Abstract
Streptococcus pneumoniae and a number of commensal streptococcal species are competent for natural genetic transformation. The natural habitat of these bacteria is multispecies biofilms in the human oral cavity and nasopharynx. Studies investigating lateral transfer of virulence and antibiotic resistance determinants among streptococci have shown that interspecies as well as intraspecies gene exchange takes place in these environments. We have previously shown that the action of a competence-specific murein hydrolase termed CbpD strongly increases the rate of gene transfer between pneumococci grown in liquid cultures. CbpD is the key component of a bacteriolytic mechanism termed the fratricide mechanism. It is secreted by competent pneumococci and mediates the release of donor DNA from sensitive streptococci present in the same environment. However, in nature, gene exchange between streptococci takes place in biofilms and not in liquid cultures. In the present study, we therefore investigated whether CbpD affects the rate of gene transfer in laboratory-grown biofilms. Our results show that the fratricide mechanism has a strong positive impact on intrabiofilm gene exchange, indicating that it is important for active acquisition of homologous donor DNA under natural conditions. Furthermore, we found that competent biofilm cells of S. pneumoniae acquire a Novr marker much more efficiently from neighboring cells than from the growth medium. Efficient lysis of target cells requires that CbpD act in conjunction with the murein hydrolase LytC. In contrast, the major autolysin LytA does not seem to be important for fratricide-mediated gene exchange in a biofilm environment.
INTRODUCTION
Bacteria that are competent for natural genetic transformation have the ability to take up naked DNA from the surrounding medium and incorporate it into their genomes by homologous recombination. In members of the genus Streptococcus, the competent state is strictly regulated. Competence induction in these bacteria depends on a quorum sensing-like mechanism as well as internal and external cues. In Streptococcus pneumoniae, the quorum sensing mechanism consists of a secreted unmodified signal peptide encoded by comC, its histidine kinase receptor ComD, and the cognate response regulator ComE (15, 31). The signal peptide (CSP) is exported by a dedicated secretion and processing apparatus termed ComAB (19). Synthesis of CSP and the ComDE two-component regulatory system is controlled by upstream regulators such as CiaRH and StkP (8, 12, 14). Extracellular CSP is sensed by ComD (16), which, upon binding of its ligand, presumably transfers a phosphoryl group to ComE. Phosphorylated ComE drives the expression of about 20 early competence genes (7, 32, 40). Among these are comX1 and comX2, two identical genes that encode the alternative sigma factor ComX (27). ComX controls transcription of the late competence genes, some of which are involved in DNA binding, uptake, and recombination (7, 32).
In addition to proteins required for uptake and processing of DNA, competent streptococci produce bacteriolytic cell wall hydrolases, so-called fratricins (for reviews, see references 3 and 5). Two major families of fratricins, designated CbpD and LytF, have been identified in the genus Streptococcus (2, 13, 22). With the exception of Streptococcus agalactiae, each species synthesizes a particular fratricin subtype that belongs to one of these families (2). Fratricins have varying but limited target ranges and are in general active against bacteria that are relatively closely related to the producer strain (3). Since they are encoded by late competence genes, fratricins are produced only when streptococci enter the competent state. S. pneumoniae and its close commensal relatives Streptococcus mitis and Streptococcus oralis produce fratricins that belong to the CbpD family (2, 6). To protect themselves against their own fratricins, these species produce an immunity protein termed ComM, which is encoded by an early competence gene (17). ComM is an integral membrane protein of unknown function that has no close homologues in the databases. No immunity protein has been identified that protects against LytF-type fratricins (2).
In mixed cultures of competent and noncompetent pneumococci, a fraction of the noncompetent cells will lyse and release DNA that can be taken up by their competent siblings. Consequently, it has been proposed that fratricide serves as a mechanism for active acquisition of homologous DNA (36). Evidence supporting this hypothesis have been obtained in experiments carried out with planktonic cells. Johnsborg et al. (21) showed that transfer of an antibiotic resistance marker in mixed cultures of competent and noncompetent pneumococci is a thousandfold more efficient when the competent attacker cells have an intact cbpD gene. A strong positive effect of CbpD was also observed in experiments where competent pneumococci were cocultivated with noncompetent S. mitis or S. oralis cells (21). The CbpD subtype produced by S. pneumoniae, S. mitis, and S. oralis consists of an N-terminal CHAP domain, one or two central SH3 domains, and a C-terminal choline-binding domain. The CHAP domain belongs to a family of cysteine- and histidine-dependent amidohydrolases/peptidases whose members function either as N-acetylmuramoyl-l-Ala amidases or as endopeptidases that cleave within the peptide part of peptidoglycan (1, 26, 34). Evidence suggests that the SH3 domain of CbpD binds to the peptidoglycan part of the cell wall, while the choline-binding domain anchors the enzyme noncovalently to choline residues decorating the wall and lipoteichoic acids of S. pneumoniae, S. mitis, and S. oralis (10, 35). In addition to CbpD, the murein hydrolases LytA and LytC are part of the fratricide mechanism in S. pneumoniae (9, 13). In liquid cultures, susceptible cells are first targeted by CbpD, which binds to and attacks their septal regions. The damage caused by CbpD activates LytA and LytC, resulting in more extensive lysis of target cells than achieved by CbpD alone (9, 10). LytA as well as LytC is constitutively synthesized by noncompetent cells. However, while the expression of LytA increases during competence (29), LytC is not part of the competence regulon in S. pneumoniae (7, 32).
In a recent study, Berg et al. (2) showed that the presence of LytF dramatically enhances the rate of gene exchange between two Streptococcus gordonii strains in mixed liquid cultures. The two strains, Challis and NCTC 7865, produce CSPs with different primary structures. Consequently, when competence is induced in strain Challis by addition of synthetic Challis-CSP, the NCTC 7865 cells remain noncompetent. To determine the effect of LytF on lateral gene transfer, the cocultivation experiments were carried out with LytF-proficient as well as LytF-deficient Challis cells. The results demonstrated that transfer of a rifampin resistance gene from noncompetent NCTC 7865 cells to competent Challis cells was a hundredfold more efficient in experiments where Challis-proficient cells were used.
The natural habitat of S. pneumoniae and many other naturally transformable species in the genus Streptococcus is complex biofilms covering mucosal surfaces in humans and animals. Studies on penicillin-resistant clinical isolates of S. pneumoniae have shown that some of them carry resistance determinants originating from Streptococcus mitis or Streptococcus oralis (4). Transfer of genetic material from these commensals to S. pneumoniae must have taken place in an environment where they come into close contact. Since all three species can be isolated from multispecies biofilms in the human naso- and oropharynx, it is reasonable to assume that they normally exchange DNA in this environment. This prompted us to investigate whether fratricins have the same positive impact on gene exchange in biofilms as previously observed in liquid cultures. To address this question, we grew different mutant strains of S. pneumoniae in mixed biofilms and estimated the rate of gene transfer between them. Our results show that competent pneumococci expressing CbpD acquire transforming DNA from their noncompetent biofilm neighbors much more efficiently than competent pneumococci lacking a functional cbpD gene. Furthermore, in accordance with previous studies performed on planktonic cells, we found that the fratricide mechanism requires the joint action of CbpD, LytA, and LytC to function optimally.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pneumoniae strains used in the present study were stored as precultures at −80°C. Precultures were made by growing the strains in C medium (25) at 37°C without shaking. At an optical density at 550 nm (OD550) of ∼0.3 cultures were mixed with glycerol (17% vol/vol) and stored at −80°C.
Construction of streptococcal mutants.
The properties of pneumococcal strains used in this work are described in Table 1, and the primers used in the construction of mutant strains are listed in Table 2. DNA was introduced into pneumococcal strains by means of natural transformation using 100 ng ml−1 of CSP-1 (15) to induce the competent state. Cultures of S. pneumoniae were grown at 37°C until they reached an OD550 of ∼0.06. Then CSP and transforming DNA were added, and the cultures were further incubated for 120 min at 37°C followed by plating on selective medium to identify transformants. S. pneumoniae transformants were selected by plating on Todd-Hewitt agar supplemented with kanamycin (400 μg ml−1), streptomycin (200 μg ml−1), novobiocin (2.5 μg ml−1), tetracycline (0.2 μg ml−1), or spectinomycin (200 μg ml−1).
Table 1.
S. pneumoniae strains
| Strains | Genotype and/or relevant feature(s) | Source or reference |
|---|---|---|
| R704 | R6 derivative; comA::ermAM Eryr | J.-P. Claverysa |
| CP1200 | mal rpsL1 Smr | 28 |
| RH1 | R704 but ebg::spc ΔcomA Eryr Spcr | 21 |
| RH2 | RH1 but hirL::pEVP3 ΔcomA Cmr Eryr Spcr | 21 |
| RH4 | RH2 but Smr by transformation with CP1200 DNA; ΔcomA Cmr Eryr Spcr Smr | 9 |
| RH5 | RH4 but ΔlytA::Janus ΔcomA Cmr Eryr Spcr Kanr | 9 |
| RH6 | RH5 but removal of Janus by transformation with appropriate PCR fragment; ΔcomA ΔlytA Cmr Eryr Spcr Smr | 9 |
| RH7 | RH6 but ΔcomE::kan ΔcomA ΔlytA Cmr Eryr Spcr Smr Kanr | 9 |
| RH10 | RH6 but ΔlytC::tet ΔcomA ΔlytA Cmr Eryr Spcr Smr Tcr | 9 |
| RH14 | RH1 but ΔlytA::kan ΔcomA Eryr Spcr Kanr | 9 |
| RH16 | RH14 but ΔlytC::tet ΔcomA Eryr Spcr Kanr Tcr | 9 |
| RH17 | RH1 but ΔcbpD::kan ΔcomA Eryr Spcr Kanr | 21 |
| RH400 | R704 but Novr | 21 |
| RH401 | RH400 but ΔcomE::kan ΔcomA Kanr Novr | 21 |
| SPH27 | RH400 but Smr by transformation with CP1200 DNA; ΔcomA Eryr Novr Smr | This study |
| SPH28 | SPH27 but ΔlytA::Janus, ΔcomA, Eryr, Novr, Kanr | This study |
| SPH29 | SPH28 but removal of Janus cassette by transformation with appropriate PCR fragment; ΔcomA ΔlytA Eryr Novr Smr | This study |
| SPH30 | SPH29 but ΔcomE::kan by transformation with RH7 DNA, ΔcomA, ΔlytA Eryr Novr Smr Kanr | This study |
| SPH147 | SPH29 but ΔlytC::tet by transformation with RH10 DNA, ΔcomA ΔlytA Eryr Novr Smr Tcr | This study |
| SPH148 | SPH147 but ΔcomE::kan by transformation with RH7 DNA; ΔcomA ΔlytA ΔlytC Eryr Novr Smr Tcr Kanr | This study |
| SPH106 | RH6 but ΔcbpD::kan ΔcomA ΔlytA Cmr Eryr Spcr Smr Kanr | This study |
| SPH149 | SPH106 but ΔlytC::tet by transformation with RH10 DNA; ΔcomA ΔlytA ΔcbpD Cmr Eryr Spcr Smr Kanr Tcr | This study |
Gift from Jean-Pierre Claverys.
Table 2.
Primers
In order to use the Janus cassette for gene replacement through negative selection in S. pneumoniae, the recipient strain must carry the rpsL1 mutant allele, which gives rise to streptomycin resistance (38). Strain RH400 (rpsL+) was therefore transformed with genomic DNA from S. pneumoniae CP1200 (rpsL1 Smr) followed by selection on agar plates containing streptomycin. This gave rise to strain SPH27. SPH28 was constructed by transforming strain SPH27 with a PCR fragment consisting of the Janus cassette flanked by ∼1,000 bp of the regions immediately upstream and downstream of the lytA gene. This fragment was amplified from the RH5 strain using the primers LytAF and LytAR. Transformants were identified by selection on agar plates containing kanamycin. Next, the Janus cassette was removed from the SPH28 strain by transformation with a PCR fragment consisting of the fused flanking regions. To make this fragment, PCR was carried out with the primers LytAF and LytAR and genomic DNA from strain RH6 as the template. The resulting transformants, designated SPH29, were selected on agar plates containing streptomycin. To delete its comE gene, SPH29 was transformed with genomic DNA from strain RH7 followed by selection on kanamycin-containing plates. This gave rise to strain SPH30. SPH147 was constructed by transforming SPH29 with a PCR fragment consisting of a tetracycline resistance gene flanked by ∼1,000 bp of the regions immediately upstream and downstream of the pneumococcal lytC gene. PCR was carried out with the primers LytCF and LytCR and with genomic DNA from strain RH10 as the template. Transformants were selected on agar plates containing tetracycline. SPH148 was constructed by transforming SPH147 with DNA from RH7 as described above. To construct SPH106, we first used PCR to amplify a fragment consisting of the Janus cassette flanked by ∼1,000 bp of the regions immediately upstream and downstream of the pneumococcal cbpD gene. PCR was carried out with the primers CbpD.1 and CbpD.4 and with genomic DNA from strain RH17 as the template. Next, this PCR fragment was used to transform strain RH6, followed by selection on kanamycin-containing plates. Finally, SPH149 was constructed by deleting the lytC gene in strain SPH106 exactly as described above for construction of SPH147.
Cultivation of S. pneumoniae R6 biofilm.
Biofilms of the different strains used in this study were grown in glass-bottom dishes (WillCo-dish; WillCo Wells B V, Amsterdam, The Netherlands) essentially as described by Moscoso et al. (30). After glycerol stocks were thawed at 37°C, cells were pelleted by centrifugation at 4°C. Pellets were resuspended in an equal volume of fresh C medium, diluted 1/10 in the same medium, and cultivated at 37°C to an OD600 of ∼0.5. Next, cell suspensions were diluted 1/100 in C medium and dispensed into glass-bottom petri dishes (2 ml per well). Dishes were incubated at various times at 37°C.
Microscopy.
To follow the kinetics of pneumococcal biofilm formation, RH1 cells were grown in separate glass-bottom dishes for 2, 4, 6, 9, 12, and 24 h at 37°C. Before microscopic examination, the culture medium was removed, and the biofilms were gently rinsed three times with prewarmed C medium (37°C) to remove nonadherent bacteria. Cells attached to the inner surface of the glass coverslip were viewed by differential interference contrast microscopy (DIC) using an inverted confocal microscope (model LSM 700; Zeiss, Heidelberg, Germany).
RH14 biofilms grown for 12 and 24 h were treated as described above and stained with FilmTracer calcein red-orange biofilm stain (Invitrogen) according to the manufacturers' specifications. After staining, biofilms were gently rinsed with sterilized water and analyzed on a Zeiss LSM 700 confocal microscope equipped with the ZEN software (Zeiss). Images were acquired using a Plan-Apochromat 63×/1.4 oil immersion objective and a 555-nm laser line for excitation of calcein red-orange.
Staining with a BacLight Live/Dead staining kit (Invitrogen) was used to investigate the effect of CbpD-mediated fratricide in biofilms. Mixed biofilms of RH14/SPH30 and SPH106/SPH30 cells were grown in glass-bottom dishes at 37°C for 4 h. After rinsing with C medium three times to remove nonadherent bacteria, 2 ml of C medium supplemented with CSP-1 (100 ng ml−1) was gently added to the edge of the dish to induce the competent state. After further incubation at 30°C for 20 min, biofilms were washed three times and stained with the BacLight kit as described by the manufacturer. Biofilm images were acquired as described above using the 555-nm laser line for excitation of propidium iodide and the 488-nm laser line for excitation of SYTO 9. Zen software (Zeiss) and Comstat2 (18) were used for biofilm analysis.
Transformation of biofilm cells by exogenously added DNA.
Glycerol stocks of RH1 and RH14 were thawed and treated as specified in the paragraph “Cultivation of S. pneumoniae R6 biofilm.” Two milliliters of diluted RH1 and RH14 cells were dispensed into separate glass-bottom dishes and incubated at 37°C for 4 h, 6 h, 8 h, and 12 h. After the biofilms were washed three times, 2 ml of C medium supplemented with CSP-1 (100 ng ml−1) and genomic DNA (1 μg ml−1) from RH401 cells was gently added to each petri dish. Uninduced samples, supplemented only with DNA, were run in parallel as negative controls. Following incubation at 30°C for 2 h, the biofilms were washed three times with prewarmed medium to remove nonadherent bacteria. Next, 1 ml prewarmed C medium was added to each dish, and biofilm cells were detached by sonication in a water bath for 15 s followed by scraping with a sterile cell scraper. Detached bacteria were collected in a Falcon tube. Then, 2 ml fresh prewarmed medium was added to the dishes, and they were scraped again to remove any remaining cells. This procedure was repeated four times. All samples originating from the same petri dish were collected in a single tube. Next, the bacteria were pelleted by centrifugation at 4,600 × g for 6 min at 4°C. After careful removal of the supernatants, pellets were resuspended in 1 ml C medium and put on ice. Then each bacterial suspension was serially diluted and plated on Todd-Hewitt agar containing spectinomycin and a combination of spectinomycin and novobiocin. The plates were incubated anaerobically at 37°C for 15 to 20 h using the Oxoid AnaeroGen system. The resulting colonies were counted and used to calculate the transformation efficiencies.
Intrabiofilm gene transfer.
Glycerol stocks of attacker and target strains were thawed and treated as described above. When they reached an OD600 of ∼0.5, they were diluted 1/50 in fresh C medium, and 1-ml portions of each were mixed in glass-bottom dishes and grown for the desired period of time at 37°C. Following washes (three times), 2 ml of C medium containing 100 ng ml−1 of CSP was gently added to the dishes to induce the competent state. Uninduced samples were run in parallel as negative controls. After incubation of the dishes at 30°C for 2 h, biofilm cells were detached and harvested as described above. To determine transformation efficiency, serially diluted bacterial samples were plated on Todd-Hewitt agar containing spectinomycin, novobiocin, and a combination of the two. The number of spectinomycin-resistant colonies was used to calculate the total number of attacking (recipient) cells present in the biofilm, while the number of novobiocin-resistant colonies was used to calculate the total number of target (donor) cells. The number of colonies growing on plates containing both spectinomycin and novobiocin was used to estimate the total number of Novr transformants obtained by intrabiofilm gene transfer.
RESULTS
Growth of S. pneumoniae R6 in glass-bottom dishes.
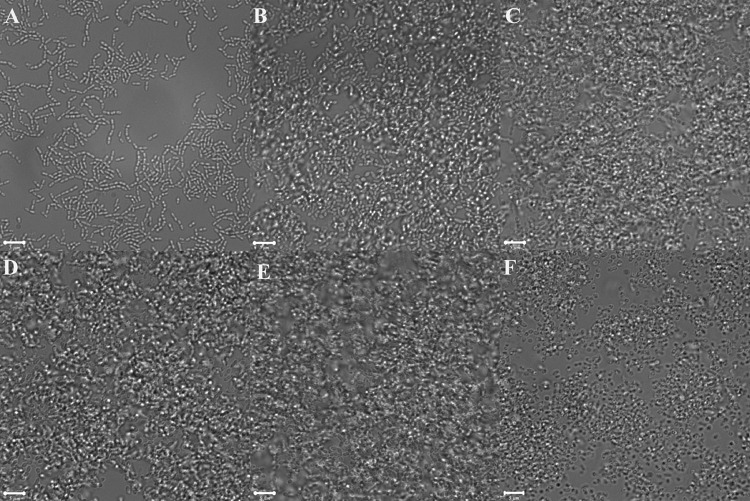
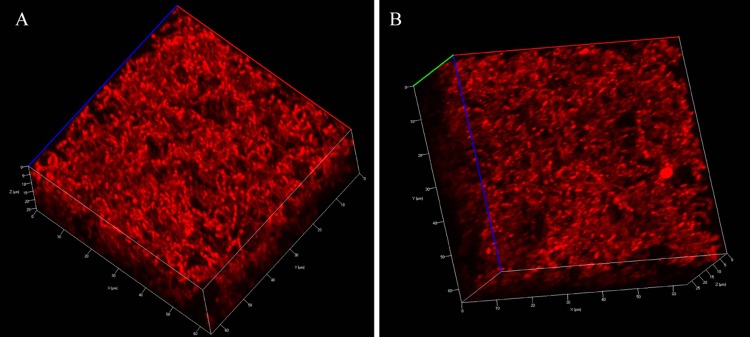
To explore the ability of the S. pneumoniae R6 derivative RH1 to grow and form biofilms in glass-bottom dishes, we grew the strain in C medium at 37°C for 2, 4, 6, 9, 12, and 24 h. Before inspection, the dishes were washed three times to remove nonattached cells. Two hours after seeding, pneumococcal cells began to attach to the glass surface (Fig. 1A). After 4 h, microcolony formation with multilayers of cells was detected (Fig. 1B). At this stage, the average thickness of the biofilm was estimated to be about 6.3 ± 0.5 μm. This estimate represents the mean of five independent measurements performed at different spots. Each spot represents a single stack of images. The biofilm continued to increase in thickness up to 12 h (Fig. 1E). However, after 24 h, the biofilm had deteriorated and the biomass was strongly reduced (Fig. 1F). This deterioration took place regardless of whether the culture medium was changed at 12 h. S. pneumoniae is prone to LytA-mediated autolysis in liquid cultures after entering stationary growth phase (39). We therefore assumed that the same phenomenon was behind the observed disappearance of the 24-h-old biofilm. To test this hypothesis, the RH14 (ΔlytA) strain of S. pneumoniae R6 was cultivated in exactly the same way as the LytA-proficient RH1 strain discussed above. In this case, biofilms examined at 12 and 24 h displayed similar structures (Fig. 2). The average thickness and biomass of an RH14 biofilm cultured for 12 h were 16.5 ± 0.6 μm and 7.9 ± 0.5 μm3 μm−2, respectively. The corresponding estimates for an RH14 biofilm grown for 24 h were 17.3 ± 0.9 μm and 8.1 ± 0.8 μm3 μm−2. These results demonstrate that, under the conditions used, LytA triggers autolysis in monospecies biofilms of S. pneumoniae 12 to 24 h after seeding.
Fig 1.
S. pneumoniae biofilm development followed over a period of 24 h. DIC micrographs were taken 2 h (A), 4 h (B), 6 h (C), 9 h (D), 12 h (E), and 24 h (F) after inoculation.
Fig 2.
Confocal microscopy of strain RH14 (ΔlytA) biofilms grown for 12 h (A) and 24 h (B) at 37°C. The biofilms were stained with calcein red-orange to visualize the entire biofilm structure.
Transformability and gene transfer in pneumococcal biofilms of different ages.
In liquid culture, CSP induces the competent state in pneumococci as long as they are actively growing. In stationary phase, however, they remain noncompetent even when subjected to CSP (15). To determine whether the stage of pneumococcal biofilm formation influences competence development, the transformability of biofilm cells was tested at various time points postinoculation. Biofilms grown for 4, 6, 8, and 12 h at 37°C were carefully washed three times in C medium and then subjected to CSP (100 ng/ml) and genomic Novr DNA (1 μg/ml). After further incubation in fresh C medium for 2 h at 30°C, the biofilms were washed again (three times), sonicated in a water bath for 15 s, and harvested by detaching the cells from the culture dishes with a scraper. Resuspended biofilm cells were serially diluted and spread on agar plates containing 2.5 μg/ml novobiocin. The number of transformants (CFU) was determined after overnight incubation at 37°C. Our results show that in the early stage of biofilm formation (4 h) the cells become competent when treated with CSP, whereas only a very small fraction of the cells were transformed in 8-h-old biofilms (Fig. 3A). These results clearly demonstrate that under the conditions used, the transformability of sessile pneumococci is strongly affected by the stage of biofilm formation.
Fig 3.
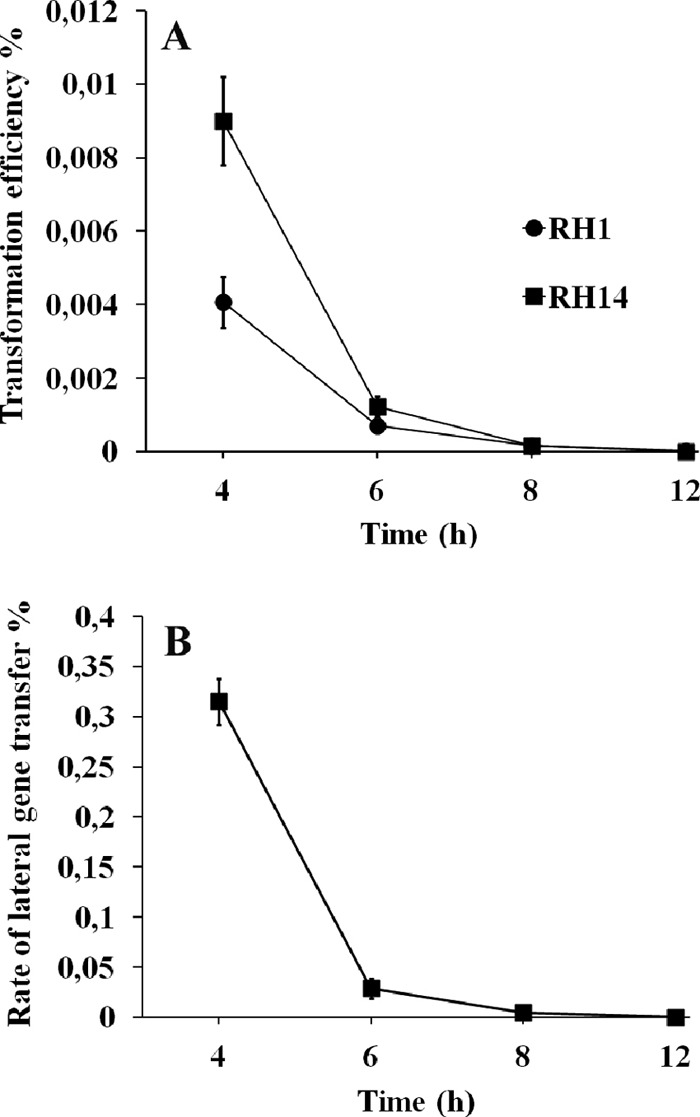
Transformation in S. pneumoniae biofilms cultivated for different periods of time. (A) Transformation efficiencies obtained in RH1 and RH14 biofilms subjected to CSP (100 ng ml−1) and purified Novr DNA (1 μg ml−1) 4, 6, 8, and 12 h postinoculation. CSP and transforming DNA were added to the biofilm medium concomitantly. (B) Intrabiofilm transfer of a Novr marker in mixed biofilms consisting of RH1 (Spcr) and RH401 (ΔcomE Novr) cells seeded at a 1:1 ratio. To induce the competent state, CSP (100 ng ml−1) was added 4, 6, 8, and 12 h postinoculation. Results are means ± standard errors from three independent experiments.
Next we wanted to establish an assay that would enable us to determine the rate of gene transfer between biofilm cells carrying different antibiotic resistance markers. RH1 (Spcr) and noncompetent RH401 (ΔcomE Novr) cells were seeded together in a 1:1 ratio in glass-bottom dishes and incubated at 37°C. After 4, 6, 8, and 12 h of incubation, the resulting biofilms were washed three times in C medium. Fresh C medium containing 100 ng/ml of CSP was then added, and the culture dishes were further incubated at 30°C for 2 h. Before being harvested by the procedure described above, the cells were washed carefully three times to remove nonattached cells. Resuspended cells were serially diluted and spread on agar plates containing the appropriate antibiotic(s). Following overnight incubation at 37°C, the transformation efficiency was calculated by dividing the number of Spcr Novr colonies by the total number of Spcr resistant colonies. Similar to the transformation experiments carried out with naked genomic DNA, intrabiofilm gene transfer was most efficient in 4-h-old biofilms (Fig. 3B). However, most interestingly, the RH1 cells acquired the Novr marker much more efficiently (about 80-fold) from neighboring RH401 cells than from the growth medium (Fig. 3).
Fratricide strongly enhances intrabiofilm gene transfer.
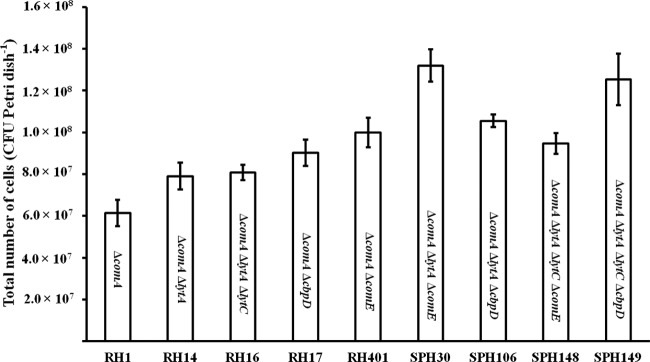
As described in the introduction, previous studies have shown that the fratricide mechanism enhances the efficiency of gene exchange between planktonic streptococci by orders of magnitude (2, 21). To determine whether this mechanism boosts gene exchange between adherent pneumococci, we compared the rate of intrabiofilm gene transfer in the presence and absence of CbpD. In biofilms containing the RH17 (ΔcbpD Spcr) and RH401 (ΔcomE Novr) mutant strains, transformation efficiency was reduced almost 50-fold compared to that in biofilms consisting of the RH1 (Spcr) and RH401 (ΔcomE Novr) strains (Table 3). In principle, this difference could be influenced by the number of RH1 and RH17 cells present in their respective biofilms during the course of the experiments. If RH17 is a very poor biofilm producer compared to RH1, it would be expected that the experiments involving RH17 and RH401 would give rise to fewer Spcr Novr transformants. To rule out this possibility, the biofilm-forming properties of RH1, RH17, and the other RH1 derivatives used in this study were compared. The results presented in Fig. 4 show that their ability to form biofilms under the conditions used is about the same. Together, these results clearly show that the presence of CbpD strongly enhances the efficiency of gene transfer between neighboring cells in pneumococcal biofilms.
Table 3.
Impact of CbpD, LytC, and LytA on lateral gene transfer in biofilms
| Strain |
No. (CFU/petri dish) of: |
Transformation efficiency (%)a | ||
|---|---|---|---|---|
| Competent attacker | Noncompetent target | Spcr attackers | Spcr Novr transformants | |
| RH1 (ΔcomA Spcr) | RH401 (ΔcomA ΔcomE Novr) | 6.7 × 107 ± 8.2 × 106 | 2.2 × 105 ± 2.1 × 104 | 0.34 ± 0.014 |
| RH17 (ΔcomA ΔcbpD Spcr) | RH401 (ΔcomA ΔcomE Novr) | 9.2 × 107 ± 9.1 × 106 | 5.9 × 103 ± 8.3 × 102 | 0.007 ± 0.001 |
| RH14 (ΔcomA ΔlytA Spcr) | SPH30 (ΔcomA ΔlytA ΔcomE Novr) | 9.3 × 107 ± 7.4 × 106 | 3.1 × 105 ± 2.7 × 104 | 0.338 ± 0.012 |
| SPH106 (ΔcomA ΔlytA ΔcbpD Spcr) | SPH30 (ΔcomA ΔlytA ΔcomE Novr) | 1.2 × 108 ± 4.1 × 106 | 1.6 × 103 ± 1.2 × 102 | 0.0015 ± 0.0001 |
| RH16 (ΔcomA ΔlytA ΔlytC Spcr) | RH401 (ΔcomA ΔcomE Novr) | 8.1 × 107 ± 3.6 × 106 | 7.4 × 104 ± 4.1 × 103 | 0.094 ± 0.009 |
| SPH149 (ΔcomA ΔlytA ΔlytC ΔcbpD Spcr) | RH401 (ΔcomA ΔcomE Novr) | 1.3 × 108 ± 1.2 × 107 | 1.1 × 104 ± 1.4 × 103 | 0.009 ± 0.0002 |
| RH16 (ΔcomA ΔlytA ΔlytC Spcr) | SPH30 (ΔcomA ΔlytA ΔcomE Novr) | 1.2 × 108 ± 2.7 × 106 | 4.7 × 104 ± 3.6 × 103 | 0.04 ± 0.003 |
| SPH149 (ΔcomA ΔlytA ΔlytC ΔcbpD Spcr) | SPH30 (ΔcomA ΔlytA ΔcomE Novr) | 1.0 × 108 ± 1.1 × 107 | 1.5 × 103 ± 3.2 × 102 | 0.0015 ± 0.0003 |
| RH16 (ΔcomA ΔlytA ΔlytC Spcr) | SPH148 (ΔcomA ΔlytA ΔlytC ΔcomE Novr) | 1.1 × 108 ± 3.7 × 106 | 7.1 × 103 ± 3.2 × 102 | 0.0064 ± 0.0002 |
| SPH149 (ΔcomA ΔlytA ΔlytC ΔcbpD Spcr) | SPH148 (ΔcomA ΔlytA ΔlytC ΔcomE Novr) | 1.2 × 108 ± 5.1 × 106 | 3.1 × 102 ± 3.3 × 101 | 2.6 × 10−4 ± 2.4 × 10−5 |
Number of Spcr Novr transformants obtained (CFU/petri dish) divided by the total number of Spcr resistant attacker cells (CFU/petri dish) multiplied by 100. Results are the means from at least three independent experiments ± standard errors.
Fig 4.
Comparison of the biofilm-forming capacities of different S. pneumoniae mutant strains used in the present study. The strains were grown in glass-bottom petri dishes for 4 h at 37°C and then transferred to 30°C for 2 h in the presence of CSP (100 ng ml−1). An identical procedure was followed in the intrabiofilm transformation experiments whose results are presented in Table 3. Results are the means ± standard errors from at least three independent experiments.
As shown in Fig. 1, LytA-mediated autolysis occurs in old biofilms. To determine whether such autolysis contributes to DNA release in 4-h biofilms, we performed the experiments described in the paragraph above with attacker and target strains lacking a functional lytA gene. The rate of Novr transfer in biofilms consisting of RH14 (ΔlytA Spcr) and SPH30 (ΔlytA ΔcomE Novr) cells was the same as in biofilms made with the LytA-positive RH1 and RH401 strains. However, in the parallel experiments performed with SPH106 (ΔlytA ΔcbpD Spcr) and SPH30 cells, 4 to 5 times fewer transformants were obtained per attacker cell than with the LytA-positive RH17 and RH401 cells (Table 3). Thus, in the absence of LytA, the effect of CbpD on intrabiofilm gene transfer is even more pronounced than in LytA-positive biofilms. This is due to LytA-mediated CbpD-independent autolysis of RH401 target cells, which increases the amount of extracellular Novr DNA in LytA-positive compared to LytA-negative biofilms.
Recently, Eldholm et al. (9) investigated the relative contributions of CbpD, LytA, and LytC to pneumococcal fratricide in liquid cultures. They found that competent pneumococci lacking CbpD are unable to lyse target cells, demonstrating that CbpD is an essential component of the fratricide mechanism under the conditions used. However, their results also revealed that CbpD is inefficient on its own and that fratricide is most effective when CbpD acts in conjunction with both LytA and LytC. Our finding that the rate of gene transfer in biofilms grown from LytA-deficient (RH14 and SPH30) and LytA-proficient (RH1 and RH401) strains is the same indicates that LytA is not required for efficient gene exchange in the biofilm environment. To further investigate the role of LytA and LytC, mixed biofilms were grown with the following combinations of strains: RH16 (ΔlytA ΔlytC Spcr)-RH401 (ΔcomE Novr), SPH149 (ΔcbpD ΔlytA ΔlytC Spcr)-RH401, RH16-SPH30 (ΔlytA ΔcomE Novr), SPH149-SPH30, RH16-SPH148 (ΔlytA ΔlytC ΔcomE Novr), and SPH149-SPH148 (Table 3). Deletion of the lytA and lytC genes in the RH16 attacker strain (combination RH16-RH401) reduced the transformation efficiency by about 70% compared to that seen with the wild-type control (RH1-RH401). When the lytA gene was deleted in the target strain as well (RH16-SPH30), the rate of transformation was further reduced. This is in accordance with previous experiments carried out with planktonic cells. Eldholm et al. (9) found that lytA- and/or lytC-deficient target cells are lysed less efficiently than lytA- and/or lytC-proficient target cells. To determine whether attacker cells are able to lyse target cells in the absence of both LytA and LytC, the rate of Novr transfer was assayed in 4-h-old biofilms consisting of strains RH16 and SPH148. Although the gene transfer efficiency was much higher in RH16-SPH148 biofilms than in the CbpD-negative control (SPH149-SPH148), the total number of transformants obtained was very small. In sum, our results show that CbpD is not able to significantly boost intrabiofilm gene transfer on its own. CbpD is essential but requires the assistance of LytA and/or LytC to function efficiently.
Visualization of fratricide in biofilm.
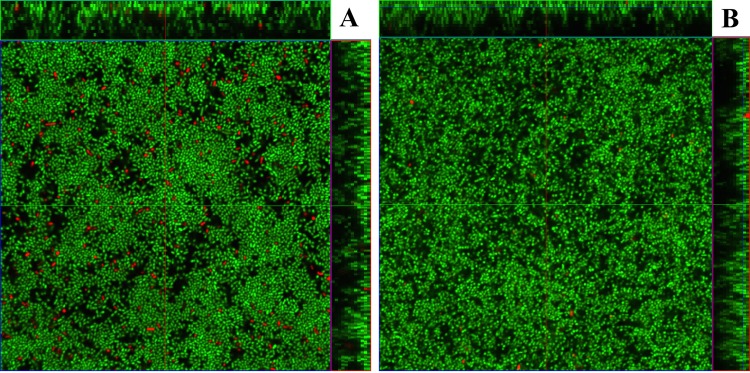
Most pneumococcal cells targeted by CbpD and the accessory muralytic enzymes LytA and LytC rupture and release their genomic DNA to the external milieu. These cells are not detected by the BacLight Live/Dead staining method (Invitrogen), which is based on two nucleic acid stains with different abilities to penetrate the cytoplasmic membrane of healthy bacteria. However, we speculated that some of the targeted cells might be killed without undergoing complete lysis and that such cells may be visible when subjected to BacLight Live/Dead staining. To test this idea, we inoculated glass-bottom dishes with equal numbers of RH14 (ΔlytA Spcr) and SPH30 (ΔlytA ΔcomE Novr) cells. In parallel, mixed biofilms were grown from equal amounts of SPH106 (ΔlytA ΔcbpD Spcr) and SPH30 cells. LytA-negative strains were chosen to prevent fratricide-independent LytA-mediated autolysis. After the biofilms were grown for 4 h at 37°C, they were washed three times in C medium. Fresh C medium was then added together with CSP (100 ng/ml), and the glass-bottom dishes were further incubated at 30°C for 20 min. BacLight Live/Dead staining of the biofilms was performed according to the manufacturer's instructions. Finally, confocal laser scanning microscopy was employed to visualize live and dead cells in the biofilms. The results showed that biofilms consisting of RH14 and SPH30 cells contained considerably more dead cells than those consisting of SPH106 and SPH30 cells (Fig. 5). To quantitate the difference, we used Comstat 2, a computer program for the analysis and treatment of biofilm images in three dimensions. The proportion of dead cells was estimated to be 1.53% ± 0.06% and 0.29% ± 0.02% in RH14/SPH30 and SPH106/SPH30 biofilms, respectively. The only difference between these biofilms was that RH14 cells, in contrast to SPH106 cells, produce and secrete CbpD when induced to competence by CSP. These results demonstrate that the fratricide mechanism is active in the biofilm environment.
Fig 5.
Fratricide in biofilm visualized by confocal laser scanning microscopy and BacLight Live/Dead staining. (A) Mixed biofilm consisting of CbpD-proficient RH14 attacker cells and noncompetent SPH30 (ΔcomE) target cells seeded in a 1:1 ratio. Competence was induced 4 h postinoculation by addition of CSP (100 ng ml−1) to the biofilm medium. After addition of CSP, the biofilm was transferred from 37°C to 30°C for 20 min. Before being examined by confocal microscopy the biofilm was stained using the BacLight Live/Dead kit, which stains live bacteria green and dead bacteria red. (B) Mixed biofilm consisting of CbpD-deficient SPH106 attacker cells and noncompetent SPH30 target cells. The biofilms in both panels were grown and treated identically. The only difference between them is that the SPH106 attacker strain in panel B is not able to produce CbpD when induced to competence by addition of CSP.
DISCUSSION
Our results show that only a minor fraction of pneumococcal cells grown in biofilms for 8 h or more are transformable when subjected to exogenous CSP (Fig. 3). This finding suggests that biofilm cells must be actively growing in order to respond to CSP. This interpretation is in accordance with the fact that planktonic pneumococci become refractory to CSP when cultures enter the stationary phase (15). Alternatively, or in addition, cells located inside the biofilm may fail to develop the competent state due to restricted diffusion of CSP into the biofilm interior. Interestingly, the results presented in Fig. 3 show that DNA is exchanged efficiently between cells within the biofilm. The number of transformants obtained when competent biofilm cells acquired Novr DNA from their neighboring cells was much higher than when Novr DNA was taken up from the surrounding medium. What could be the reason for this? As the purified Novr DNA used for transformation had been isolated from the RH401 (Novr) cells used in the intrabiofilm gene transfer experiments, the nature of the donor DNA could not have influenced the transformation efficiency. It is also unlikely that the concentration of externally added Novr DNA (1 μg/ml) was a limiting factor with respect to the number of transformants obtained. From our own experience, we know that this amount of transforming DNA is well above the saturation level in planktonic cultures. Most likely, externally added genomic DNA is not able to penetrate into the biofilm and is therefore available only to competent cells that are exposed to the growth medium.
The efficient gene transfer observed in 4 h biofilms, consisting of competent RH1 (Spcr) and noncompetent RH401 (Novr) cells (Fig. 3B), shows that extracellular Novr DNA is present within the biofilm during the period of competence. To become available to the competent RH1 cells, this DNA must have been released from the RH401 cells. To determine whether CbpD-mediated fratricide plays a role in the release process, we compared the rate of Novr transfer in RH1-RH401 and RH17-RH401 biofilms. A 50-fold reduction in novobiocin-resistant transformants was obtained when the CbpD-deficient RH17 strain was used as the attacker instead of the CbpD-proficient RH1 strain. Since our data indicate that long DNA strands do not pass easily through the biofilm, this finding suggests that competent cells lyse their noncompetent neighbors within the biofilm. Due to the close proximity between DNA donor and recipient, released DNA is quickly taken up by the competent cells. This reduces the chance that extracellular donor DNA is degraded by nucleases.
Recent advances in our understanding of the role of lateral gene transfer in S. pneumoniae and other streptococci strongly indicate that natural genetic transformation is important for recombinational DNA repair in these bacteria (37). Such repair depends on the uptake of homologous DNA. How do competent pneumococci and their commensal relatives get access to homologous DNA in the multispecies biofilms that constitute their natural habitat? In natural biofilms, bacteria have been found to be embedded in an extracellular matrix consisting of macromolecules such as polysaccharides, proteins, lipids, lipopolysaccharides, and DNA (11). It is therefore likely that competent streptococci inhabiting natural biofilms are exposed to matrix DNA. Biofilms in the oral cavity and nasopharynx are home to hundreds of different species, most of which are not related to streptococci. It is known that naturally transformable streptococci such as S. gordonii, S. oralis, and Streptococcus sanguinis coaggregate with species such as Porphyromonas gingivalis, Fusobacterium nucleatum, and some members of the genus Veillonella. Streptococci are also known to exhibit intrageneric and intraspecies coaggregation, presumably via specific interactions (24, 33). Thus, depending on whether streptococci primarily grow in close contact with related or unrelated bacteria, they will be embedded in homologous or foreign matrix DNA. To what degree matrix DNA serves as a source of DNA for competent streptococci under natural conditions can only be a matter of speculation at present. However, to solely rely on matrix DNA as a substrate for recombinational repair would seem like an error-prone and risky strategy.
Before entering the competent state, S. pneumoniae and other naturally transformable streptococci secrete the competence-stimulating peptide (CSP) to probe their environment. A large variety of CSP pheromones is produced by different strains and species of streptococci (20). Streptococci that produce CSPs with the same primary structure (i.e., that belong to the same pherotype) are in general closely related (23). To trigger development of the competent state, the external concentration of CSP must reach a critical level (about 10 ng ml−1 in liquid cultures). Why is competence development in streptococci controlled by the concentration of an external peptide? One possibility is that the CSP pheromone is utilized by individual cells to sense the presence of potential DNA donors in the immediate neighborhood. The concentration of external CSP depends on two opposing factors: the rate of CSP synthesis and secretion and the rate of CSP loss through diffusion and advection. In order to reach the critical external concentration required for competence induction, the rate of CSP synthesis must exceed the rate of loss. This should be more easily achieved by clusters of CSP-producing streptococci than by individual cells. However, if the CSP threshold level is reached in a cluster of cells producing the same CSP, they will synthesize ComM and become immune to CbpD. Hence, no donor DNA will be released. It is possible, though, that the cells in a cluster experience different levels of stress and therefore are heterogeneous with respect to ComCDE expression. If so, the competent cells will attack and lyse their noncompetent siblings residing in the same cluster.
An alternative scenario is that there are clusters or microcolonies inside biofilms in the oral cavity and nasopharynx that consist of different strains and/or species of streptococci that are heterogeneous with respect to the CSP type they produce. In such an environment, streptococci belonging to the same pherotype will collaborate and coordinate their attack on streptococci belonging to different pherotypes. In principle, this could lead to release of relatively nonhomologous DNA from distantly related streptococci. However, since the target range of fratricins is restricted, cells lysed by these murein hydrolases will in general be relatively closely related to the competent attacker cells. Thus, streptococci appear to use two different strategies to increase their chances of acquiring homologous DNA during competence. They use strain-specific CSPs for competence regulation and fratricins directed against related streptococci for the release of transforming DNA.
In the present study, we have shown that CbpD strongly enhances the rate of gene transfer between pneumococci in laboratory-grown biofilms. This finding makes it reasonable to assume that the fratricide mechanism is highly important for streptococcal gene exchange under natural conditions. To determine which, if any, of the hypotheses discussed in the above paragraph is correct, future work should focus on unraveling the structure and composition of the intrabiofilm communities in which streptococcal gene exchange takes place.
ACKNOWLEDGMENTS
This work was supported by The Research Council of Norway.
We thank Claus Sternberg at the Technical University of Denmark for giving us access to the Comstat2 software.
Footnotes
Published ahead of print 15 June 2012
REFERENCES
- 1. Bateman A, Rawlings ND. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234–237 [DOI] [PubMed] [Google Scholar]
- 2. Berg KH, Solheim Ohnstad H, Håvarstein LS. 2012. LytF, a novel competence-regulated murein hydrolase in the genus Streptococcus. J. Bacteriol. 194:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berg KH, Biørnstad TJ, Johnsborg O, Håvarstein LS. 2012. Properties and biological role of streptococcal fratricins. Appl. Environ. Microbiol. 78:3515–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chi F, Nolte O, Bergmann C, Ip M, Hakenbeck R. 2007. Crossing the barrier: evolution and spread of a major class of mosaic pbp2x in Streptococcus pneumoniae, S. mitis and S. oralis. Int. J. Med. Microbiol. 297:503–512 [DOI] [PubMed] [Google Scholar]
- 5. Claverys JP, Håvarstein LS. 2007. Cannibalism and fratricide: mechanisms and the raisons d'être. Nat. Rev. Microbiol. 5:219–229 [DOI] [PubMed] [Google Scholar]
- 6. Claverys JP, Martin B, Håvarstein LS. 2007. Competence-induced fratricide in streptococci. Mol. Microbiol. 64:1423–1433 [DOI] [PubMed] [Google Scholar]
- 7. Dagkessamanskaia A, et al. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071–1086 [DOI] [PubMed] [Google Scholar]
- 8. Echenique J, Kadioglu A, Romao S, Andrew PW, Trombe MC. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72:2434–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eldholm V, Johnsborg O, Haugen K, Solheim Ohnstad H, Håvarstein LS. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155:2223–2234 [DOI] [PubMed] [Google Scholar]
- 10. Eldholm V, et al. 2010. Pneumococcal CbpD is a murein hydrolase that requires a dual cell-envelope binding-specificity to kill target cells during fratricide. Mol. Microbiol. 76:905–917 [DOI] [PubMed] [Google Scholar]
- 11. Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 12. Guenzi E, Gasc AM, Sicard MA, Hakenbeck R. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505–515 [DOI] [PubMed] [Google Scholar]
- 13. Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of non-competent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U. S. A. 102:8710–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Halfmann A, Kovács M, Hakenbeck R, Brückner R. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66:110–126 [DOI] [PubMed] [Google Scholar]
- 15. Håvarstein LS, Coomaraswami G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Håvarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863–869 [DOI] [PubMed] [Google Scholar]
- 17. Håvarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. 2006. New insights into the pneumococcal fratricide: relationships to clumping and identification of a novel immunity factor. Mol. Microbiol. 59:1297–1307 [DOI] [PubMed] [Google Scholar]
- 18. Heydorn A, et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 19. Hui FM, Morrison DA. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173:372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnsborg O, Blomqvist T, Kilian M, Håvarstein LS. 2007. Biologically active peptides in streptococci, p 25–59 In Hakenbeck R, Chhatwal S. (ed), Molecular biology of streptococci. Horizon Scientific Press, Wymondham, United Kingdom [Google Scholar]
- 21. Johnsborg O, Eldholm V, Bjørnstad ML, Håvarstein LS. 2008. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae. Mol. Microbiol. 69:245–253 [DOI] [PubMed] [Google Scholar]
- 22. Kausmally L, Johnsborg O, Lunde M, Knutsen E, Håvarstein LS. 2005. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 187:4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kilian M, et al. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 8:471–480 [DOI] [PubMed] [Google Scholar]
- 25. Lacks S, Hotchkiss RD. 1960. A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39:508–518 [DOI] [PubMed] [Google Scholar]
- 26. Layec S, Decaris B, Leblond-Bourget N. 2008. Diversity of Firmicutes peptidoglycan hydrolases and specificities of those involved in daughter cell separation. Res. Microbiol. 159:507–515 [DOI] [PubMed] [Google Scholar]
- 27. Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morrison DA, Lacks SA, Guild WR, Hageman JM. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mortier-Barrière I, de Saizieu A, Claverys JP, Martin B. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27:159–170 [DOI] [PubMed] [Google Scholar]
- 30. Moscoso M, García E, López R. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188:7785–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pestova EV, Håvarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853–862 [DOI] [PubMed] [Google Scholar]
- 32. Peterson SN, et al. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051–1070 [DOI] [PubMed] [Google Scholar]
- 33. Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94–100 [DOI] [PubMed] [Google Scholar]
- 34. Rigden DJ, Jedrzejas MJ, Galperin MY. 2003. Amidase domains from bacterial and phage autolysins define a family of γ-D,L-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230–234 [DOI] [PubMed] [Google Scholar]
- 35. Sánchez-Puelles JM, Sanz JM, García JL, García E. 1990. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene 89:69–75 [DOI] [PubMed] [Google Scholar]
- 36. Steinmoen H, Knutsen E, Håvarstein LS. 2002. Induction of competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. U. S. A. 99:7681–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stevens KE, Chang D, Zwack EE, Sebert ME. 2011. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2:e00071–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tomasz A. 1981. Surface components of Streptococcus pneumoniae. Rev. Infect. Dis. 3:190–211 [DOI] [PubMed] [Google Scholar]
- 40. Ween O, Gaustad P, Håvarstein LS. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817–827 [DOI] [PubMed] [Google Scholar]