low concentration of O2 is required by most diazotrophs in order to fix nitrogen. Under laboratory conditions, the maintenance of continuously low oxygen concentrations either in liquid or on solid medium represents a challenge. While the continuous uptake of O2 through high rates of microbial respiration reduces the availability of this gas in liquid, the high O2 concentration in air (approximately 21%) can irreversibly damage the nitrogenase, when cells are grown on solid medium. Only a limited number of studies have reported on the nitrogen fixation activity by microaerophilic diazotrophs from terrestrial ecosystems in liquid medium (8, 24, 26), and none have reported on nitrogen fixation activity on solid medium.
Considering the limitations described above, Döbereiner and her colleagues (10) developed a semisolid N-free malate medium (NFM) for the isolation of free-living N2-fixing microorganisms. The semisolid nature of the medium, achieved through a low agar concentration, provided the microaerophilic condition, which, combined with a lack of N source, selectively favored the growth of diazotrophs. This method was based upon the traditional enrichment strategy, in which repeated series of soil dilutions were transferred or subcultured on the N-free medium before plating on N-containing medium. This classical approach has led to the isolation of a number of diazotrophic strains (2, 6, 12, 21, 34). However, the final outcome of any bacterial enrichment-based method is known for being influenced by differential growth rates and negative interactions among microorganisms (45). When the process of subculturing was performed several times, only a few strains could be isolated, and these lacked representation of the diversity of the source community (13, 49, 52).
In order to overcome the limitations of the above-described methods and increase the diversity of N2-fixing microorganisms, we developed an alternative method for the isolation of free-living diazotrophs. The objectives of the current study were (i) to establish a means for the direct isolation of diazotrophs based on a combination of plating on solid medium and controlled hypoxia, (ii) to compare the diversity of the N2-fixing bacteria obtained from soil under three different isolation strategies and highlight some of the potential biases that occur during preenrichment or subculturing before isolation, and (iii) to obtain culturable diazotrophs from the Amazon forest.
MATERIALS AND METHODS
Soil collection, inoculation, and diazotroph isolation.
A sample of soil was collected from the Amazon rainforest at Fazenda Nova Vida in the state of Rondonia, Brazil (10°10′18.71″S, 62°47′15.67″W) in 2009. The intact soil core (5-cm diameter by 10-cm depth) was transported on ice within 3 days to the laboratory and immediately homogenized with a 2 mm sieve. Soil (10 g) was added to 100 ml of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7), vigorously mixed for 10 min, and diluted in PBS. Serial dilutions from 10−3 to 10−7 were used for all subsequent procedures. Nitrogen-free combined carbon (NFCC) medium was prepared as previously described (10, 11) with addition of the following carbon sources (0.25% of each): 14 mM glucose, 23 mM sodium pyruvate, and 30 mM sodium acetate (all carbon sources were purchased from Sigma Co., St. Louis, MO). Three different incubation procedures were used for isolation of free-living diazotrophs (Fig. 1). In procedure A, solid NFCC medium (2% gellan gum) prepared on petri dishes (150 by 15 mm) was inoculated with 200 μl of soil serial dilutions and incubated under hypoxic conditions (2% O2 and 98% N2) in a vinyl hypoxic chamber fitted with an automated oxygen sensor and controller (Coy Laboratory Products, Grass Lake, MI) for 21 days. In procedure B, semisolid NFCC medium (0.15% gellan gum) prepared in culture tubes (14 ml) was inoculated with soil dilutions and incubated under atmospheric conditions (21% O2) for 7 days. Samples were 10-fold serially diluted and plated on the same medium, followed by incubation under hypoxic conditions (2% O2) for 21 days. In procedure C, semisolid NFCC medium prepared in culture tubes was inoculated with previously used soil dilutions and incubated under 21% O2 for 7 days, followed by two other transfers of 1 ml of inoculum to fresh medium. After the third incubation, plating was carried out on solid NFCC medium containing 50 mM NH4Cl. Plates were incubated under atmospheric conditions for 21 days. Three replicate plates or tubes were used per treatment, with a minimum of 80 bacterial colonies isolated from each plate.
Fig 1.
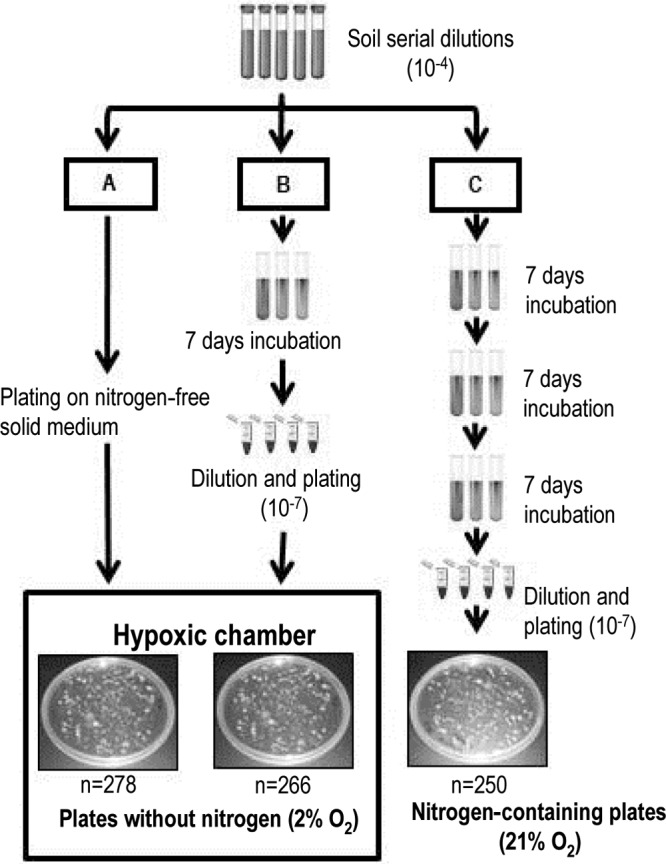
Different strategies used for the isolation of free-living diazotrophs from an Amazon forest soil. (A) Strategy A, direct plating on solid N-free medium incubated inside a hypoxic chamber (2% O2); (B) strategy B, inoculation into semisolid N-free medium with incubation in air for a week, followed by serial dilution and plating on solid N-free medium and incubation inside a hypoxic chamber 2% O2; (C) strategy C, inoculation into semisolid N2-free medium with incubation in air with three subsequent transfers in the same semisolid medium before plating on solid medium amended with NH4Cl. The boxed area represents the 2% O2 condition in the hypoxic chamber. The total number of isolates (n) obtained with each method is provided.
DNA extraction.
Cells were grown in liquid NFCC medium containing nitrogen (50 mM NH4Cl) for 5 days. One milliliter of the cell culture was harvested by centrifugation at 14,000 rpm for 2 min. DNA extractions were carried out using the bead-beating method as described previously (31). DNA was purified by sequential phenol, phenol-chloroform, and chloroform extractions, followed by isopropanol precipitation, a 70% ethanol wash, and resuspension in 100 μl of sterile H2O. DNA samples were stored at −20°C until use.
Genomic fingerprinting patterns.
PCR-based fingerprinting patterns were generated from genomic DNA samples using the BoxA1R primer (CTACGGCAAGGCGACGCTGACG) (50). PCR was performed in a total volume of 25 μl containing 0.8 mM each deoxynucleoside triphosphate, 2 μM primer, 1× PCR buffer, 3 mM MgCl2, 0.16 ng/μl of bovine serum albumin, 50 ng of DNA as the template, and 2 U of Taq DNA polymerase (GenScript, Piscataway, NJ) that was added after an initial incubation at 96°C for 10 min. PCR conditions were identical to those previously described (33). Fingerprinting profiles were analyzed on 2% agarose gels after staining with ethidium bromide (0.5 μg ml−1). Isolates were grouped on the basis of the identical profiles, and a representative strain of each unique group was identified by sequencing of the 16S rRNA gene.
16S rRNA gene amplification and sequencing.
The 16S rRNA gene of the bacterial isolates was amplified using primers 8F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT) (28) in a reaction volume of 50 μl containing 0.2 mM each deoxynucleoside triphosphate, 0.4 μM each primer, 1× PCR buffer, 2 mM MgCl2, 50 ng of template DNA, and 2 U of Taq DNA polymerase (GenScript, Piscataway, NJ). The PCR conditions were 2 min of denaturation, followed by 35 rounds of temperature cycling (95°C for 30 s, 52°C for 30 s, and 72°C for 45 s) and a final extension at 72°C for 7 min. Aliquots (5 μl) of PCR products were observed on ethidium bromide-stained 1% agarose gels. Amplified PCR products were treated with ExoSAP-IT (USB, Cleveland, OH) and subsequently sequenced with the 8F primer. Sequencing was performed with a 3130XL genetic analyzer (Applied Biosystems, Foster City, CA) at the University of Texas Arlington Genomics Core Facility.
nifH gene amplification.
Isolates were tested for the presence of the nifH gene with either direct or nested PCRs. For the direct PCR, primers PolF (TGCGAYCCSAARGCBGACTC) and PolR (ATSGCCATCATYTCRCCGGA) (38) were used with the reagents and PCR conditions described above for the 16S rRNA gene, but an annealing temperature of 59°C was used. Samples that yielded negative results with the above-described nifH primers were subjected to a nested PCR approach in which the first PCR was performed with the forward primer NifHforA (GCIWTITAYGGNAARGGNGG) and the reverse primer NifHrev (GCRTAIABNGCCATCATYTC) (53). The product of the first PCR was purified using an Ultra Clean 15 DNA purification kit (Mo Bio, Carlsbad, CA) and diluted 1:10 with sterile H2O. Two microliters of the diluted PCR product was used as the template for a nested PCR with the forward primer PolF and the reverse primer NifHrev. All reagents and PCR conditions for both reactions were the same as those described earlier, with the exception of the use of annealing temperatures of 52°C and 56°C for the first and nested reactions, respectively. Twenty randomly selected amplified products of the nested PCR were sequenced to confirm amplification of the nifH gene.
Phylogenetic analyses.
A total of 210 sequences of 16S rRNA genes were aligned and trimmed to 950 bp using the Sequencher (version 4.2.2; Gene Codes Corp., Ann Arbor, MI), CLUSTAL_X (48), and MacClade (version 4.05) (29) programs. Sequences of 60 pure culture isolates from close relatives were obtained from GenBank and used as placement guides. Aligned sequences were analyzed with four different phylogenetic methods: neighbor joining (NJ), maximum parsimony (MP), Bayesian, and maximum likelihood (ML). The software MEGA (version 4) (47) was used for the NJ and MP methods to generate unrooted consensus trees with 5,000 bootstrap replicates. Bayesian analysis was completed using MrBayes (version 3.0) software (25), and the majority consensus tree for each of the resulting tree files was constructed in PAUP* (version 4.0b10). Maximum likelihood analysis was performed using PHYML (version 3.0) software (19, 20) with the settings previously described elsewhere (32). The software TreeView (Win16) was used to display the trees (37), and MEGA (version 4) was used to generate unrooted consensus trees and compress the sequences at the genus level (47). The fine-scale grouping, i.e., individual sequences at the subgroup level, was also analyzed with all the above-described methods and is presented in Fig. SF1A to F in the supplemental material.
Acetylene reduction assay (ARA).
Isolates representing unique genomic fingerprinting profiles were grown in triplicate using the liquid NFCC medium containing nitrogen (50 mM NH4Cl) for 3 days. Cell cultures (1 ml) were centrifuged, washed twice with PBS, and suspended in 100 μl of the same solution, before being inoculated (25 μl) into 6 ml of solid NFCC medium slants. Cultures were incubated under a hypoxic atmosphere (2% O2) for 7 days and made airtight with serum stoppers while they were maintained inside the hypoxic chamber. Ten percent of the headspace air was exchanged with an equal volume of acetylene gas, and the cultures were incubated for 14 days. The concentration of ethylene (C2H4), the product of acetylene reduction, was measured with a gas chromatograph (GC-2014; Shimadzu) equipped with a hydrogen flame detector and a 3-m Porapak N column. Cultures of Herbaspirillum seropedicae (ATCC 35892) were used as positive controls, whereas noninoculated tubes with and without medium (in triplicate), but injected with acetylene, served as a negative controls.
Competition experiment.
Six isolates of different genera (Sinorhizobium sp. strain AMF3700, Phyllobacterium sp. strain AMF4046, Burkholderia sp. strain AMF3694, Pseudomonas sp. strain AMF3038, Enterobacter sp. strain AMF3712, and Bacillus sp. strain AMF3664) were grown individually in liquid NFCC medium containing nitrogen (50 mM NH4Cl). When the optical density at 600 nm (OD600) reached 0.3, cells were harvested by centrifugation, washed twice, and suspended with PBS. Equal numbers of cells from each culture (107 cells ml−1) were combined, and the mixture (100 μl) was used for the inoculation of the nitrogen-free semisolid tubes in triplicate. Combined cultures were transferred three times on the same medium before isolation on plates. A total of 22 colonies were randomly selected and identified by comparing their genomic fingerprinting profiles to those of pure cultures previously used for inoculation. In addition, cells were tested for their ability to grow in the N-free semisolid tubes.
Statistical analysis.
The numbers of bacterial genera detected per plate for each treatment were analyzed by single-factor analysis of variance using the software package R, version 2.9.2 (www.R-project.org).
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences obtained in this study were deposited in GenBank under the accession numbers JQ316215 through JQ316433.
RESULTS
Isolation of free-living diazotrophs.
Three different strategies were used for the isolation of free-living N2-fixing microorganisms from soil: direct plating (strategy A), enrichment (strategy B), and enrichment followed by transfers in the N-free medium before plating (strategy C) (Fig. 1). The 10−4 dilution was used for final isolations, as individual colonies could be retrieved after 21 days of incubation. For comparison purposes, the same dilution was selected for samples under isolation strategies B and C; however, a 10−7 dilution was required from semisolid to solid medium. Plates inoculated with lower dilutions showed colonies too numerous to count (Fig. 1). A total of 794 colonies were randomly selected with the three different methods: 278, 266 and 250 colonies with isolation strategies A, B, and C, respectively.
Diversity of isolates.
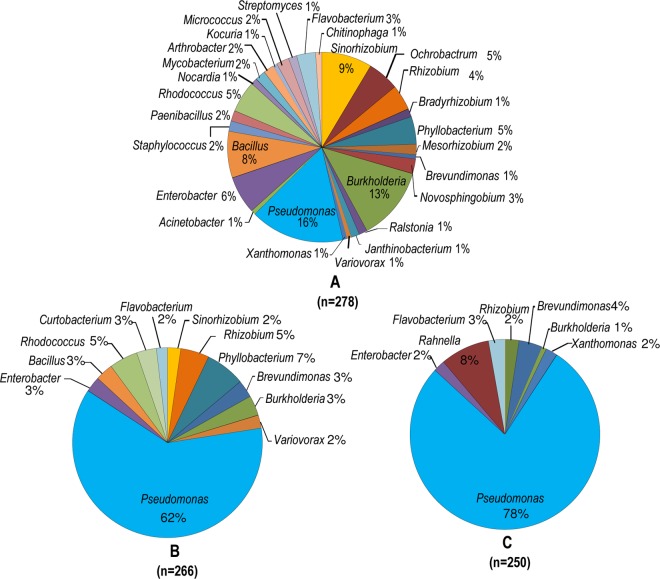
Purified colonies from each isolation strategy were differentiated on the basis of their genomic fingerprinting profiles, and isolates representing unique profiles (total of 210) were identified through sequencing of the 16S rRNA gene. The direct plating method had a significantly higher (F2,6 = 53.41, P = 0.0001) number of bacterial genera in isolation in comparison to the semisolid enrichment methods (Fig. 2). A total of 28 different bacterial genera were isolated under strategy A, whereas strategies B and C yielded 12 and 8 genera, respectively. Approximately 96% of culturable diazotrophs obtained in this study were isolated through direct plating on N-free solid medium, with 59% of them being unique to this strategy. Members of the genus Rahnella were isolated only after enrichment, followed by two sequential transfers in the N-free semisolid method (Fig. 2), and were not observed in the other two procedures.
Fig 2.
Percentage distribution of bacterial genera isolated from the Amazon forest soil using three different isolation strategies: direct plating (A), enrichment in semisolid N-free medium before plating (B), and enrichment followed by subculturing in semisolid N-free medium before plating (C). Isolates were identified on the basis of sequencing of the 16S rRNA gene. The total number of isolates (n) obtained with each method is provided.
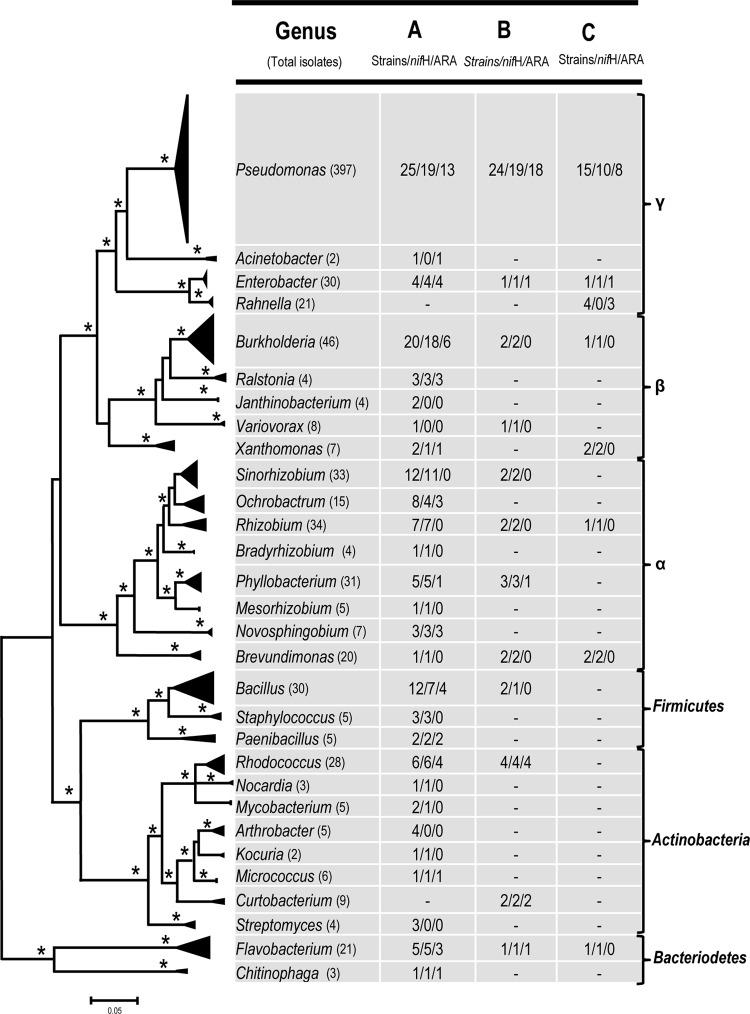
The isolates were grouped into four different bacterial phyla on the basis of 16S rRNA gene sequences: Proteobacteria, Firmicutes, Actinobacteria, and Bacteriodetes. A majority of isolates (84%) belonged to Proteobacteria, followed by Actinobacteria, with 17 and 8 different bacterial genera, respectively (Fig. 3; see Fig. SF1A to F in the supplemental material). The most abundant bacterial genera observed with isolation strategy A were Pseudomonas (16%), Burkholderia (13%), Sinorhizobium (9%), and Bacillus (8%). The percentage of Pseudomonas isolates increased four times when semisolid medium-based strategies B (62%) and C (78%) were used. This genus was the dominant group among isolates, followed by Phyllobacterium and Rahnella, being the only genera in which the percentages of isolates were above 5% for strategies B and C, respectively (Fig. 2). Approximately 80% of all isolates detected with strategies B and C were also observed with isolation strategy A.
Fig 3.
Maximum likelihood phylogenetic tree based on partial sequences of the 16S rRNA gene of the 210 unique pure cultures obtained through three different isolation methods. Sequences belonging to the same bacterial genera were clustered together, and branches within clusters were collapsed to show the overall relationships of the clusters to one another. The size of the triangle is proportionate to the number and variation of the sequences within a cluster. Asterisks at the nodes reflect bootstrap support values above 70% or posterior probability values from at least three of four phylogenetic methods: maximum likelihood, maximum parsimony, neighbor joining, and Bayesian analyses. Numbers corresponding to each bacterial genus represent number of the unique strains within the genus/number of strains that yielded positive amplification for the nifH gene/number of strains that showed acetylene reduction activity. Clustering at the phylum level is shown on the right.
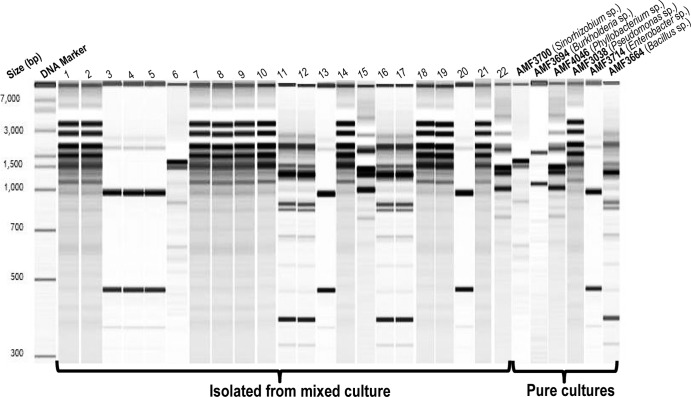
The diversity of isolates was also assessed at the strain level by performing genomic fingerprinting with BoxA1R primers. Isolation strategy A yielded the highest number of unique genomic fingerprinting patterns, with a total of 114, followed by 27 and 17 fingerprints from strains isolated with semisolid medium-based strategies B and C, respectively (see Fig. SF1A to F and SF2 in the supplemental material). As an example, we used the genus Burkholderia to compare isolates between strategies A and B. Among 33 Burkholderia isolates obtained through direct plating, 20 showed unique fingerprinting profiles (isolate-to-fingerprint ratio = 1.65). In contrast, the nine Burkholderia isolates captured with the semisolid medium-based method B belonged to only two unique profiles (isolate-to-fingerprint ratio = 4.5; see Fig. SF1B in the supplemental material). All other bacterial genera showed values similar to those observed for the genus Burkholderia, with exception of the genus Rahnella (Fig. 4).
Fig 4.
Genomic fingerprint profiles of 22 bacterial isolates obtained from enrichment and subculturing in semisolid N-free medium of six strains grown together. Lanes on the right represent results for individual strains used for the inoculation. Fragment sizes on the left indicate DNA size markers.
Nitrogen fixation.
All isolates were evaluated for their potential for biological N2 fixation with three consecutive growth transfers on N-free medium. Consistent with our isolation strategy, isolates were able to grow on plates under hypoxic condition (2% O2) but showed poor or no growth on plates maintained under 21% O2.
Genomic DNA from one representative of each fingerprinting profile was PCR amplified with nifH-specific primers and was further tested for acetylene reduction activity (ARA) under hypoxic conditions (2% O2) on solid medium. A total of 164 isolates (78%) showed positive PCR amplification results, confirmed by the presence of bands of the expected sizes. Twenty randomly selected amplified products were sequenced and confirmed to be partial sequences of the nifH gene. Ethylene was detected for 39% of the 210 unique representatives (Fig. 3; see Table ST1 in the supplemental material). Representatives of eight genera showed the presence of the nifH gene, but their nitrogenase activity was not detected. They belonged to the Alphaproteobacteria (Rhizobium, Bradyrhizobium, Mesorhizobium, Brevundiomonas), Actinobacteria (Nocardia, Mycobacterium, Kocuria), and Firmicutes (Staphylococcus). Bacterial isolates belonging to the Rahnella, Acinetobacter, and Xanthomonas genera had ARA, but the nifH gene was not amplified under the conditions tested (Fig. 3; see Table ST1 in the supplemental material). Representative isolates from four bacterial genera, Janthinobacterium, Variovorax, Arthrobacter, and Streptomyces, yielded negative results for both nifH and ARA.
Competition experiment.
When equal numbers of cells from six isolates were mixed together, inoculated into the semisolid NFCC medium, and later recovered after three consecutive transfers, the number of Pseudomonas colonies was approximately 45% of the total. Random colonies were selected for DNA isolation and genomic fingerprint profiling (Fig. 4). Pseudomonas was the dominant group, followed by Enterobacter and Bacillus strains. Isolates belonging to the genus Burlkholderia and Sinorhizobium yielded only two and one fingerprint profiles, respectively. No fingerprint profile was observed for the Phyllobacterium isolate.
DISCUSSION
Biological N2 fixation is the most important process for the uptake of N in natural forests. It is also a widespread genetic trait among microorganisms, but the extent of its diversity has not been consistently examined. In the current study, a combination of carbon sources, solid N-free medium, and hypoxia was used for the isolation of free-living diazotrophs. First, gellan gum was selected as a solidifying agent with a 2-fold purpose: (i) advantageous physical properties in culturing free-living N2-fixing bacteria, as reported previously (23), and (ii) increase of N2 fixation activity. Hara and coworkers (21) reported 10- to 200-fold increases in acetylene reduction activity for nitrogen fixers grown in semisolid medium with gellan gum in comparison to the activity for the same isolates grown in N-free medium containing 0.2% agar. Second, different carbon sources were combined in order to increase the diversity of isolates, as suggested elsewhere (49). Finally, the O2 concentration used for incubation was maintained at 2%. The selection of the O2 concentration was based on the highest nitrogenase activity detected for a pure culture of the well-known free-living diazotroph Herbaspirillum seropedicae ATCC 35892 (data not shown). These three basic modifications to the current isolation strategies for N2 fixers increased the number of unique isolates by 62.6%.
The diversity of N2 fixers obtained with isolation method A was higher than that observed with the other methods, and this result was significant, indicating that direct plating was an effective strategy for obtaining isolates of different genera. Most of the bacterial isolates obtained with this strategy were closely related to the diazotrophic strains that have previously been reported to be endophytes and were isolated from several plant species (12, 27, 51). In contrast to these studies, we isolated N2 fixers directly from a complex environment such as soil without using any plant trapping-based methods, which are influenced by the plant selectivity (33). To our knowledge, this is the first report of a solid N-free medium being used for this purpose and the direct isolation of highly diverse groups of diazotrophic strains from a soil sample.
It is possible that the success of this strategy is likely due to the circumvention of the possible effects of differential bacterial growth rates and negative interactions among microorganisms growing together. In the semisolid medium, fast-growing microorganisms tend to outcompete slow-growing ones, and subsequent isolations from a high dilution (10−7) increase the likelihood of retrieving fast-growing strains. This has been observed for multiple species grown together in liquid medium, with only the fastest-growing species being retrieved (5, 42). A second plausible explanation is the possibility that microbial species growing in proximity could directly inhibit or reduce the growth of others through antibiotic production or other growth-inhibiting substances (15, 17, 22, 44). In contrast, direct plating or solid medium-based methods can reduce the effect of differential growth rates or species interactions (positive or negative) owing to the spatial separation between microbial cells. Accordingly, others have reported on the advantages of direct plating methods for the isolation of a diverse microbial population compared to enrichment- or subculturing-based methods (13, 45, 49, 52).
Pseudomonas isolates were the most abundant group after enrichment and subculturing for isolation strategies B (62%) and C (78%), indicating an exceptional ability of this genus to outcompete other microorganisms under laboratory conditions (Fig. 2). Recently, Garbeva and coauthors (18) reported on the production of broad-spectrum antibiotics by Pseudomonas species when grown in combination with other Gram-negative microorganisms. Antibiotic production not only was triggered by the presence of Rhizobium, Brevundimonas, and Pedobacter but also significantly reduced the growth of Gram-positive microorganisms when grown in a carbon-limited medium. We tested whether Pseudomonas could outcompete other diazotrophs in an N-free semisolid medium with conditions identical to those used for isolation strategy C. The overwhelming dominance of Pseudomonas in a simple competition experiment (Fig. 4) highlights the potential bias that might occur during attempts to isolate N2 fixers using traditional methods, regardless of their dominance in the environment. Previously, Hara and coworkers (21) reported that 80 to 100% of the N2-fixing isolates from surface and subsurface soil samples from a Siberian forest belonged to the genus Pseudomonas. We reason that the high dominance of this group might be associated with the use of a semisolid N-free medium.
Our study underscores the importance of the appropriate hypoxic condition for direct isolation of free-living diazotrophs. It is well established that a low O2 concentration is required for the optimum functioning of the nitrogenase. However, the requirement of O2 concentration varies among different species, as Pseudomonas requires from 2 to 4% O2, whereas the optimum concentrations for methanotrophs varies from 0.5 to 10% (7, 24, 26, 36, 46). Owing to the harmful effects of O2 to nitrogenase, we selected a concentration that provided the highest acetylene reduction activity for H. seropedicae, a well-known free-living diazotroph. We acknowledge that the establishment of a single hypoxic condition might have prevented the growth of free-living diazotrophs requiring even lower O2 concentrations. Nonetheless, the breadth of diversity obtained with our procedure represents a step toward a practical goal of cultivating novel N2 fixers.
All isolates from the solid-based method were able to grow on N-free medium under a 2% O2 concentration after multiple transfers, suggesting their ability to maintain N2 fixation activity. It has been reported that non-nitrogen-fixing microorganisms can grow with N traces present in the medium and have been detected as false positives (30, 39). In order to screen for the presence of false positives, we grew all strains on the same N-free medium and incubated them under 2 and 21% O2 concentrations. All isolates showed growth under 2% O2, but no to very poor growth was observed under 21% O2. All strains showed growth on the same medium amended with NH4Cl as a nitrogen source (50 mM) under a 21% O2 concentration. It is noteworthy to observe that only growth on N-free medium does not warrant recognition as a true diazotroph, and experimental evidence determined through acetylene reduction activity or expression of the nitrogen fixation genes or from results of an assimilation assay with 15N-labeled N2 gas should be obtained.
A large proportion of the isolates (79%) showed the presence of the nifH gene, an important gene encoding the nitrogenase reductase (component II or Fe protein of nitrogenase) (Fig. 3; see Table ST1 in the supplemental material). First, the difference may be explained by the sequence variability of the nifH gene and lack of PCR primers that are suitable for capturing the diversity of all isolates (54). In contrast to the molecular assessment results, only 39% of isolates displayed acetylene reduction activity. Previously, it has been reported that the N2-fixing ability of the different diazotrophs varied depending upon a number of factors, such as incubation time and temperature, growth medium, O2 concentration, and type of solidifying agent (7, 8, 14, 21, 24, 26). Therefore, it is likely that the absence of nitrogenase activity can be attributed to the experimental conditions used in this study. Second, not all the functional nitrogenases can reduce acetylene to ethylene, as reported in the case of Streptomyces thermoautotrophicus (16, 40). Third, it is also possible that the acetylene reduction activity or rate of nitrogen fixation (due to the slow-growing nature of some of the organisms) is below the detection limit of the technique (<50 pmol/ml) (35). The percentages of total isolates that tested negative for either the presence of the nifH gene or ARA were 19.7%, 7%, and 18.5% for isolation strategies A, B, and C, respectively. Although we have not found evidence that solid N-free medium under hypoxic conditions substantially increases the possibility of false positives, these isolates certainly require further testing with a diverse set of primers for amplification of the nifH gene and growth under different conditions for ARA measurements. Our results are in accordance with those of several previous studies on N2-fixing isolates, demonstrating their ability to grow on N-free medium and the presence of the nifH gene but negative tests for acetylene reduction (1, 3, 4, 9, 12, 51). Accordingly, the majority of the isolates which possessed the nifH gene, but no ARA, were from the order Rhizobiales: the Rhizobium, Sinorhizobium, Bradyrhizobium, Mesorhizobium, Ochrobactrum, and Phyllobacterium genera. In spite of being well characterized for their N2 fixation when in symbiotic association with legumes, these genera show ARA only when inside the root nodules (27).
The Amazon forest is one of the most important terrestrial ecosystems on Earth sequestering CO2, which occurs through the process of photosynthesis. As biogeochemical cycles are linked together, one would expect that biological N2 fixation plays an important role in the ecosystem balance. Our results underscore a high and yet to be explored diversity of free-living diazotrophs in the Amazon forest. Currently, this successful method has been employed for the isolation of diazotrophic communities from primary forest, pastures of different ages, and a secondary forest. In future, the diversification of the growth media and oxygen concentrations may contribute to the isolation of diverse diazotrophs from rare biosphere environments.
Supplementary Material
ACKNOWLEDGMENTS
We express our gratitude to Thomas H. Chrzanowski for providing access to the gas chromatography facility and undergraduate students Neda Jooya, Thao Nguyen, and Maria Bhatti for their assistance with isolations. We thank the ARMO team members for their assistance with sampling.
This work was supported by grant CSREES 2008-04556 from the United States Department of Agriculture.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Achouak W, Normand P, Heulin T. 1999. Comparative phylogeny of rrs and nifH genes in the Bacillaceae. Int. J. Syst. Bacteriol. 49:961–967 [DOI] [PubMed] [Google Scholar]
- 2. Beneduzi A, Peres D, Vargas LK, Bodanese-Zanettini MH, Passaglia LMP. 2008. Evaluation of genetic diversity and plant growth promoting activities of nitrogen-fixing Bacilli isolated from rice fields in South Brazil. Appl. Soil Ecol. 39:311–320 [Google Scholar]
- 3. Berg G, et al. 2002. Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl. Environ. Microbiol. 68:3328–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brighigna L, Montaini P, Favilli F, Trejo AC. 1992. Role of the nitrogen-fixing bacterial microflora in the epiphytism of Tillandsia (Bromeliaceae). Am. J. Bot. 79:723–727 [Google Scholar]
- 5. Button DK, Schut F, Quang P, Martin R, Robertson BR. 1993. Viability and isolation of typical marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caballero-Mellado J, Onofre-Lemus J, Estrada-de los Santos P, Martínez-Aguilar L. 2007. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 73:5308–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dedysh SN, Ricke P, Liesack W. 2004. NifH and NifD phylogenies: an evolutionary basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 150:1301–1313 [DOI] [PubMed] [Google Scholar]
- 8. Desnoues N, et al. 2003. Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology 149:2251–2262 [DOI] [PubMed] [Google Scholar]
- 9. Ding Y, Wang J, Liu Y, Chen S. 2005. Isolation and identification of nitrogen-fixing Bacilli from plant rhizospheres in Beijing region. J. Appl. Microbiol. 99:1271–1281 [DOI] [PubMed] [Google Scholar]
- 10. Döbereiner J, Marriel I, Nery M. 1976. Ecological distribution of Spirillum lipoferum Beijerinck. Can. J. Microbiol. 22:1464–1473 [DOI] [PubMed] [Google Scholar]
- 11. Döbereiner J, Urquiaga S, Boddey RM. 1995. Alternatives for nitrogen nutrition of crops in tropical agriculture. Fertil. Res. 42:339–346 [Google Scholar]
- 12. Doty SL, et al. 2009. Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 47:23–33 [Google Scholar]
- 13. Eilers H, Pernthaler J, Amann R. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Estrada-De Los Santos P, Bustillos-Cristales R, Caballero-Mellado J. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fong KP, Chung WO, Lamont RJ, Demuth DR. 2001. Intra and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans luxS. Infect. Immun. 69:7625–7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gadkari D, Morsdorf G, Meyer O. 1992. Chemolithoautotrophic assimilation of dinitrogen by Streptomyces thermoautotrophicus UBT1 identification of an unusual N2-fixing system. J. Bacteriol. 174:6840–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garbeva P, de Boer W. 2009. Inter-specific interactions between carbon-limited soil bacteria affect behavior and gene expression. Microb. Ecol. 58:36–46 [DOI] [PubMed] [Google Scholar]
- 18. Garbeva P, Silby MW, Raaijmakers JM, Levy SB, de Boer W. 2011. Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J. 5:973–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 20. Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559 doi:10.1093/nar/gki352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hara S, Hashidoko Y, Desyatkin RV, Hatano R, Tahara S. 2009. High rate of N2 fixation by East Siberian cryophilic soil bacteria as determined by measuring acetylene reduction in nitrogen-poor medium solidified with gellan gum. Appl. Environ. Microbiol. 75:2811–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harrison F, Paul J, Massey RC, Buckling A. 2008. Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J. 2:49–55 [DOI] [PubMed] [Google Scholar]
- 23. Hashidoko Y, Tada M, Osaki M, Tahara S. 2002. Soft gel medium solidified with gellan gum for preliminary screening for root-associating, free-living nitrogen-fixing bacteria inhabiting the rhizoplane of plants. Biosci. Biotechnol. Biochem. 66:2259–2263 [DOI] [PubMed] [Google Scholar]
- 24. Hatayama K, Kawai S, Shoun H, Nakamura A. 2005. Pseudomonas azotofigens sp. nov., a novel nitrogen-fixing bacterium isolated from a compost pile. Int. J. Syst. Evol. Microbiol. 55:1539–1544 [DOI] [PubMed] [Google Scholar]
- 25. Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 26. Khadem AF, Pol A, Jetten MSM, den Camp HJMO. 2010. Nitrogen fixation by the verrucomicrobial methanotroph ‘Methylacidiphilum fumariolicum ’ SolV. Microbiology 156:1052–1059 [DOI] [PubMed] [Google Scholar]
- 27. Kuklinsky-Sobral J, et al. 2004. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 6:1244–1251 [DOI] [PubMed] [Google Scholar]
- 28. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 29. Maddison WP, Maddison DR. 1999. MacClade: analysis of phylogeny and character evolution. Sinauer, Sunderland, MA: [DOI] [PubMed] [Google Scholar]
- 30. Martinez-Romero E. 2006. Dinitrogen-fixing prokaryotes, p 793–817 In Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E. (ed), The prokaryotes, a handbook of the biology of bacteria, 3rd ed, vol 2 Springer, New York, NY [Google Scholar]
- 31. Mirza BS, Welsh A, Hahn D. 2009. Growth of Frankia strains in leaf litter-amended soil and the rhizosphere of a nonactinorhizal plant. FEMS Microbiol. Ecol. 70:132–141 [DOI] [PubMed] [Google Scholar]
- 32. Mirza BS, Welsh AK, Rieder JP, Paschke MW, Hahn D. 2009. Diversity of frankiae in soils from five continents. Syst. Appl. Microbiol. 32:558–570 [DOI] [PubMed] [Google Scholar]
- 33. Mirza BS, et al. 2009. Variation in Frankia populations of the Elaeagnus host infection group in nodules of six host plant species after inoculation with soil. Microb. Ecol. 58:384–393 [DOI] [PubMed] [Google Scholar]
- 34. Montanez A, Abreu C, Gill PR, Hardarson G, Sicardi M. 2009. Biological nitrogen fixation in maize (Zea mays L.) by 15N isotope-dilution and identification of associated culturable diazotrophs. Biol. Fertil. Soils 45:253–263 [Google Scholar]
- 35. Montoya JP, Voss M, Kahler P, Capone DG. 1996. A simple, high-precision, high-sensitivity tracer assay for N2 fixation. Appl. Environ. Microbiol. 62:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murrell JC, Dalton H. 1983. Nitrogen-fixation in obligate methanotrophs. J. Gen. Microbiol. 129:3481–3486 [Google Scholar]
- 37. Page RDM. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 38. Poly F, Monrozier LJ, Bally R. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152:95–103 [DOI] [PubMed] [Google Scholar]
- 39. Postgate J. 1988. The ghost in the laboratory. New Sci. 117:49–52 [Google Scholar]
- 40. Ribbe M, Gadkari D, Meyer O. 1997. N2 fixation by Streptomyces thermoautotrophicus involves a molybdenum-dinitrogenase and a manganese-superoxide oxidoreductase that couple N2 reduction to the oxidation of superoxide produced from O2 by a molybdenum-CO dehydrogenase. J. Biol. Chem. 272:26627–26633 [DOI] [PubMed] [Google Scholar]
- 41. Robson RL, Postgate JR. 1980. Oxygen and hydrogen in biological nitrogen-fixation. Annu. Rev. Microbiol. 34:183–207 [DOI] [PubMed] [Google Scholar]
- 42. Roussanov B, Hawkins DM, Tatini SR. 1996. Estimating bacterial density from tube dilution data by a Bayesian method. Food Microbiol. 13:341–363 [Google Scholar]
- 43. Rudnick P, Meletzus D, Green A, He LH, Kennedy C. 1997. Regulation of nitrogen fixation by ammonium in diazotrophic species of proteobacteria. Soil Biol. Biochem. 29:831–841 [Google Scholar]
- 44. Ryan RP, Dow JM. 2008. Diffusible signals and interspecies communication in bacteria. Microbiology 154:1845–1858 [DOI] [PubMed] [Google Scholar]
- 45. Schoenborn L, Yates PS, Grinton BE, Hugenholtz P, Janssen PH. 2004. Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl. Environ. Microbiol. 70:4363–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takeda K. 1988. Characteristics of a nitrogen-fixing methanotroph, Methylocystis T-1. Antonie Van Leeuwenhoek J. Microbiol. 54:521–534 [DOI] [PubMed] [Google Scholar]
- 47. Tamura K, Dudley J, Nei M, Kumar S. 2007. Molecular evolutionary genetics analysis (MEGA) software version 4. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 48. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uphoff HU, Felske A, Fehr W, Wagner-Dobler I. 2001. The microbial diversity in picoplankton enrichment cultures: a molecular screening of marine isolates. FEMS Microbiol. Ecol. 35:249–258 [DOI] [PubMed] [Google Scholar]
- 50. Versalovic J, de Bruijn FJ, Lupski JR. 1998. Repetitive sequence-based PCR (Rep.-PCR) DNA fingerprinting of bacterial genomes, p 437–454 In de Bruijn FJ, Lupski JR, Weinstock GM. (ed), Bacterial genomes: physical structure and analysis. Chapman and Hall, New York, NY [Google Scholar]
- 51. Videira SS, de Araujo JLS, Rodrigues LS, Baldani VLD, Baldani JI. 2009. Occurrence and diversity of nitrogen-fixing Sphingomonas bacteria associated with rice plants grown in Brazil. FEMS Microbiol. Lett. 293:11–19 [DOI] [PubMed] [Google Scholar]
- 52. Watanabe K, Teramoto M, Futamata H, Harayama S. 1998. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Widmer F, Shaffer BT, Porteous LA, Seidler RJ. 1999. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl. Environ. Microbiol. 65:374–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zehr JP, Crumbliss LL, Church MJ, Omoregie EO, Jenkins BD. 2003. Nitrogenase genes in PCR and RT-PCR reagents: implications for studies of diversity of functional genes. Biotechniques 35:996–1005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.