Abstract
Magnetotactic bacteria have the unique capacity of being able to swim along geomagnetic field lines. They are Gram-negative bacteria with diverse morphologies and variable phylogenetic relatedness. Here, we describe a group of uncultivated marine magnetococci collected from intertidal sediments of Huiquan Bay in the Yellow Sea. They were coccoid-ovoid in morphology, with an average size of 2.8 ± 0.3 μm by 2.0 ± 0.2 μm. Differential interference contrast microscopy, fluorescence microscopy, and transmission electron microscopy revealed that each cell was apparently composed of two hemispheres. The cells synthesized iron oxide-type magnetosomes that clustered on one side of the cell at the interface between the two hemispheres. In some cells two chains of magnetosomes were observed across the interface. Each cell had two bundles of flagella enveloped in a sheath and displayed north-seeking helical motion. Two 16S rRNA gene sequences having 91.8% identity were obtained, and their authenticity was confirmed by fluorescence in situ hybridization. Phylogenetic analysis revealed that the magnetococci are affiliated with the Alphaproteobacteria and are most closely related to two uncultured magnetococci with sequence identities of 92.7% and 92.4%, respectively. Because they display a >7% sequence divergence to all bacteria reported, the bean-like magnetococci may represent two novel genera.
INTRODUCTION
Magnetotactic bacteria (MTB) were first discovered independently by Bellini in 1964 and Blakemore in 1975 (3–5, 9). They are a morphologically, metabolically, and phylogenetically diverse assemblage of motile prokaryotes that can orient and navigate along geomagnetic field lines (2, 28). MTB contain intracellular membrane-bound, nano-sized, single-domain crystals termed magnetosomes, which usually consist of iron oxide (magnetite, Fe3O4) or iron sulfide (greigite, Fe3S4) (2). Magnetosome formation is genetically controlled, and the magnetic crystals have species-specific morphologies and specific arrangements within the cell (2). Magnetosomes usually organize in chains and form a magnetic dipole moment in the cell. This makes it possible for the cell to align to the Earth's magnetic field, which enables the bacterium to find and maintain an optimum position in the oxygen and chemical gradient (10, 11). Magnetotactic bacteria are ubiquitous in the water column and sediments of freshwater and marine habitats and are believed to play an important role in iron cycling (2, 7).
MTB comprise a variety of morphological types (including coccoid, spiral, vibroid, rod-like, or aggregated) (8, 27, 29) and have a great phylogenetic diversity. MTB have been identified in Proteobacteria and Nitrospirae, and a recent study indicates the existence of large ovoid-shaped MTB belonging to candidate division OP3 (1, 14, 18–20). The coccoid is the dominant morphology among magnetotactic bacteria in most habitats (21, 22, 24, 25, 31, 32), and the interactions of bacterial species in a given habitat with the prevailing environmental conditions are likely to shape the dominant phylogenetic pattern. In a previous study we reported the almost homogeneous occurrence of magnetotactic cocci in sediments from an intertidal sampling site (25). However, magnetotactic cocci appear to be extremely diverse. We report here a novel group of bean-like magnetococci collected from the intertidal sediments of Huiquan Bay in the Yellow Sea, China. We observed a hemispheric morphology, and phylogenetic analysis suggests that they belong to two novel genera of Alphaproteobacteria.
MATERIALS AND METHODS
Sampling and collection of MTB.
Surface sediments and water (approximate ratio, 1:1) were collected during low tide from the Huiquan Bay (36°03′ N, 120°21′ E) of the Yellow Sea and were stored in 500-ml plastic bottles. The sampling site is an intertidal sandy area with a water salinity of 35‰ and a pH of 7.6. As previously reported, magnetotactic bacteria in the samples were magnetically enriched by attaching the south pole of permanent magnets outside the bottles (25) and were purified using the racetrack method (34).
Optical and electron microscopy.
Cell morphology and motility were analyzed by optical microscopy (Olympus BX51, equipped with a DP71 camera system) using the hanging-drop method in an applied magnetic field, and the motility track was recorded using dark-field microscopy.
For scanning electron microscopy (SEM), the samples were fixed in 2.5% glutaraldehyde for more than 3 h at 4°C, filtered onto polycarbonate filters with high-density pores 0.2 μm in diameter (Whatman), and then dehydrated with ethanol and isoamyl acetate. The dried and gold-coated samples were examined using a KVKV-2800B SEM operating at 25 kV.
For transmission electron microscopy (TEM), the samples were drawn onto Formvar-coated copper grids and washed with distilled water prior to analysis using a Hitachi H8100 transmission electron microscope (operating at 75 kV) and a JEM 2100 high-resolution transmission electron microscope ([HRTEM] operating at 200 kV) equipped for energy-dispersive X-ray spectroscopy (EDXS). The length and width of magnetosomes were measured using Adobe Photoshop software. The size and shape factor of the magnetosomes were calculated as (length + width)/2 and width/length, respectively (22).
Sequence analysis of the 16S rRNA gene.
The 16S rRNA genes of collected bacterial cells were amplified with the universal primers 27f (5′-AGA GTY TGA TCC TGG CTC AG-3′) and 1492r (5′-GGT TAC CTT GTT ACG ACT T-3′) using PCR (16). The PCR amplification and construction of a 16S rRNA gene clone library were performed as previously reported (25). After eliminating four false-positive clones from library of 200 clones, we further screened the other clones using restriction fragment length polymorphism (RFLP) analysis. Clones with identical patterns were defined as an operational taxonomic unit (OTU), and then representative clones of each OTU were randomly selected for sequencing (21). After we eliminated the sequences for chimeras by using the Pintail program (http://www.bioinformatics-toolkit.org/Web-Pintail/), the sequences obtained were analyzed using BLAST (NCBI website [http://www.ncbi.nlm.nih.gov/BLAST/]). The related sequences were aligned using CLUSTAL W multiple alignment. Identity was calculated using the BioEdit program, and phylogenetic trees were constructed using MEGA, version 4.0, applying a neighbor-joining method. Bootstrap values were calculated with 1,000 replicates.
Fluorescence in situ hybridization (FISH).
The specific oligonucleotide probes p-3 (5′-TCT TTG AGG AGG GAG CCG TTG-3′; nucleotide positions 1381 to 1402) and p-9 (5′-TGG ATG ACC TGC CCT GAG ATG G-3′; nucleotide positions 112 to 133) were designed using the probe design tool in Primer Premier, version 5.0 software. The probes were labeled with Cy3 as the fluorescent dye, and the general probe EUB338 (5′-GCT GCC TCC CRT AGG AGT-3′; nucleotide positions 338 to 355) was labeled with 6-carboxyfluorescein (FAM) and used as the positive control in the hybridization. Escherichia coli Top 10 cells and magnetotactic spirillum QH-2 from Huiquan Bay were used as negative controls in the hybridization with specific probes.
FISH was carried out according to protocols reported by Pan et al. (25) and Pernthaler et al. (26). The racetrack-purified samples were fixed with 4% paraformaldehyde for 3 h at 4°C, washed in phosphate-buffered saline (PBS), and then stored in ethanol-PBS (1:1) at −20°C. The samples were dried on prepared glass slides, dehydrated in an ethanol series, immersed in hybridization buffer for 2 h at 46°C, and then washed in washing buffer for 10 min at 48°C. The hybridizations were analyzed by fluorescence microscopy (Olympus BX51 fluorescence microscope).
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA genes in clones 1-3 and 1-9 were deposited in GenBank under accession numbers JF421219 and JF421220, respectively.
RESULTS AND DISCUSSION
Morphology and motility of the bean-like magnetococci.
The magnetococci collected from intertidal sediments were highly homogeneous in morphology and numbered up to 103 to 104 cells/cm3. The cells were coccoid-ovoid and had an average size of 2.8 ± 0.3 μm by 2.0 ± 0.2 μm (n = 562), determined using bright-field microscopy. They survived in an aquarium for more than 1 year under laboratory conditions.
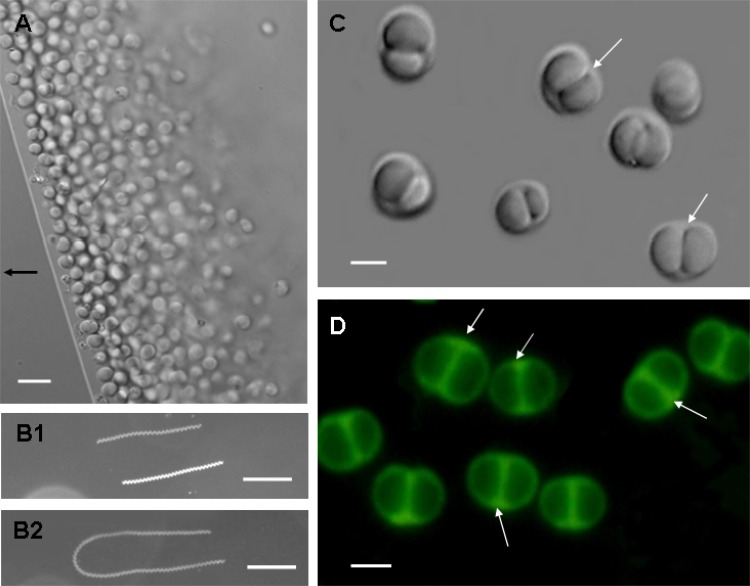
In the presence of an applied magnetic field, the freshly collected magnetococci in a hanging drop displayed north-seeking taxis (Fig. 1A). Using long-time-exposure photography of the swimming magnetococci, we observed a clockwise helical trajectory during forward swimming. The cells completed approximately 20 rotations/200 ms (Fig. 1B1) and displayed a U-like track in response to reversal of the magnetic field (Fig. 1B2). The velocity varied from 35 to 160 μm/s, with an average of 87.46 μm/s (n = 398) and a Gaussian speed distribution (data not shown). This velocity is faster than that of magnetotactic spirillum QH-2 (20 to 50 μm/s) isolated from Huiquan Bay (36) but slower than MO-1 (approximately 250 μm/s) from the Mediterranean Sea (17).
Fig 1.
Morphology and motility of bean-like magnetococci (BMC) based on optical microscopy. Differential interference contrast (DIC) images of BMC are shown in panels A and C. In panel A, the arrow indicates the direction of the magnetic field. For panels B1 and B2, motility tracks of BMC cells in a magnetic field were recorded using dark-field microscopy. BMC cells display a clockwise helical trajectory during forward swimming (B1) and a U-like track in response to reversal of the magnetic field (B2). The exposure time was 3 s for panel B1 and 5 s for panel B2. Panel D shows the fluorescence of BMC exposed to blue light (wavelength 450 to 480 nm). Scale bars, 5 μm (A), 100 μm (B), and 2 μm (C and D).
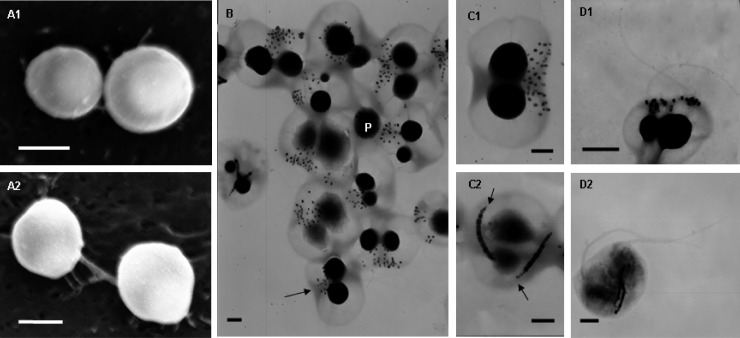
Fig 2.
Characteristics of BMC cells determined by SEM and TEM. (A) Morphology of BMC cells by SEM as shown by a top view (A1) and a lateral view (A2). The two types of cells were observed in the examples shown in panel B: BMC with two chains of magnetosomes (C1) and flagella (D1) and BMC with clustered magnetosomes (C2) and flagella (D2). Scale bars, 2 μm (A) and 500 nm (B to D).
Differential interference contrast (DIC) microscopy revealed that in lateral view most cells had an isometric double-hemisphere morphology (similar to that of a bean) with a space between the two hemispheres (Fig. 1C, white arrows). Fluorescence microscopy of cells fixed in glutaraldehyde revealed membrane-like septa between the two hemispheres (Fig. 1D, white arrows), consistent with the observed space under DIC microscopy. In view of their remarkable morphology, we termed them bean-like magnetococci (BMC). We used SEM and TEM to analyze their morphology. SEM revealed an overall spherical morphology without an obvious double hemisphere (Fig. 2A1 and 2A2). In contrast, the hemispherical morphology was evident using TEM, which revealed two large electron-dense granules (Fig. 2B, labeled P), a groove around the center confirming that the cell comprised two parts (Fig. 2B, black arrow), and an intracellular hemispherical architecture. We hypothesize that the intracellular hemispherical architecture of the BMC cells may be a consequence of the distribution of substances of differing electron densities or fluorescence properties. Similar structures had been observed in MTB from the Itaipu Lagoon, Brazil (32), and from a salt pond in Falmouth, MA (30).
Intracellular features of the bean-like magnetococci.
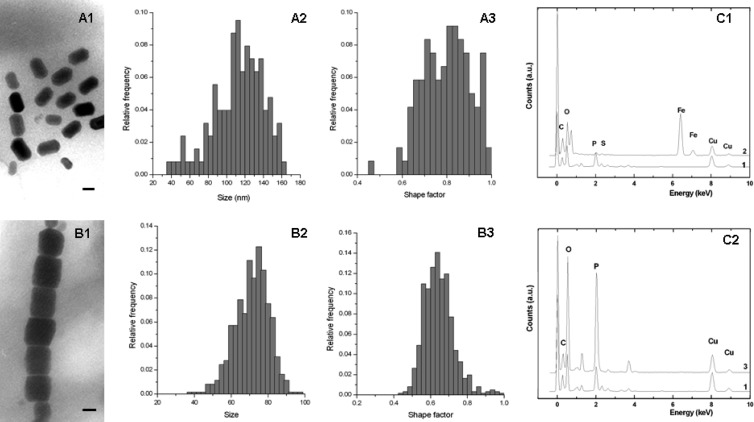
Despite similarity in the morphologies of the magnetococci, TEM revealed two magnetosome arrangements (Fig. 2B). Most cells in the population contained approximately 32 magnetosomes with a rectangular projected shape in a disorganized cluster on one side of the cell at the interface between the two hemispheres (Fig. 2C1). Despite the apparent disorganization of the magnetosomes, the cells displayed the capacity of north-seeking magnetotaxis, implying that the magnetosomes still form an overall magnetic dipole moment. The size of the magnetosome crystals varied from 47 to 145 nm, and the shape factor was 0.60 (average length and width, 102 ± 20 nm and 60 ± 9 nm, respectively; n = 313) (Fig. 3A). Ten percent of the BMC cells contained an average of 22 ± 4 (n = 64) magnetosomes arranged in two orderly chains across the interface between the hemispheres (Fig. 2C2). Each magnetosome had a rectangular projected shape of length 125 ± 32 nm and width 99 ± 25 nm (n = 118). This produced a shape factor of approximately 0.80 ± 0.11 (Fig. 3B), which is similar to that of the magnetotactic coccus QHL, collected from the low-tide zone in Huiquan Bay, China (25). Relative to the distribution of magnetosomes in BMC cells that had magnetosomes arranged in a disorganized cluster, distribution of magnetosomes in cells having magnetosomes in chains was broad, and their size distribution was narrow. TEM indicated that the crystals at ends of the chains were much smaller and had lower contrast relative to the background than the other crystals in the chains (Fig. 2C2, black arrows). The smaller crystals may be early-stage immature magnetosomes. A similar distribution of magnetosomes has been observed for the cultivated magnetotactic spirillum strains AMB-1 and MSR-1 (15, 33).
Fig 3.
Intracellular features of the bean-like magnetococci. (A) Characteristics of cluster-forming magnetosomes: morphology (A1), size histograms (A2), and shape factor distribution (A3). (B) Characteristics of chain-forming magnetosomes: morphology (B1), size histograms (B2), and shape factor distribution (B3). Energy dispersive X-ray (EDX) analysis of magnetosomes (C1) and granules (C2) is also shown. Line 1, cell; line 2, magnetosomes (note the peaks of iron and oxygen); line 3, granules (note the peaks of phosphorus and oxygen). Scale bars, 50 nm.
TEM analysis revealed that each bean-like magnetococcus had two flagellar bundles approximately 80 nm diameter (Fig. 2D), which are substantially larger than the diameter of 10 to 30 nm observed for single flagella (12), and dispersed individual flagella had been observed (data not shown). These results suggested the presence of a sheath often observed for marine magnetotactic coccoid-ovoid bacteria (17, 35).
The magnetosome crystals of each cell morphotype are likely to be single-domain crystals, according to theoretical predictions (6). Energy-dispersive X-ray spectroscopy (EDXS) analysis indicated that the magnetosome crystals in both groups were composed of iron and oxygen (Fig. 3C1). This is consistent with the results of high-resolution TEM (HRTEM) analysis, which identified the magnetosome crystals as magnetite (data not shown). In addition to magnetosomes, two (occasionally one) large electron-dense granules were observed in each group of BMC cells. EDXS analysis revealed that these were rich in phosphorus and oxygen (Fig. 3C2) and may be polyphosphate, as reported for other MTB (17, 23, 36).
Fluorescence in situ hybridization and phylogenetic analysis.
Nearly full-length 16S rRNA genes from the BMC cells were amplified and cloned. After four false-positive clones were eliminated from the clone library, 13 OTUs were identified by RFLP analysis and sequenced. Two dominant OTUs (107 clones) were identified after the putative non-MTB (nine OTUs, 87 clones) and putative chimeras (two OTUs, 2 clones) were eliminated. Clones 1-3 and 1-9 are representatives of the two dominant OTUs (including 61 clones and 46 clones, respectively), and they exhibited 91.8% sequence identity.
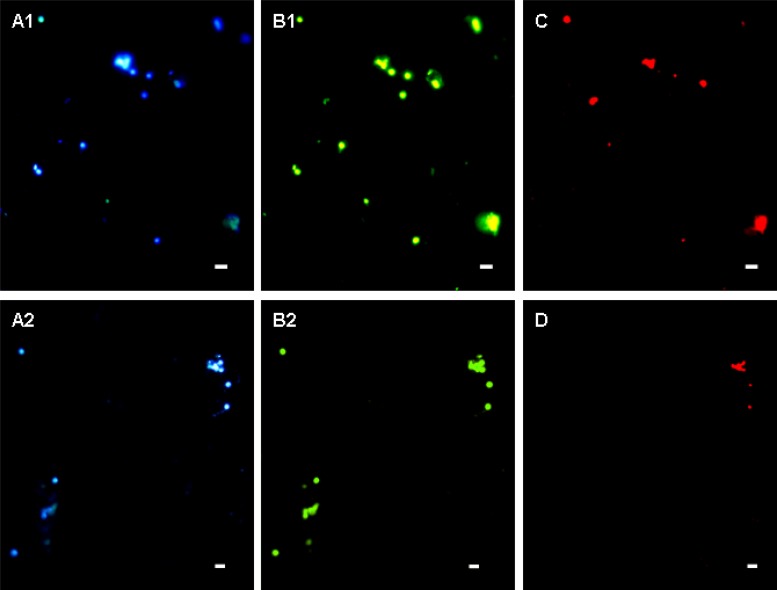
To confirm that the two sequences were associated with the BMC cells, two specific probes (p-3 and p-9) were designed and used in FISH analysis. Fluorescence microscopy revealed a strong signal for all collected cocci following hybridization with the universal bacterial probe EUB338 (Fig. 4B1 and B2). In contrast, the specific probes p-3 and p-9 hybridized with only some of the cells (Fig. 4C and D), confirming the presence of two types of BMC cells.
Fig 4.
Fluorescence in situ hybridization analyses of magnetically enriched BMC cells. The same microscopic field is shown following staining with 4′,6′-diamidino-2-phenylindole (A1 and A2), hybridization with the 5′-FAM-labeled bacterial universal probe EUB338 (B1 and B2), and hybridization with 5′-Cy3-labeled probe p-3 (C) and p-9 (D). Scale bars, 5 μm.
TEM analysis revealed either the clustered or chain-forming organization of the magnetosomes, whereas FISH experiments distinguished BMC species hybridizing with p-3 or p-9 in the collected samples. It was technically infeasible to directly relate magnetosome organization to the phylogenetic affiliation of the cells, and as magnetosomes display species specificity (2), it was possible to make a connection based on the occurrence frequency. RFLP analysis showed that the 16S rRNA gene sequence 1-3 represented 61 clones while the 1-9 represented 46 clones. In addition, most cells had cluster-type magnetosomes (Fig. 2B), and the p-3 probe hybridized with more cells (45%) than did the p-9 probe (38%) (Fig. 4C and D). Therefore, it may be possible that the 1-3 16S rRNA gene sequence is associated with cells that have clustered magnetosomes and hybridize with the p-3 probe. This hypothesis cannot be fully assessed until pure cultures of the BMC bacteria are obtained or until successful separation of single cells by various approaches, such as micromanipulation, is achieved.
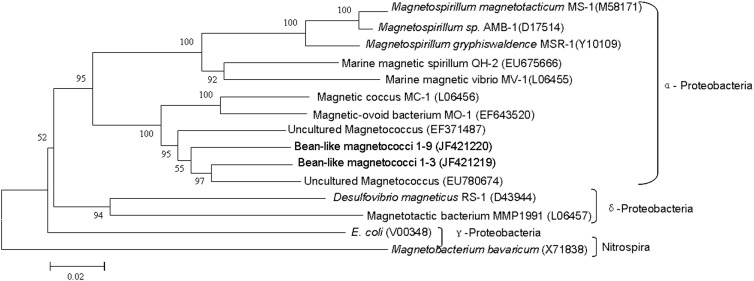
Among all 16S rRNA gene sequences in GenBank, the 1-3 16S rRNA gene sequence showed a maximum sequence identity (92.7%) with uncultured magnetococci collected from sediments of a freshwater lake (Miyun Lake, China; accession number EU780674), while clone 1-9 16S rRNA gene sequence showed 92.4% sequence identity with uncultured magnetococci from intertidal sediments of Huiquan Bay (accession number EF371487). Phylogenetic analysis revealed that the BMC cells were affiliated with the Alphaproteobacteria, and their 16S rRNA gene sequences had >7% divergence from all previously reported bacteria (Fig. 5). Therefore, the bean-like magnetococci may represent two new genera of magnetotactic bacteria (13). This finding highlights the remarkable biodiversity of magnetotactic bacteria in general and suggests that many novel magnetococcus species remain to be discovered.
Fig 5.
Phylogenetic tree showing the relationship between the BMC and related magnetotactic bacteria. The tree is constructed based on neighbor-joining analysis using the sequence region from position 27 to 1492 using E. coli numbering. The sequences determined in this study are shown in bold. GenBank accession numbers of the sequences used are indicated in parentheses. The scale bar is 0.02 substitutions per nucleotide position.
ACKNOWLEDGMENTS
We thank Jianhong Xu for assistance in sampling.
This work was supported by the National Natural Science Foundation of China (NSFC 40906069, 41106135, and 40776094), the Special Construction Engineering Foundation for Taishan Scholar, and the KC Wong Education Foundation.
Footnotes
Published ahead of print 1 June 2012
REFERENCES
- 1. Amann R, Peplies J, Schüler D. 2006. Diversity and taxonomy of magnetotactic bacteria, p 25–36 In Schüler D. (ed), Microbiology monographs: magnetoreception and magnetosomes in bacteria. Springer Press, Heidelberg, Germany [Google Scholar]
- 2. Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217–230 [DOI] [PubMed] [Google Scholar]
- 3. Bellini S. 2009. Further studies on “magnetosensitive bacteria.” Chin. J. Oceanol. Limnol. 27:6–12 [Google Scholar]
- 4. Bellini S. 2009. On a unique behavior of freshwater bacteria. Chin. J. Oceanol. Limnol. 27:3–5 [Google Scholar]
- 5. Blakemore RP. 1975. Magnetotactic bacteria. Science 190:377–379 [DOI] [PubMed] [Google Scholar]
- 6. Butler RF, Banerjee SK. 1975. Theoretical single-domain grain size range in magnetite and titanomagnetite. J. Geophys. Res. 80:4049–4058 [Google Scholar]
- 7. Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875–4898 [DOI] [PubMed] [Google Scholar]
- 8. Farina M, Lins de Barros JG, de Esquivel MS, Danon J. 1983. Ultrastructure of a magnetotactic microorganism. Biol. Cell 48:85–88 [Google Scholar]
- 9. Frankel RB. 2009. The discovery of magnetotactic/magnetosensitive bacteria. Chin. J. Oceanol. Limnol. 27:1–2 [Google Scholar]
- 10. Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frankel RB, Williams TJ, Bazylinski DA. 2006. Magneto-aerotaxis, p 2–24 In Schüler D. (ed), Microbiology monographs: magnetoreception and magnetosomes in bacteria. Springer, Heidelberg, Germany [Google Scholar]
- 12. Galkin VE, et al. 2008. Divergence of quaternary structures among bacterial flagellar filaments. Science 320:382–385 [DOI] [PubMed] [Google Scholar]
- 13. Hagström R, Pinhassi J, Zweifel UL. 2000. Biogeographical diversity among marine bacterioplankton. Aquat. Microb. Ecol. 21:231–244 [Google Scholar]
- 14. Kolinko S, et al. 18 October 2011. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within candidate division OP3. Environ. Microbiol. doi:10.1111/j.1462-2920.2011.02609.x [DOI] [PubMed] [Google Scholar]
- 15. Komeili A. 2007. Molecular mechanisms of magnetosome formation. Annu. Rev. Biochem. 76:351–366 [DOI] [PubMed] [Google Scholar]
- 16. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Wiley & Sons Press, Chichester, United Kingdom [Google Scholar]
- 17. Lefèvre CT, Bernadac A, Yu-Zhang K, Pradel N, Wu L-F. 2009. Isolation and characterization of a magnetotactic bacterial culture from the Mediterranean Sea. Environ. Microbiol. 11:1646–1657 [DOI] [PubMed] [Google Scholar]
- 18. Lefèvre CT, Frankel RB, Abreu F, Lins U, Bazylinski DA. 2011. Culture-dependent characterization of a novel, uncultivated magnetotactic member of the Nitrospirae phylum. Environ. Microbiol. 13:538–549 [DOI] [PubMed] [Google Scholar]
- 19. Lefèvre CT, et al. 2011. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science 334:1720–1723 [DOI] [PubMed] [Google Scholar]
- 20. Lefèvre CT, et al. 2012. Novel magnetite-producing magnetotactic bacteria belonging to the Gammaproteobacteria. ISME J. 6:440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin W, Li JH, Schüler D, Jogler C, Pan YX. 2009. Diversity analysis of magnetotactic bacteria in Lake Miyun, northern China, by restriction fragment length polymorphism. Syst. Appl. Microbiol. 32:342–350 [DOI] [PubMed] [Google Scholar]
- 22. Lin W, Pan YX. 2009. Uncultivated magnetotactic cocci from Yuandadu Park in Beijing, China. Appl. Environ. Microbiol. 75:4046–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lins U, Farina M. 1999. Phosphorus-rich granules in uncultured magnetotactic bacteria. FEMS Microbiol. Lett. 172:23–28 [Google Scholar]
- 24. Mann S, Sparks NH, Board RG. 1990. Magnetotactic bacteria: microbiology, biomineralization, palaeomagnetism and biotechnology. Adv. Microb. Physiol. 31:125–181 [DOI] [PubMed] [Google Scholar]
- 25. Pan HM, et al. 2008. Characterization of a homogeneous taxonomic group of marine magnetotactic cocci within a low tide zone in the China Sea. Environ. Microbiol. 10:1158–1164 [DOI] [PubMed] [Google Scholar]
- 26. Pernthaler J, Glöckner FO, Schönhuber W, Amann R. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes, p 207–226 In Paul JH. (ed), Methods in microbiology: marine microbiology. Academic Press, London, United Kingdom [Google Scholar]
- 27. Schüler D. 1999. Formation of magnetosomes in magnetotactic bacteria. J. Mol. Microbiol. Biotechnol. 1:79–86 [PubMed] [Google Scholar]
- 28. Schüler D, Frankel RB. 1999. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl. Microbiol. Biotechnol. 52:464–473 [DOI] [PubMed] [Google Scholar]
- 29. Simmons SL, Bazylinski DA, Edwards KJ. 2006. South-seeking magnetotactic bacteria in the Northern Hemisphere. Science 311:371–374 [DOI] [PubMed] [Google Scholar]
- 30. Simmons SL, Sievert SM, Frankel RB, Bazylinski DA, Edwards KJ. 2004. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl. Environ. Microbiol. 70:6230–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spring S, et al. 1994. Phylogenetic analysis of uncultured magnetotactic bacteria from the alpha-subclass of Proteobacteria. Syst. Appl. Microbiol. 17:501–508 [Google Scholar]
- 32. Spring S, et al. 1998. Phylogenetic affiliation and ultrastructure of uncultured magnetic bacteria with unusually large magnetosomes. Arch. Microbiol. 169:136–147 [DOI] [PubMed] [Google Scholar]
- 33. Tanaka M, Arakaki A, Matsunaga T. 2010. Identification and functional characterization of tubulation protein from magnetotactic bacteria. Mol. Microbiol. 76:480–488 [DOI] [PubMed] [Google Scholar]
- 34. Wolfe RS, Thauer RK, Pfennig N. 1987. A “capillary racetrack” method for isolation of magnetotactic bacteria. FEMS Microbiol. Ecol. 45:31–35 [Google Scholar]
- 35. Zhang WJ, et al. 2012. Complex spatial organization and flagellin composition of flagellar propeller from marine magnetotactic ovoid strain MO-1. J. Mol. Biol. 416:558–570 [DOI] [PubMed] [Google Scholar]
- 36. Zhu KL, et al. 2010. Isolation and characterization of a marine magnetotactic spirillum axenic culture QH-2 from an intertidal zone of the China Sea. Res. Microbiol. 161:276–283 [DOI] [PubMed] [Google Scholar]