Abstract
Kasugamycin (KSM), a unique aminoglycoside antibiotic, has been used in agriculture for many years to control not only rice blast caused by the fungus Magnaporthe grisea but also rice bacterial grain and seedling rot or rice bacterial brown stripe caused by Burkholderia glumae or Acidovorax avenae subsp. avenae, respectively. Since both bacterial pathogens are seed-borne and cause serious injury to rice seedlings, the emergence of KSM-resistant B. glumae and A. avenae isolates highlights the urgent need to understand the mechanism of resistance to KSM. Here, we identified a novel gene, aac(2′)-IIa, encoding a KSM 2′-N-acetyltransferase from both KSM-resistant pathogens but not from KSM-sensitive bacteria. AAC(2′)-IIa inactivates KSM, although it reveals no cross-resistance to other aminoglycosides. The aac(2′)-IIa gene from B. glumae strain 5091 was identified within the IncP genomic island inserted into the bacterial chromosome, indicating the acquisition of this gene by horizontal gene transfer. Although excision activity of the IncP island and conjugational gene transfer was not detected under the conditions tested, circular intermediates containing the aac(2′)-IIa gene were detected. These results indicate that the aac(2′)-IIa gene had been integrated into the IncP island of a donor bacterial species. Molecular detection of the aac(2′)-IIa gene could distinguish whether isolates are resistant or susceptible to KSM. This may contribute to the production of uninfected rice seeds and lead to the effective control of these pathogens by KSM.
INTRODUCTION
The aminoglycoside kasugamycin (KSM) is produced by Streptomyces kasugaensis isolated from soil at Kasuga Shrine in Nara, Japan (49). KSM is absolutely effective on rice blast caused by the fungus Magnaporthe grisea and has been widely used in agriculture since 1965 (22). KSM also has good activities against plant bacterial diseases and has been used to control rice bacterial grain and seedling rot caused by the proteobacterium Burkholderia glumae (17) and fire blight of apple and pear caused by Erwinia amylovora (29). Unlike other aminoglycosides, KSM has a unique structure: it is composed of two sugars, a d-chiro-inositol moiety and a kasugamine moiety (2,4-diamino-2,3,4,6-tetradeoxy-d-arabino-hexose), with a carboxy-imino-methyl group (11). KSM inhibits protein synthesis in prokaryotes and M. grisea (27, 46), but it fails to induce translational misreading due to lack of the deoxystreptamine moiety common among aminoglycosides (20). KSM is known to bind the bacterial ribosome 30S subunit in the region of the mRNA-binding tunnel in the E-site and P-site and indirectly inhibits tRNA binding at the P-site by perturbing the mRNA-tRNA codon-anticodon interaction during translational initiation (35, 36).
Bacterial resistances to aminoglycoside antibiotics are conferred by several mechanisms such as decreased drug uptake, modification of the ribosomal target, efflux of drugs, and enzymatic modification of drugs (50). In the clinical scene, modifications of the amino or hydroxyl groups of antibiotics are primary mechanisms of bacterial resistance and are mediated by three major groups of modifying enzymes: N-acetyltransferases (AACs), O-adenyltransferases, and O-phosphotransferases (50).
As with other aminoglycosides, resistance mutations for KSM have been found in bacteria. First, a dimethyltransferase, the ksgA gene was mapped in Escherichia coli (38). Loss of methylation of the two adjacent nucleotides A1518 and A1519 in the helix 45 of the 16S rRNA due to ksgA mutation indirectly influenced the conformation of helix 24, which is involved in KSM binding (10, 15, 16). The mutations of ksgA then confer modest resistance to KSM (15). Higher resistance to KSM is known to result from the mutations of A794, G926, or A1519 in 16S rRNA (52). Recent studies by a chemical probing assay and structural analysis of the KSM-30S subunit complex show that universally conserved residues A794 and G926 are involved in binding KSM (35, 36, 53). Furthermore, mutations in three other genes (ksgB, ksgC, and ksgD) in E. coli cause resistance to KSM independently (13, 39, 54). Mutation of ksgC alters the amount of ribosomal protein S2 (54). Cell-free protein synthesis with ribosomes from ksgA and ksgC mutants show resistance to KSM; in contrast, ribosomes from ksgB mutant are sensitive to KSM in vitro, indicating that a mutation of ksgB results in altered cell membrane permeability to KSM (39). However, detailed molecular mechanisms of KSM resistance in ksgB, ksgC, and ksgD mutants have not been elucidated yet.
The emergence of two phytopathogenic bacteria resistant to KSM in rice plants is known in Japan. KSM-resistant field isolates of Proteobacteria, Acidovorax avenae subsp. avenae, which is the causal agent of rice bacterial brown stripe, appeared in 1990 (43). Fourteen years later, KSM-resistant B. glumae was isolated (19). Because these are seed-borne diseases, bacterial pathogens infest rice seedlings in nursery boxes and result in serious losses in yield. Treating rice seeds in nursery boxes with KSM granules (0.3 to 0.4 g as the active ingredient per box [30 cm by 60 cm by 3 cm]) or with KSM liquid (0.125 to 0.25 g as the active ingredient per box) is highly efficacious in the control of KSM-sensitive rice bacterial stripe and bacterial seedling rot. Unfortunately, KSM fails to control KSM-resistant isolates of A. avenae and B. glumae (19, 43). Moreover, the mechanism of resistance to KSM in A. avenae and B. glumae remains obscure.
The aim in the present study was to clarify the genetic determinants of KSM-resistance in these rice-pathogenic bacteria. Our results demonstrate that acetylation of the 2′-amino residue of the KSM by the novel acetyltransferase, AAC(2′)-IIa, encoded by the IncP genomic island confers specific resistance to KSM.
MATERIALS AND METHODS
Bacterial strains.
Eighteen KSM-resistant B. glumae isolates (isolated from grain or diseased seedlings) and three KSM-susceptible B. glumae isolates (one of them showing resistance to oxolinic acid) were used in the present study, as were nine KSM-resistant A. avenae subsp. avenae isolates (isolated from grain or diseased seedlings) and three KSM-susceptible A. avenae isolates (one of them showing resistance to oxolinic acid). All isolates were supplied by Hokko Chemical Industry Co., Ltd., collections in Japan (Table 1). Liquid cultures of B. glumae and A. avenae strains were grown aerobically in Luria-Bertani (LB) broth at 28°C with shaking at 120 rpm. E. coli DH5α was grown either on LB agar or in LB broth at 37°C except for the antibiotic susceptibility assay.
Table 1.
Bacterial strains used in this study
| Strain | MIC (μg/ml)a |
Susceptibilityb |
Presence of aac(2′)-IIac | Source | |||||
|---|---|---|---|---|---|---|---|---|---|
| KSM | OX | CuCl2·3Cu(OH)2 | Cu(OH)2 | TMTD | KSM | OX | |||
| Burkholderia glumae | |||||||||
| 210 | 12.5 | 0.8 | 25 | 25 | 100 | S | S | – | This study |
| 216 | 25 | 0.4 | 25 | 25 | 50 | S | S | – | This study |
| 3 | 25 | 100 | 25 | 50 | 50 | S | R | – | This study |
| 5091 | 3,200 | 0.4 | 25 | 50 | 50 | R | S | + | This study |
| 5096 | 3,200 | R | S | + | This study | ||||
| 5321 | 3,200 | R | S | + | This study | ||||
| 2-1 | 3,200 | R | S | + | This study | ||||
| 4-1-1 | 3,200 | 0.4 | 50 | 50 | 50 | R | S | + | This study |
| 4-1-2 | 3,200 | R | S | + | This study | ||||
| 4-1-3 | 3,200 | R | S | + | This study | ||||
| 4-1-4 | 3,200 | R | S | + | This study | ||||
| 4-1-5 | 3,200 | R | S | + | This study | ||||
| 4-1-6 | 1,600 | R | S | + | This study | ||||
| 4-1-8 | 1,600 | R | S | + | This study | ||||
| 4-1-9 | 1,600 | 0.4 | 50 | 50 | 50 | R | S | + | This study |
| 4-2-1 | 3,200 | R | S | + | This study | ||||
| 4-2-2 | 3,200 | R | S | + | This study | ||||
| 9-1-1 | 3,200 | 0.4 | 50 | 50 | 50 | R | S | + | This study |
| 9-1-2 | 3,200 | R | S | + | This study | ||||
| 11-1-1 | 1,600 | 0.4 | 25 | 50 | 50 | R | S | + | This study |
| 11-1-2 | 1,600 | R | S | + | This study | ||||
| Acidovorax avenae subsp. avenae | |||||||||
| 161 | 6.3 | 0.8 | S | S | – | This study | |||
| 199 | 6.3 | 0.4 | S | S | – | This study | |||
| 5 | 6.3 | >100 | S | R | – | This study | |||
| 83 | 3,200 | 0.4 | R | S | + | This study | |||
| 1-1 | 800 | 0.4 | R | S | + | This study | |||
| 2-1 | 800 | 0.4 | R | S | + | This study | |||
| 3-2 | 800 | 0.4 | R | S | + | This study | |||
| 4-2 | 800 | 0.4 | R | S | + | This study | |||
| 5-1 | 800 | 0.4 | R | S | + | This study | |||
| 213 | 800 | 100 | R | R | + | This study | |||
| 214 | 800 | 100 | R | R | + | This study | |||
| 215 | 800 | 100 | R | R | + | This study | |||
The MIC was determined by using the agar streak method with peptone-glucose medium. KSM, kasugamycin; OX, oxolinic acid.
S, susceptible; R, resistant.
The presence (+) or absence (–) of the aac(2′)-IIa gene is indicated.
Chemicals.
Kasugamycin hydrochloride, semisynthetic 2′-acetyl-kasugamycin, oxolinic acid, and bis(dimethylthiocarbamyl) disulfide (TMTD) were supplied from Hokko Chemical Industry Co., Ltd. Oxolinic acid was dissolved in 100 mM sodium hydroxide, which, at a concentration of 1 mM in the medium, did not affect bacterial growth. The copper compounds copper chloride oxide hydrate [CuCl2·3Cu(OH)2] and copper (II) hydroxide [Cu(OH)2] were used from the agrichemical products Germany-Bord'eaux A and KOCIDE, respectively. Streptomycin sulfate, kanamycin sulfate, gentamicin sulfate, chloramphenicol, and tetracycline hydrochloride were obtained from Wako Pure Chemical Industries, Ltd. Neomycin trisulfate, paromomycin sulfate, ribostamycin sulfate, tobramycin sulfate, sisomicin sulfate, and spectinomycin dihydrochloride were obtained from Sigma-Aldrich Corp.
Antibiotic susceptibility test.
The MICs of antibiotics and antibacterial compounds were determined by the agar dilution method. Briefly, 2-fold serial dilutions of drugs were plated in peptone-glucose medium (peptone, 5 g; glucose, 5 g; agar, 18 g; deionized water, 1 liter [pH 6.8]) or Muller-Hinton medium (Eiken Chemical Co., Ltd.). Bacterial suspensions were then streaked on each medium, and the results were observed after 2 days of incubation at 28°C for B. glumae and A. avenae or after overnight incubation at 37°C for E. coli. The lowest concentration of each drug, which completely inhibited bacterial growth, was defined as the MIC.
DNA techniques.
Procedures for general DNA techniques, such as E. coli transformation, plasmid extraction, restriction enzyme digestion, ligation, and agarose gel electrophoresis were performed essentially as described previously (5). Genomic DNA of B. glumae and A. avenae was prepared using a Qiagen RNA/DNA kit (Qiagen) or a Bacteria GenomicPrep Mini Spin kit (GE Healthcare).
The sequences of ksgA and 16S rRNA genes from B. glumae were determined directly by using Ksg-F (5′-ACCGCGGGCATGACGG-3′) and Ksg-831R (5′-TTCGTTTGATTCTTTCGATGTCGA-3′) primers for the ksgA gene and S705-F (5′-ATGTGGAGGAATACCGATGG-3′) and S1536-R (5′-AAAGGAGGTGATCCAGCC-3′) primers for the 16S rRNA gene with a BigDye Terminator cycle sequencing kit (Applied Biosystems, Inc.). DNA sequences were performed on a 3100 DNA sequencer (Applied Biosystems, Inc.). Sequence alignment was conducted using the CLUSTAL W program (47).
Cloning of the KSM acetyltransferase aac(2′)-IIa gene.
The KSM acetyltransferase gene from B. glumae 5091 was cloned from libraries of EcoRI-digested genomic DNA constructed in vector pBluescript II SK(+). The ligation products were transformed into E. coli DH5α by heat shock. Then, KSM- and ampicillin-resistant transformants were selected on LB agar containing 200 μg of KSM/ml and 50 μg of ampicillin/ml. The recombinant plasmid contains a 5.6-kb insert. DNA fragment analysis confirmed that the responsible gene for KSM-resistance was present in the 5.6-kb insert at the HindIII site.
Nucleotide sequences of 5.6-kb DNA fragments from three independent KSM-resistant clones were determined by the primer-walking method. Sequence analyses were conducted by using CLC Sequence Viewer version 6.5.1 (CLC Bio) and BLASTN and BLASTP algorithms (4). The phylogenetic positions of AAC(2′)-IIa and other N-acetyltransferases were determined using the software MEGA 5.05 with the neighbor-joining method (45).
The aac(2′)-IIa gene was amplified by PCR using 5′-phosphorylated primers aac(2′)IIa-2F (5′-CAATGAAAGACAGATCCCATGA-3′) and aac(2′)IIa-R (5′-AACAATTTCAGCCTATCAGGC-3′) or the primers aac(2′)IIa-92F (5′-AGATTTGGATTGTGCAATCC-3′) and aac(2′)IIa-R, and the amplicons were cloned into the HincII site of pUC118 (TaKaRa Bio, Inc.) to produce KSM acetyltransferase without the 91-bp promoter region of the aac(2′)-IIa gene [pUC118-aac(2′)-IIa P−] or with the promoter region (P+). To verify whether the aac(2′)-IIa gene was present among KSM-resistant B. glumae and A. avenae isolates, the following primers were used for PCR analysis: aac(2′)IIa-57F (5′-GAACCACTGGCTGAAGACCGATTAT-3′) and aac(2′)IIa-710R (5′-ATGTTGTCTCGGTCCGTGCTGTAG-3′). PCR amplification was performed with 1 cycle of 94°C for 2 min, followed by 30 cycles of 98°C for 10 s, 60°C for 30 s, and 68°C for 1 min.
Detection of the acetyltransferase activity.
E. coli BL21(DE3) harboring pUC118-aac(2′)-IIa P+ was grown to the mid-log phase. The bacterial pellet was resuspended in 100 mM sodium phosphate buffer (pH 7.6). Bacterial cells were disrupted by ultrasonication, and insoluble materials were removed by centrifugation at 30,000 × g for 1 h. Then, the soluble fraction was prepared by using PD-10 columns (GE Healthcare).
Acetyltransferase assays were carried out by measuring the increase in absorbance at 412 nm due to the formation of 5-thio-2-nitrobenzoate (TNB; ε412 = 14,150 M−1 cm−1), generated by the reaction between sulfhydryl group of CoA-SH, the product of the acetyltransferase reaction, and 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB). The reaction mixture (1 ml) contained 0.2 mM acetyl coenzyme A (acetyl-CoA), 1 mM DTNB, 100 mM sodium phosphate (pH 7.6), and cell extract (600 μg of total protein), and the reaction was initiated by the addition of 1 mM antibiotics at room temperature. The degree of enzymatic reaction was calculated by subtracting from that of a control reaction that lacked antibiotic substrate. For high-performance liquid chromatography (HPLC) analysis, the reactions were performed in 100 μl of a reaction mixture of 2 mM acetyl-CoA, 1 mM KSM, 50 mM sodium phosphate (pH 7.6), and cell extract (300 μg of total protein). After 2 h of incubation at 30°C, heating at 80°C for 10 min terminated the reactions. HPLC analyses were performed by utilizing a YMC-pack ODS-AQ column (4.6 by 250 mm) with UV detection (230 nm) and a mobile phase of acetonitrile-water (20:80 [vol/vol]) at pH 3.5 containing 0.1% sodium decane sulfonate as the ion-pairing agent.
Cloning of the IncP island.
To clone the full length of the IncP island, long PCR DNA amplification was performed using KOD plus DNA polymerase (Toyobo Co., Ltd.) with 1 cycle of 94°C for 2 min, followed by 35 cycles of 10 s at 98°C (DNA denaturation), then 10 min at 74, 72, and 70°C for 5 cycles each, and finally 10 min at 68°C for 20 cycles (primer annealing and DNA synthesis). guaA-77F (5′-ACGTCTATTGCGAGATTCATCCGAACGAT-3′) and E1-404R (5′-GGTACCGCTGGTATGGATTTATGAGGCATT-3′) primers were used to clone the IncP island from B. glumae 5091. These 5′-phosphorylated primers were designed based on the complete nucleotide sequence of B. glumae BGR1 (26) and on the partial sequence of the IncP island from 5091 strain. A 9.6-kb fragment of the IncP island was cloned into pUC118 and sequenced by the primer-walking method. Comparative nucleotide sequence analyses were conducted using Mauve 2.3.1 (7).
Filter-mating assay.
KSM-resistant B. glumae 5091 was used as the donor strain in filter matings with the recipient B. glumae strain 3 or A. avenae strain 5, which were KSM-sensitive and oxolinic acid-resistant strains, respectively. Conjugational transfers were conducted by mixing a donor and recipient strain in a 1:1 ratio, and mixed cultures were filtered using a 0.45-μm-pore-size filter (Millipore). After 8 h of incubation on an antibiotic-free peptone-glucose plate, the cells were resuspended from the filter in 3 ml of 50 mM potassium chloride. The transconjugants that could grow on peptone-glucose plates with 100 μg of KSM/ml and 10 μg of oxolinic acid/ml were counted. Conjugation frequencies were expressed as the number of transconjugants per number of donor cells recovered after 2 days of incubation at 28°C.
Detection of the circular forms containing aac(2′)-IIa.
Inverse nested PCR was used to analyze the circular intermediates containing the aac(2′)-IIa and bglu_5091g13 genes from genomic DNA of B. glumae or A. avenae. First, inverse PCR was conducted using the aac(2′)IIa-308R (5′-AGTGAAAGATAACGTTCAAGATGCT-3′) and aac(2′)IIa-451F (5′-AGAGTACCGGAAGCTTTGTTTGAG-3′) primers, but no detectable PCR product was obtained. Second, inverse nested PCR was carried out using the aac(2′)IIa-240R (5′-TTCTTCGAGCTTTAGTGCTATGTCA-3′) and hypo-F (5′-TTCATCTGCATTCTGACGACTT-3′) primers from the diluted DNA amplified by the first PCR. Then, the attP sequences in the obtained PCR products were analyzed.
Reverse transcription-PCR (RT-PCR) analysis.
Total RNA was isolated as described previously (2). First-strand cDNA was synthesized from 500 ng of DNase-treated RNA using a gene-specific primer, hypo-R (5′-GCCTGTCCATCGGTAACTAAAA-3′), and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. After first-strand cDNA synthesis, RNA template was removed by treatment with RNase H. PCR amplification was conducted using aac(2′)IIa-57F and aac(2′)IIa-710R primers for the aac(2′)-IIa gene and hypo-F and hypo-R primers for the bglu_5091g13 gene.
Nucleotide sequence accession number.
The nucleotide sequence of the IncP island and surrounding region from B. glumae 5091 was deposited in the GenBank nucleotide sequence database under the accession number AB669090.
RESULTS
Antibacterial susceptibility of KSM-resistant isolates.
To clarify the mechanism of bacterial KSM-resistance in field isolates, we collected KSM-resistant bacterial pathogens from Japan. First, we examined the antibiotic susceptibility of KSM-sensitive and -resistant isolates of B. glumae. Although KSM-sensitive strains (strains 210, 216, and 3) exhibited MICs from 12.5 to 25 μg/ml to KSM, 18 KSM-resistant strains exhibited MICs from 1,600 to 3,200 μg/ml to KSM (Table 1). These KSM-resistant isolates caused rice seedling rot in nursery boxes, even after treatment with KSM granules or KSM liquid, which is consistent with a previous report (19). We also examined the susceptibility to other ribosome-targeting antibiotics such as other aminoglycosides, chloramphenicol (inhibition of peptidyl transferase activity), and tetracycline (inhibition of binding aminoacyl-tRNA). Although strain 4-1-9 was more susceptible to other aminoglycoside antibiotics than other strains, there was no significant difference between the susceptibility of KSM-sensitive and -resistant B. glumae to streptomycin, neomycin, paromomycin, ribostamycin, kanamycin, tobramycin, gentamicin, sisomicin, and spectinomycin, as well as chloramphenicol and tetracycline (Table 2). KSM resistance was also not correlated with susceptibility to other agrichemicals such as copper, oxolinic acid, and TMTD, which are used to control bacterial plant diseases in Japan (Table 1). These results indicated that KSM-resistant B. glumae isolates were resistant to KSM specifically, revealing no cross-resistance to other antibiotics or agrichemicals.
Table 2.
MICs of various antibiotics for KSM-sensitive or -resistant Burkholderia glumae
| B. glumae strain | MIC (μg/ml)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycoside |
CP | TC | ||||||||||
| KSM | SM | NM | PM | RSM | KM | TOB | GM | SISO | SPC | |||
| 210 | 12.5 | 1.6 | 25 | 25 | 3.1 | 1.6 | 3.1 | 3.1 | 6.3 | 3.1 | 100 | 3.1 |
| 216 | 25 | 1.6 | 25 | 12.5 | 3.1 | 0.8 | 3.1 | 3.1 | 6.3 | 3.1 | 100 | 3.1 |
| 3 | 25 | 1.6 | 25 | 25 | 3.1 | 1.6 | 3.1 | 6.3 | 6.3 | 3.1 | 50 | 3.1 |
| 5091 | 3,200 | 1.6 | 25 | 25 | 3.1 | 0.8 | 3.1 | 6.3 | 6.3 | 3.1 | 50 | 3.1 |
| 4-1-1 | 3,200 | 1.6 | 25 | 25 | 3.1 | 0.8 | 3.1 | 6.3 | 6.3 | 3.1 | 50 | 3.1 |
| 4-1-9 | 1,600 | 0.8 | 12.5 | 12.5 | 0.8 | 0.2 | 0.8 | 0.8 | 3.1 | 0.8 | 50 | 3.1 |
| 9-1-1 | 3,200 | 3.1 | 25 | 25 | 3.1 | 1.6 | 3.1 | 3.1 | 6.3 | 3.1 | 50 | 3.1 |
| 11-1-1 | 1,600 | 3.1 | 25 | 25 | 3.1 | 1.6 | 3.1 | 6.3 | 6.3 | 3.1 | 50 | 3.1 |
The MIC was determined using the agar streak method with peptone-glucose medium. SM, streptomycin; NM, neomycin; PM, paromomycin; RSM, ribostamycin; KM, kanamycin; TOB, tobramycin; GM, gentamicin; SISO, sisomicin; SPC, spectinomycin; CP, chloramphenicol; TC, tetracycline.
Mutation of the KSM interacting site.
Bacterial resistance to KSM by inactivation of the dimethyltransferase gene (ksgA) is the most frequent mechanism of spontaneous mutation in vitro (29, 33). We detected nucleotide variations, but not changes in amino acids, in ksgA gene sequences between KSM-susceptible B. glumae strains 210 and 3 (see Fig. S1 in the supplemental material). The sequence of ksgA in strain 210 was identical to that of strain BGR1, whose genome has already been sequenced (26). In contrast, the nucleotide sequence of the ksgA gene in KSM-resistant B. glumae (strains 5091, 4-1-9, and 9-1-1) differed from that of KSM-sensitive strains (strains 210 and BGR1) only by a silent C-to-T substitution at position 282 (see Fig. S1 in the supplemental material).
Previously, it had been reported that KSM-resistance is conferred by three independent 16S rRNA mutations (A794G, G926A, and A1519C; E. coli numbering) (52), whose residues are involved in binding the drug (35, 36). The 3′ sides of 16S rRNA gene sequences, including these KSM binding sites of KSM-resistant B. glumae (strains 5091, 4-1-9, and 9-1-1), were identical to those of KSM-sensitive strains (strains 210 and 3) and strain BGR1 (data not shown), indicating that KSM's interacting sites on 16S rRNA are not involved in KSM resistance in B. glumae.
Isolation of the KSM acetyltransferase gene.
To identify the gene(s) that confer KSM resistance to B. glumae, a plasmid library constructed from EcoRI-digested genomic DNA of KSM-resistant 5091 strain was introduced into KSM-susceptible E. coli DH5α, and three KSM-resistant transformants were isolated. These three transformants harbored an identical 5.6-kb DNA fragment in the plasmid vector. DNA fragment analysis with HindIII revealed that the gene responsible for KSM resistance was present at the HindIII site. Sequence analysis of transformants revealed that there was an open reading frame (ORF) encoding a general control nonrepressed 5-related N-acetyltransferase (GNAT) gene (51) at the HindIII site (Fig. 1). As described below, we denoted this N-acetyltransferase gene as aac(2′)-IIa because of the regiospecificity of the modification and the specificity of the substrate.
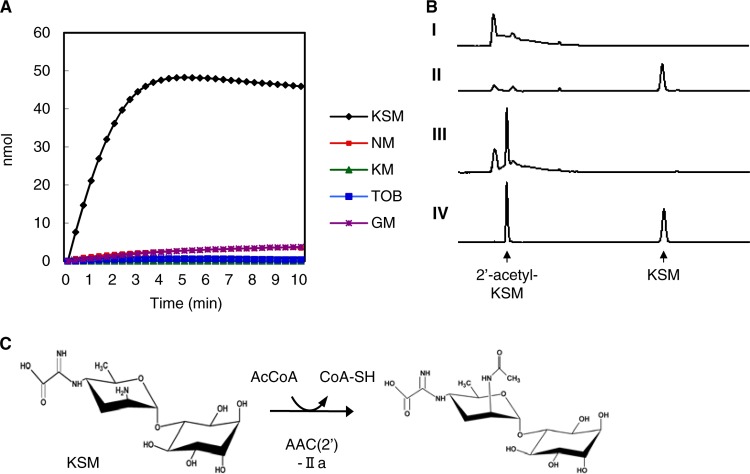
Fig 1.
Comparison of the AAC(2′)-IIa protein from B. glumae 5091 with other N-acetyltransferases. (A) Comparison of the AAC(2′)-IIa with Kac273 from Streptomyces kasugaensis 273. The closed arrowhead reveals the amino acid substitution site from serine to threonine of AAC(2′)-IIa from Acidovorax avenae subsp. avenae 83. The HindIII restriction site in aac(2′)-IIa is shown by an open arrowhead. GNAT domains are boxed. Symbols: asterisk (*), identical amino acid; colon (:), strongly similar; period (.), weakly similar. (B) Phylogenetic tree of the AAC enzymes, including SMc03107 from Sinorhizobium meliloti 1021 (NC_003047), BCAM0938 from Burkholderia cenocepacia J2315 (NC_011001), and Bcep18194_B2186 from Burkholderia sp. strain 383 (NC_007411). AAC accession numbers: AAC(2′)-Ia from Providencia stuartii, L06156; AAC(2′)-Ib from Mycobacterium fortuitum, U41471; AAC(2′)-Ic from M. tuberculosis, U72714; AAC(2′)-Id from M. smegmatis, U72743; AAC(3)-Ia from Pseudomonas aeruginosa, X15852; AAC(3)-Ib from P. aeruginosa, L06157; AAC(3)-Ic from P. aeruginosa, AJ511268; AAC(3)-IIa from Enterobacter cloacae, X51534; AAC(3)-IIb from Serratia marcescens, M97172; AAC(3)-IIc from E. coli, X54723; AAC(3)-IIIa from P. aeruginosa, X55652; AAC(3)-IIIb from P. aeruginosa, L06160; AAC(3)-IIIc from P. aeruginosa, L06161; AAC(3)-IVa from E. coli, X01385; AAC(3)-VIa from E. cloacae, M88012; AAC(3)-VIIa from Streptomyces rimosus, M22999; AAC(6′)-Ia from Citrobacter diversus, M18967; AAC(6′)-Ib from Klebsiella pneumoniae, M21682; AAC(6′)-Ic from S. marcescens, M94066; AAC(6′)-If from E. cloacae, X55353; AAC(6′)-Ig from Acinetobacter haemolyticus, L09246; AAC(6′)-Is from Acinetobacter sp., AF031327; AAC(6′)-It from Acinetobacter sp., AF031328; AAC(6′)-IIa from P. aeruginosa, M29695; AAC(6′)-IIb from P. fluorescens, L06163; and AAC(6′)-IIc from P. aeruginosa, AF162771.
To determine whether the aac(2′)-IIa gene was responsible for resistance to KSM, we transformed E. coli DH5α with the aac(2′)-IIa-expressed plasmid lacking the 91-bp promoter region of the aac(2′)-IIa gene [pUC118-aac(2′)-IIa P−] or including the promoter region of the aac(2′)-IIa gene [pUC118-aac(2′)-IIa P+]. Compared to E. coli DH5α, which harbored the control plasmid (pUC118), aac(2′)-IIa-expressed DH5α showed a notable reduction in susceptibility to KSM, except for the plasmid, which harbored an opposite direction of the aac(2′)-IIa gene with the lac promoter (Fig. 2). It was indicated that the 91-bp promoter region of aac(2′)-IIa functioned in E. coli constitutively. Susceptibility to neomycin, kanamycin, tobramycin, and gentamicin was unaffected (Fig. 2). Thus, the aac(2′)-IIa gene could contribute to KSM resistance by inactivating its target.
Fig 2.
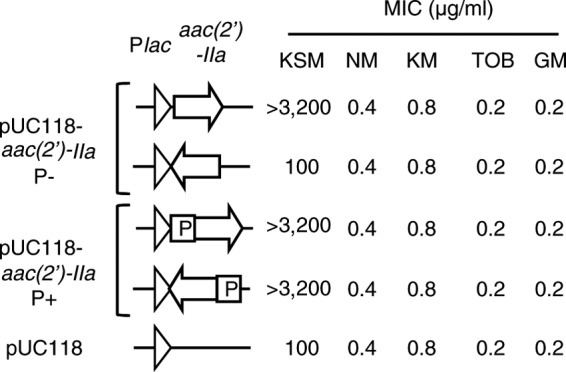
Susceptibility of E. coli DH5α expressing the aac(2′)-IIa gene to aminoglycosides. The 91-bp promoter region (P) of aac(2′)-IIa gene is not contained (P−) or contained (P+) in pUC118 vector. NM, neomycin; KM, kanamycin; TOB, tobramycin; GM, gentamicin.
Acetylation of KSM by AAC(2′)-IIa.
The aac(2′)-IIa gene consists of 783 nucleotides and encodes a 260-amino-acid protein. AAC(2′)-IIa shares sequence similarity with the GNAT superfamily of acetyltransferases, which includes the aminoglycoside N-acetyltransferases. We analyzed the acetyltransferase activity of AAC(2′)-IIa by a spectrophotometric assay that quantifies the conversion of acetyl-CoA to CoA-SH. The cell extract from AAC(2′)-IIa-expressed E. coli converted KSM into an acetylated form (Fig. 3A). On the other hand, neomycin, kanamycin, tobramycin and gentamicin were not modified by AAC(2′)-IIa (Fig. 3A). The reaction mixture that lacked acetyl-CoA showed clear antibiotic activity against KSM-sensitive B. glumae. Conversely, when a complete reaction with KSM was carried out, no antibacterial activity was observed (data not shown), indicating the inactivation of KSM. HPLC elution peaks of products from the AAC(2′)-IIa reaction were identical to those of standard 2′-acetyl-KSM (Fig. 3B).
Fig 3.
Enzymatic properties of AAC(2′)-IIa. (A) Time course of the production of acetylated antibiotics by the cell extract from E. coli expressing the AAC(2′)-IIa protein. NM, neomycin; KM, kanamycin; TOB, tobramycin; GM, gentamicin. (B) HPLC elution profiles of the product of the acetyltransferase reaction of KSM. I, no KSM control; II, no acetyl-CoA control; III, acetyltransferase reaction; IV, standard 2′-acetyl-KSM and KSM. (C) AAC(2′)-IIa catalyzes the transfer reaction of an acetyl group to the 2′-N-kasugamine moiety of KSM.
Although AAC(2′)-IIa catalyzed the same substrate as Kac273, namely, KSM acetyltransferase from Streptomyces kasugaensis (18, 21), the deduced amino acid sequences were quite distinct from each other (11.9% identity) (Fig. 1). AAC(2′)-IIa shares sequence similarity with several GNAT acetyltransferases in Sinorhizobium meliloti 1021, Burkholderia cenocepacia J2315, and Burkholderia sp. strain 383 [deduced amino acid sequence identity to AAC(2′)-IIa is 65.8, 56.3, and 55.9%, respectively], and yet their substrates are unknown (Fig. 1B). Moreover, AAC(2′)-IIa has low homology with other classes of the AAC enzymes. On the basis of sequence similarity and enzymatic properties, the aac(2′)-IIa gene is a novel KSM-specific 2′-N-acetyltransferase (Fig. 3C).
Distribution of the aac(2′)-IIa gene among KSM-resistant rice-pathogenic bacteria.
To clarify the distribution of the aac(2′)-IIa gene among KSM-resistant B. glumae, we examined whether this gene is present or absent by PCR analysis. Of the 18 resistant isolates analyzed, all of them harbored the aac(2′)-IIa gene. In contrast to the resistant isolates, the gene was absent in KSM-sensitive strains (Table 1). Because the acetylation of KSM seemed to be major cause for KSM resistance in field isolates of B. glumae, we examined whether the aac(2′)-IIa gene is present in KSM-resistant A. avenae subsp. avenae, which causes rice bacterial brown stripe. In addition to KSM-resistant B. glumae, of the nine isolates of KSM-resistant A. avenae analyzed, all of them harbored the aac(2′)-IIa gene. In contrast to the resistant isolates, the gene was absent in KSM-sensitive strains (Table 1).
The nucleotide sequences of the aac(2′)-IIa gene from B. glumae 4-1-9, 9-1-1, and 11-1-1 are identical to that of B. glumae 5091. Furthermore, the nucleotide sequences of aac(2′)-IIa from KSM-resistant A. avenae strain 1-1, 3-2, 4-2, 5-1, and 213 were identical to that of B. glumae 5091. On the other hand, KSM-resistant A. avenae strain 83 contains three substitutions of a nucleotide sequence at position 234 from C to G, at position 258 from T to G, and at position 436 from T to A within the coding region. T436A leads to the substitution of an amino acid from serine to threonine at position 146 in AAC(2′)-IIa (Fig. 1A). MIC of KSM was relatively higher in A. avenae strain 83 (3,200 μg/ml) than in other strains 1-1, 3-2, 4-2, 5-1, and 213 (800 μg/ml) (Table 1). We are currently investigating whether the substitution from serine to threonine at position 146 in AAC(2′)-IIa contributes to an increased MIC to KSM.
Identification of the IncP Island.
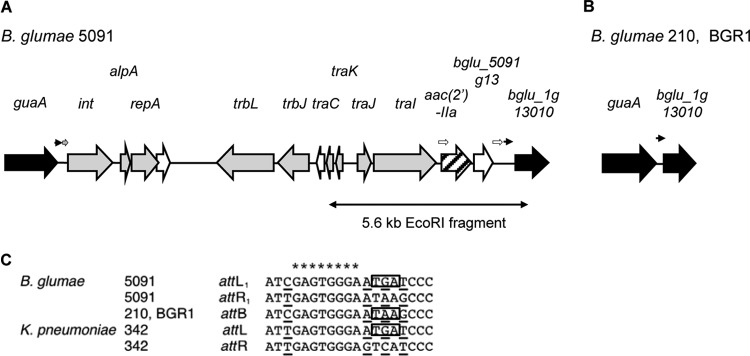
The genetic organization of regions flanking the aac(2′)-IIa gene on the 5.6-kb EcoRI fragment is shown in Fig. 4. At the right end of the sequence is a part of the hypothetical gene (bglu_1g13010), which was identified in a genome sequence of B. glumae BGR1 chromosome 1 (26). Although the upstream region of bglu_1g13010 contains the guaA gene encoding GMP synthase in BGR1, the 5′ upstream of aac(2′)-IIa in strain 5091 encodes two ORFs, oriented in the same direction as aac(2′)-IIa: the putative traJ gene and the traI gene (Fig. 4). In addition to these genes, a part of the putative traC gene was located on the left end of the 5.6-kb EcoRI fragment. These tra genes are known to be involved in horizontal gene transfer and originated from phages or plasmids (1, 9). A comparison of the B. glumae BGR1 and 5091 genomes indicated that a genetic element carrying the aac(2′)-IIa gene was inserted between the guaA and bglu_1g13010 genes.
Fig 4.
Structure of the IncP island from KSM-resistant B. glumae 5091 (A) and the guaA-hypothetical (bglu_1g13010) gene region of the KSM-sensitive strain 210 and the genome-sequenced strain BGR1 (B). Genes related to the phage or plasmid origin, hypothetical genes in the IncP island, and aac(2′)-IIa are indicated by gray arrows, open arrows, and a slashed arrow, respectively. The small black arrows above the gene map indicate the 19-bp direct repeats at the boundaries of the IncP island, whereas small open arrows indicate the 36- to 37-bp direct repeats flanking the aac(2′)-IIa and bglu_5091g13 genes. The small gray arrow indicates the 52-bp inverted repeat sequence which functions as an att site for Brucella suis 1330 (25). (C) Comparison of the site-specific integration sites (19-bp direct repeats) of the IncP islands of B. glumae 5091 with Klebsiella pneumoniae 342. Asterisks indicate a consensus sequence, which is the putative excision site of the guaA-associated genomic islands (37). Open boxes indicate the translational termination codon of the guaA gene.
To identify the genetic element carrying aac(2′)-IIa, the region between guaA and traC was amplified by PCR based on the BGR1 and 5091 genome sequences. A 9.6-kb fragment was amplified from KSM-resistant B. glumae 5091; however, no PCR product was observed when genomic DNA from the KSM-sensitive strain 210 was used as a template. The insertion in the B. glumae 5091 genome is 12,724 bp and contains 13 ORFs (bglu_5091g01 to bglu_5091g13) between guaA and the hypothetical (bglu_1g13010) gene (Fig. 4). The protein encoded by bglu_5091g01 is a member of the Int family of site-specific recombinase. CP4-like integrases are found in temperate bacteriophages, integrative plasmids, and other mobile genomic islands (24). The proteins encoded by bglu_5091g02 to 11 are putative AlpA (transcription regulator), putative RepA (replication protein), hypothetical protein, putative TrbL (conjugative transfer protein), putative TrbJ (conjugal transfer/entry exclusion protein), hypothetical protein, putative TraC (primase), putative TraK (auxiliary protein for relaxase), putative TraJ (auxiliary protein for relaxase), and putative TraI (relaxase). TrbL and TrbJ are components of the mating pair formation apparatus, which forms a channel between the donor and recipient cells (1). TraI recognizes the DNA strand at an origin of transfer (oriT) with TraJ and TraK, yielding a single-stranded DNA break for DNA transfer to the recipient cell via the conjugation machinery (1, 9). The genetic organization from the integrase gene to the traI gene in B. glumae 5091 reveals high similarity to the IncP island in the genome of Brucella suis 1330 and Klebsiella pneumoniae 342 (12, 25) (see Fig. S2 in the supplemental material). These IncP islands are integrated into the 3′ end of the guaA gene, which has been recognized as a hot spot for the insertion of genomic islands (37). While the G+C content of chromosome 1 of B. glumae BGR1 is 68.11% (26), that of the IncP island of B. glumae 5091 is 57.24%. The difference in G+C content from the surrounding chromosome suggests an acquisition of the IncP island of B. glumae 5091 by horizontal gene transfer.
Integration site of the IncP island and excision activity.
Genomic islands are often flanked by short (16- to 20-bp) almost-perfect direct repeats, called attachment sites (atts) (24). Direct repeats are usually produced by the site-specific recombination of genomic islands into the insertion sites. Integrases control both functions of the integration into and the excision from the genome. Site-specific integration of DNA into an attB site in a bacterial chromosome generates two junctions (attL and attR). Upon comparison of the att sequence of the IncP islands from B. glumae 5091 to those from B. suis 1330 and K. pneumoniae 342, short direct repeats (19 bp) are observed on either side of the IncP islands from B. glumae 5091 and K. pneumoniae 342 (12) (Fig. 4). These 19-bp direct repeats contain a consensus sequence (5′-GAGTGGGA-3′), which is the putative excision site of the guaA-associated genomic islands (37). In contrast, the 52-bp almost-perfect direct repeats are used for att sites in the IncP island of B. suis 1330 (25). The IncP island of B. glumae 5091 contains the same 52-bp nucleotide sequence on the left side only, which exists inside of the 19-bp short border sequence (Fig. 4).
To investigate whether integrase could mediate the recombination in B. glumae 5091 between attL1 and attR1 sites, we tried to detect the excision of the IncP island from its genome, the first step of the gene transfer. Whereas excision of the IncP island was previously reported in B. suis 1330 (25), its excision was not detected in B. glumae 5091. The excision and transfer of some genomic islands is facilitated by environmental stress or growth conditions (3, 14, 48). However, in our experimental conditions, and even in other bacterial strains (4-1-1, 4-1-9, 9-1-1, and 11-1-1), excision of IncP islands from KSM-resistant B. glumae was not observed (see Fig. S3 in the supplemental material). Furthermore, in a mating assay, transfer of the IncP island from the donor B. glumae 5091 to the recipient, B. glumae 3 or A. avenae 5 (oxolinic acid-resistant strains) was not detected (the transfer frequency from B. glumae 5091 to B. glumae 3 or to A. avenae 5 was <2.2 × 10−9 or <1.3 × 10−10 per donor, respectively).
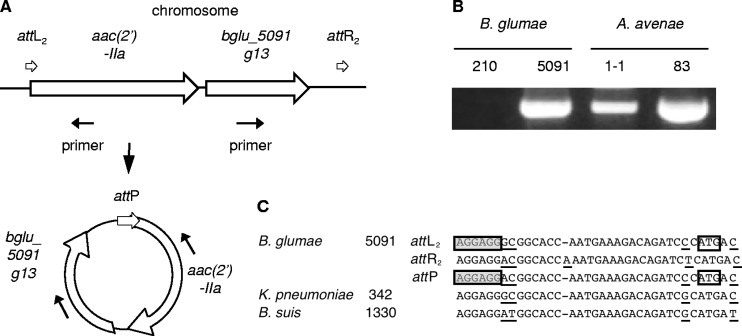
Generation of the circular intermediates containing the aac(2′)-IIa and bglu_5091g13 genes.
Compared to the IncP islands from B. glumae 5091 with those from B. suis 1330 and K. pneumoniae 342, the 5′ sequences (from integrase to the traC gene) in the IncP island are conserved among the three species; however, the remaining 3′ sequences are quite different to each other (see Fig. S2 in the supplemental material). The aac(2′)-IIa and hypothetical genes (bglu_5091g13) exist in the IncP island of B. glumae 5091 only. We found that the second 36- to 37-bp direct repeats (attL2 and attR2) were observed on either side of the aac(2′)-IIa and bglu_5091g13 genes (Fig. 4 and Fig. 5C). Interestingly, the closed circular forms containing the aac(2′)-IIa and bglu_5091g13 genes were detected in B. glumae 5091, A. avenae 1-1, and 83 by inverse PCR, and the attP sequences were generated by joining attL2 to attR2 (Fig. 5). In addition, we determined whether aac(2′)-IIa and bglu_5091g13 were polycistronically transcribed by using RT-PCR. As shown in Fig. 6, the aac(2′)-IIa and bglu_5091g13 genes were amplified from a single transcript. These results indicate that the cassette of aac(2′)-IIa and bglu_5091g13 genes are inserted into the IncP island by another recombination and that the recombination activity may still be active.
Fig 5.
Circularization of the aac(2′)-IIa and hypothetical genes (bglu_5091g13). (A) Generation of the closed circular form of the aac(2′)-IIa and bglu_5091g13 genes at 36- to 37-bp direct repeats (att). PCR primer positions are shown under the gene map. (B) Detection of the circularization by PCR. (C) Comparison of the 36- to 37-bp direct repeats in B. glumae 5091 with other similar sequences in the IncP islands from K. pneumoniae 342 and Brucella suis 1330. The open boxes indicate the translational initiation codon of the aac(2′)-IIa gene. The gray boxes indicate the putative Shine-Dalgarno (SD) sequence of the aac(2′)-IIa gene. Underlined letters show differences in nucleotides in the direct repeats.
Fig 6.
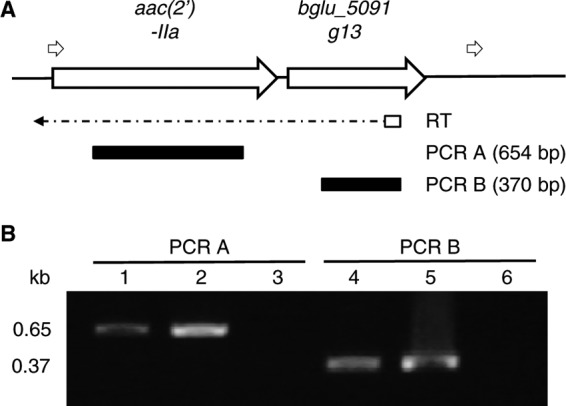
Polycistronic transcription of the aac(2′)-IIa and hypothetical genes (bglu_5091g13) from B. glumae 5091. (A) Organization of the aac(2′)-IIa and hypothetical genes. The small open arrows above the gene map indicate the 36- to 37-bp direct repeats. The dotted arrow below the gene map represents the direction of the reverse transcription (RT) reaction. The black thick bars below the RT arrow indicate the PCR products from the corresponding RT reactions. (B) Analysis of the RT-PCR products of aac(2′)-IIa (PCR A) and bglu_5091g13 (PCR B). Lanes 1 and 4, RT-PCR products; lanes 2 and 5, PCR products from the DNA template; lanes 3 and 6, PCR products from the RNA template.
DISCUSSION
To elucidate the mechanism of KSM resistance, we first investigated spontaneous mutations in the ksgA gene of KSM-resistant B. glumae isolates. Previous in vitro studies showed that spontaneous KSM-resistant mutants of E. amylovora, E. coli, and Bacillus subtilis harbored mutations in the ksgA methyltransferase gene (29, 33). However, our results clarified that a nucleotide substitution of the ksgA gene is not involved in KSM resistance in field isolates of B. glumae. Moreover, the possibility that ksgA-related KSM-resistant bacteria emerged in the field is low because of decreased fitness in the growth rate in vitro and virulence to plants (29, 33). Vila-Sanjurjo et al. identified KSM-resistant mutations (A794G, G926A, and A1519C) in the single 16S rRNA gene of genetically engineered E. coli (52). However, most bacteria harbor multiple copies of rRNA genes in their chromosomes; B. glumae BGR1 also harbors five copies of the rRNA operons (26). These multiple copies of rRNA genes would mask a mutation in the single rRNA gene. Indeed, the nucleotide sequences of 16S rRNA genes in KSM-resistant B. glumae isolates (strains 5091, 4-1-9, and 9-1-1) are identical to those of KSM-susceptible strains (strains 210 and 3).
Resistance to aminoglycoside antibiotics is often conferred by enzymatic modifications. The aminoglycoside N-acetyltransferase (AAC) can be divided into four groups, based on the regiospecificity of the modification: AAC(1), AAC(2′), AAC(3), and AAC(6′) (50). These enzymes utilize acetyl-CoA as the donor of the acetyl group to modify drugs at positions 1 and 3 of the 2-deoxystreptamine ring and positions 2′ and 6′ of the amino-sugar ring. The majority of AACs are encoded by plasmids, which permits the transfer of resistance genes among bacteria. On the other hand, representatives of the AAC(2′)-I subclass of aminoglycoside acetyltransferase are unique because they are chromosomally encoded (34). In phytopathogenic bacteria, the aminoglycoside streptomycin and KSM are usually used to control them. Streptomycin-resistant isolates of E. amylovora, Pseudomonas syringae, and Xanthomonas campestris harbor the strA-strB gene pair encoding phosphotransferase, APH(3′)-Ib-APH(6)-Ic, on the transposon (variants of Tn5393) in the large conjugative plasmid, or on the small nonconjugative plasmid RSF1010 (28, 32, 40, 50). As well as clinical pathogens, the primary mechanism of streptomycin resistance in phytopathogenic bacteria involves the acquisition of streptomycin modifying enzyme genes via horizontal gene transfer.
In the present study, we isolated the aac(2′)-IIa gene encoding the KSM 2′-N-acetyltransferase from B. glumae and A. avenae subsp. avenae. AAC(2′)-IIa could not react on other aminoglycoside antibiotics tested, indicating the KSM-specific enzyme. The substrate specificity of AAC(2′)-IIa is consistent with the unique structure of KSM. The amino acid sequence of AAC(2′)-IIa exhibits low homology with AAC(2′)-Ia in Providencia stuartii (deduced amino acid sequence identity to AAC(2′)-IIa is 9.4%) and AAC(2′)-Ib, -Ic, and -Id in mycobacterial species (13.0, 11.7, and 4.9%, respectively). Also, AAC(2′)-IIa exhibits low homology with Kac273, a self-resistant KSM acetylating enzyme of a KSM-producer, Streptomyces kasugaensis (18, 21). Based on the regiospecificity of the modification and the specificity of the substrate, the aac(2′)-IIa gene is classified as a novel subclass of aac(2′) (Fig. 1).
In agriculture, the available antibiotics are limited to inhibit the occurrence and spread of antibiotic resistance in clinically important pathogens (8). KSM possesses the least potential to affect any mechanism of aminoglycoside resistance in their emergence and spread in human and animal pathogenic bacteria due to its unique structure and low antibacterial activity against clinical pathogens (44). Our results also indicate that if aac(2′)-IIa genes are transferred from phytopathogenic bacteria to clinical pathogens, they would not influence the efficacy of clinically used antibiotics.
Generally, the intensive use of drugs increases the risk of the development of their resistance. The accumulation of resistant pathogens will be enhanced by frequent use of drugs, by the presence of larger pathogen populations before the application of drugs, and by greater descendant production (e.g., large number of spore production in fungi) and shorter generation times in the pathogen (6). Resistance of the fungus M. grisea to KSM in the field was found in 1971 in Yamagata prefecture, where KSM had been used intensively for several years (30). Although the mycelial growth, sporulation, and virulence of these resistant isolates were almost the same as for the sensitive ones (31), the competitive ability of the resistant isolates was inferior to sensitive ones (23). When the application of the antibiotic was stopped, the population of KSM-resistant M. grisea decreased rapidly (31). Currently, the combinations of KSM plus melanin biosynthesis inhibitors (MBIs) are used effectively to control rice blast. Ascospore analyses revealed that the occurrence of resistance to KSM in M. grisea is controlled by at least three independent chromosome loci (41, 42).
In plant bacterial pathogens, KSM-resistant A. avenae subsp. avenae emerged in some areas of Japan in 1990 (43). In these regions, to hasten germination and control rice bacterial diseases, rice seeds had been incubated at a high temperature in a solution of KSM with air circulation for 1 day before sowing seeds. These conditions during seed disinfection were also suitable for bacterial growth, leading to the occurrence of KSM-resistant A. avenae. In our research, all KSM-resistant rice-pathogenic bacteria (B. glumae; 18 isolates, A. avenae; 9 isolates) harbored the aac(2′)-IIa gene whose molecular detection can distinguish whether bacterial strains are resistant or susceptible to KSM. Furthermore, our studies reveal that the aac(2′)-IIa gene of B. glumae 5091 is located in the IncP genomic island of the bacterial chromosome. Acquiring the aac(2′)-IIa gene may result from the horizontal gene transfer by activity of integrase and tra genes in the IncP island. Lavigne et al. reported that the IncP island in the genome of Brucella suis 1330 is excised from its chromosome and forms circular intermediates (25). Although no excision activity of the IncP island in KSM-resistant B. glumae was observed, the circular forms containing the aac(2′)-IIa and bglu_5091g13 genes were generated at the second att recombination site, suggesting that the aac(2′)-IIa gene had been integrated into the IncP island of a donor bacterial species. Because the ability of IncP island transfer in KSM-resistant B. glumae may be tightly regulated, the occurrence of KSM-resistant isolates in various parts of the country occurs at a relatively low frequency. It is important to inhibit the spread of KSM-resistant pathogens through the distribution of rice seeds.
In conclusion, we demonstrated that the mechanism of KSM resistance in rice-pathogenic bacteria in the fields is mainly due to acetylation of the drug and that KSM-resistant isolates can be detected whether the KSM acetyltransferase, aac(2′)-IIa gene, is present or absent. The rapid distinction between KSM-sensitive and -resistant isolates by PCR analysis will serve to inhibit the spread of KSM-resistant rice seed-borne diseases and spur the effective use of the drug. To characterize the detailed function of the aac(2′)-IIa gene will provide hopeful information to combat KSM-resistant pathogens in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank Masanori Yamaguchi for supplying 2′-acetyl-KSM and for technical advice. We thank Yuichi Kita for technical support in analyzing acetyl-KSM.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Adamczyk M, Jagura-Burdzy G. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50:425–453 [PubMed] [Google Scholar]
- 2. Aiba H, Adhya S, de Crombrugghe B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905–11910 [PubMed] [Google Scholar]
- 3. Almagro-Moreno S, Napolitano MG, Boyd EF. 2010. Excision dynamics of Vibrio pathogenicity island-2 from Vibrio cholerae: role of a recombination directionality factor VefA. BMC Microbiol. 10:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ausubel FM, et al. 1994. Current protocols in molecular biology, vol 1 Greene Publishing Associates/John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 6. Brent KJ, Hollomon DW. 2007. Fungicide resistance in crop pathogens: how can it be managed? FRAC Monograph 1. Fungicide Resistance Action Committee, Basel, Switzerland [Google Scholar]
- 7. Darling AE, Mau B, Perna NT. 2010. Progressive mauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375–382 [DOI] [PubMed] [Google Scholar]
- 9. de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34:18–40 [DOI] [PubMed] [Google Scholar]
- 10. Demirci H, et al. 2010. Modification of 16S rRNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA 16:2319–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flatt PM, Mahmud T. 2007. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat. Prod. Rep. 24:358–392 [DOI] [PubMed] [Google Scholar]
- 12. Fouts DE, et al. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4:e1000141 doi:10.1371/journal.pgen.1000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fouts KE, Barbour SD. 1981. Transductional mapping of ksgB and a new Tn5-induced kasugamycin resistance gene, ksgD, in Escherichia coli K-12. J. Bacteriol. 145:914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guerin E, et al. 2009. The SOS response controls integron recombination. Science 324:1034. [DOI] [PubMed] [Google Scholar]
- 15. Helser TL, Davies JE, Dahlberg JE. 1971. Change in methylation of 16S rRNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat. New Biol. 233:12–14 [DOI] [PubMed] [Google Scholar]
- 16. Helser TL, Davies JE, Dahlberg JE. 1972. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 235:6–9 [DOI] [PubMed] [Google Scholar]
- 17. Hiramatsu M, Wada T, Kanda M, Yamamura H. 1989. Effect of kasugamycin on the control of bacterial diseases, p 219–224 Abstr. Proc. 7th Int. Conf. Plant Pathol. Bacteriol [Google Scholar]
- 18. Hirasawa K, Tamamura K, Takeuchi T, Hamada M. 1993. Kasugamycin acetyltransferase gene and the DNA fragment including kasugamycin acetyltransferase gene. Jpn. Kokai Tokkyo Koho JP A-05-23187. (In Japanese.) [Google Scholar]
- 19. Hori T, Kuroda T, Ishikawa K. 2007. Occurrence of kasugamycin-resistant Burkholderia glumae. Ann. Phytopathol. Soc. Japan 73:278 (In Japanese.) [Google Scholar]
- 20. Ikekawa T, Umezawa H, Iitaka Y. 1966. The structure of kasugamycin hydrobromide by X-ray crystallographic analysis. J. Antibiot. (Tokyo) 19:49–50 [PubMed] [Google Scholar]
- 21. Ikeno S, et al. 1998. A 7.6-kb DNA region from Streptomyces kasugaensis M338-M1 includes some genes responsible for kasugamycin biosynthesis. J. Antibiot. (Tokyo) 51:341–352 [DOI] [PubMed] [Google Scholar]
- 22. Ishiyama T, et al. 1965. Studies on the preventive effect of kasugamycin on rice blast. J. Antibiot. (Tokyo) 18:115–119 [PubMed] [Google Scholar]
- 23. Ito I, Yamaguchi T. 1979. Competition between sensitive and resistant strains of Pyricularia oryzae Cav. against kasugamycin. Ann. Phytopathol. Soc. Japan 45:40–46 (In Japanese.) [Google Scholar]
- 24. Juhas M, et al. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33:376–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lavigne JP, Vergunst AC, Bourg G, O'Callaghan D. 2005. The IncP island in the genome of Brucella suis 1330 was acquired by site-specific integration. Infect. Immun. 73:7779–7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim J, et al. 2009. Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 191:3758–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Masukawa H, Tanaka N, Umezawa H. 1968. Inhibition by kasugamycin of protein synthesis in Pyricularia oryzae. J. Antibiot. (Tokyo) 21:73–74 [DOI] [PubMed] [Google Scholar]
- 28. McGhee GC, et al. 2011. Genetic analysis of streptomycin-resistant (Smr) strains of Erwinia amylovora suggests that dissemination of two genotypes is responsible for the current distribution of Smr E. amylovora in Michigan. Phytopathology 101:182–191 [DOI] [PubMed] [Google Scholar]
- 29. McGhee GC, Sundin GW. 2011. Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora. Phytopathology 101:192–204 [DOI] [PubMed] [Google Scholar]
- 30. Miura H, Ito K, Kimura K, Takahashi S. 1973. Occurrence of resistant strains of Pyricularia oryzae to kasugamycin as a cause of the diminished fungicidal activity. Ann. Phytopathol. Soc. Japan 39:239–240 (In Japanese.) [Google Scholar]
- 31. Miura H, Katagiri M, Yamaguchi T, Uesugi Y, Ito H. 1976. Mode of occurrence of kasugamycin resistant rice blast fungus. Ann. Phytopathol. Soc. Japan 42:117–123 [Google Scholar]
- 32. Nakajima M, et al. 1995. Similarity of streptomycin resistance gene(s) in Pseudomonas syringae pv. actinidiae with strA and strB of plasmid RSF1010. Ann. Phytopathol. Soc. Japan 61:489–492 [Google Scholar]
- 33. Ochi K, et al. 2009. Inactivation of KsgA, a 16S rRNA methyltransferase, causes vigorous emergence of mutants with high-level kasugamycin resistance. Antimicrob. Agents Chemother. 53:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rather PN, Orosz E, Shaw KJ, Hare R, Miller G. 1993. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J. Bacteriol. 175:6492–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schluenzen F, et al. 2006. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat. Struct. Mol. Biol. 13:871–878 [DOI] [PubMed] [Google Scholar]
- 36. Schuwirth BS, et al. 2006. Structural analysis of kasugamycin inhibition of translation. Nat. Struct. Mol. Biol. 13:879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song L, Pan Y, Chen S, Zhang X. 2012. Structural characteristics of genomic islands associated with GMP synthases as integration hot spot among sequenced microbial genomes. Comput. Biol. Chem. 36:62–70 [DOI] [PubMed] [Google Scholar]
- 38. Sparling PF. 1970. Kasugamycin resistance: 30S ribosomal mutation with an unusual location on the Escherichia coli chromosome. Science 167:56–58 [DOI] [PubMed] [Google Scholar]
- 39. Sparling PF, Ikeya Y, Elliot D. 1973. Two genetic loci for resistance to kasugamycin in Escherichia coli. J. Bacteriol. 113:704–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sundin GW, Bender CL. 1995. Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl. Environ. Microbiol. 61:2891–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taga M, Nakagawa H, Tsuda M, Ueyama A. 1978. Ascospore analysis of kasugamycin resistance in the perfect stage of Pyricularia oryzae. Phytopathology 68:815–817 [Google Scholar]
- 42. Taga M, Nakagawa H, Tsuda M, Ueyama A. 1979. Identification of three different loci controlling kasugamycin resistance in Pyricularia oryzae. Phytopathology 69:463–466 [Google Scholar]
- 43. Takeuchi T, Tamura O. 1991. Occurrence of kasugamycin-resistant Acidovorax avenae subsp. avenae. Ann. Phytopathol. Soc. Japan 57:117–118 (In Japanese.) [Google Scholar]
- 44. Tamamura T, Sato K. 1999. Comparative studies on in vitro activities of kasugamycin and clinically used aminoglycoside antibiotics. Jpn. J. Antibiot. 52:57–67 [PubMed] [Google Scholar]
- 45. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony. Methods Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanaka N, Yoshida Y, Sashikata K, Yamaguchi H, Umezawa H. 1966. Inhibition of polypeptide synthesis by kasugamycin, an aminoglycosidic antibiotic. J. Antibiot. (Tokyo) 19:65–68 [PubMed] [Google Scholar]
- 47. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ubeda C, et al. 2005. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 56:836–844 [DOI] [PubMed] [Google Scholar]
- 49. Umezawa H, Hamada M, Suhara Y, Hashimoto T, Ikekawa T. 1965. Kasugamycin, a new antibiotic. Antimicrob. Agents Chemother. 5:753–757 [PubMed] [Google Scholar]
- 50. Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vetting MW, et al. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212–226 [DOI] [PubMed] [Google Scholar]
- 52. Vila-Sanjurjo A, Squires CL, Dahlberg AE. 1999. Isolation of kasugamycin resistant mutants in the 16S rRNA of Escherichia coli. J. Mol. Biol. 15:. 293:1–8 [DOI] [PubMed] [Google Scholar]
- 53. Woodcock J, Moazed D, Cannon M, Davies J, Noller HF. 1991. Interaction of antibiotics with A- and P-site-specific bases in 16S rRNA. EMBO J. 10:3099–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoshikawa M, Okuyama A, Tanaka N. 1975. A third kasugamycin resistance locus, ksgC, affecting ribosomal protein S2 in Escherichia coli K-12. J. Bacteriol. 122:796–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.