Abstract
A prominent feature of most cancers including Barrett’s adenocarcinoma (BAC) is genetic instability, which is associated with development and progression of disease. In this study, we investigated the role of recombinase (hsRAD51), a key component of homologous recombination (HR)/repair, in evolving genomic changes and growth of BAC cells. We show that the expression of RAD51 is elevated in BAC cell lines and tissue specimens, relative to normal cells. HR activity is also elevated and significantly correlates with RAD51 expression in BAC cells. The suppression of RAD51 expression, by short hairpin RNA (shRNA) specifically targeting this gene, significantly prevented BAC cells from acquiring genomic changes to either copy number or heterozygosity (P<0.02) in several independent experiments employing single-nucleotide polymorphism arrays. The reduction in copy-number changes, following shRNA treatment, was confirmed by Comparative Genome Hybridization analyses of the same DNA samples. Moreover, the chromosomal distributions of mutations correlated strongly with frequencies and locations of Alu interspersed repetitive elements on individual chromosomes. We conclude that the hsRAD51 protein level is systematically elevated in BAC, contributes significantly to genomic evolution during serial propagation of these cells and correlates with disease progression. Alu sequences may serve as substrates for elevated HR during cell proliferation in vitro, as they have been reported to do during the evolution of species, and thus may provide additional targets for prevention or treatment of this disease.
Keywords: Barrett’s adenocarcinoma, recombination, evolution, alu, hsRad51
Introduction
Carcinogenesis is understood to be a complex and stepwise process (Farber, 1984) in which a normal cell undergoes a variety of genetic alterations and acquires a set of unique characteristics that enable it to progress from normal to benign-cancer and then to malignant phenotypes. Ongoing and evolving genomic changes progressively lead to development of more aggressive and drug-resistant states. The genomes of normal diploid cells are quite stable, whereas genetic alterations at both the nucleotide-sequence and chromosomal-rearrangement levels are frequently seen in cancer (Pathak et al., 1994; Lengauer et al., 1997, 1998). The types of abnormalities observed at diagnosis continue to accumulate with progression of disease (Lengauer et al., 1998), indicating a striking and ongoing genomic evolution. The evaluation of genomic changes in premalignant polyp and sporadic colorectal cancer cells has indicated that genetic instability is probably a cause rather than a consequence of oncogenic transformation (Stoler et al., 1999). Ongoing genomic evolution not only enables cancer cells to progress to more malignant stages of disease but also makes both diagnosis and treatment challenging tasks (Ye et al., 2007).
Most human cancers show multiple mutations, arising as independent events (Neiman and Hartwell, 1991), frequently observed in genes, such as c-myc, APC, c-K-ras, DCC, MCC, p53, Rad51, BRCA1, BRCA2 and related genes. For example, gastric cancer is commonly associated with mutations in c-K-ras (Miki et al., 1991), APC (Horii et al., 1992) and p53 (Yamada et al., 1991), whereas hereditary nonpolyposis colorectal cancer is known to feature mutations in DNA mismatch-repair genes, such as hMLH1, hPMS1, hMSH2 and hMSH6 (Lynch and Lynch, 1998; Lynch et al., 1998). Development of colorectal carcinoma (Tsancheva, 1997) appears to require the accrual of seven or more such mutations. Progression from a benign tumor to colorectal cancer may require as long as two decades, whereas further progression to metastasis can occur in <2 more years (Jones et al., 2008). According to a recent report, changes in up to 20 genes are required for oncogenic transformation of a normal cell (Beerenwinkel et al., 2007). With availability of new genome-wide analytical tools, it is now becoming obvious that genomic evolution in cancer cells is more widespread than previously thought. For example, it has been accepted for many years that chronic lymphocytic leukemia is associated with specific molecular and cytogenetic abnormalities, and genomic changes in this cancer do not often evolve during the course of disease. However, Stilgenbauer et al. (2007) recently demonstrated that a subgroup of chronic lymphocytic leukemia patients displays an ongoing genomic instability, associated with poor clinical outcome and shorter survival of these patients.
Barrett’s esophageal adenocarcinoma is a cancer associated with gastroesophageal reflux disease. The cancer develops gradually and progressively from specialized intestinal metaplasia of Barrett’s esophagus (BE) (Spechler and Goyal, 1986), a pre-malignant lesion, and passes through stages of low-grade to highgrade dysplasia, before finally emerging as adenocarcinoma (Spechler and Goyal, 1986). Data from a number of laboratories indicate that genetic instability arises early, at the BE stage, and gradually intensifies—leading to a series of genomic changes, some of which underlie progression through successive stages of dysplasia. Rabinovitch et al. evaluating specimens of Barrett’s adenocarcinoma (BAC) for DNA amounts, proposed in 1989 that aneuploidy caused by genetic instability in a subset of cells is associated with progression of BE to adenocarcinoma. Finley et al. (2006) using probes against centromeres and specific regions of chromosomes 9, 11 and 17, demonstrated that chromosomal instability arises early in the etiology of BE. Consistent with these observations, genome-wide analyses of single-nucleotide polymorphisms (SNPs) also identified multiple alterations in a majority of both the BE and BAC specimens examined (Akagi et al., 2009). Changes in length of short repetitive DNA sites, indicative of microsatellite instability, as well as loss of heterozygosity (LOH), have also been reported in all stages of BAC including pre-malignant BE (Cai et al., 2008). A comprehensive study, utilizing image cytometry to evaluate DNA content in a large number of specimens of normal esophagi and various stages of BAC, indicated that aneuploidy is detected in a subset of BE specimens, and increases in frequency and severity from BE to low-grade dysplasia (LGD), high-grade dysplasia (HGD) and then to BAC. Evaluation of the genome by comparative genomic hybridization (CGH) indicated that relatively large deletions and amplifications were rare in early stages of disease but more frequent and spanned larger chromosomal regions in high-grade dysplasia and adenocarcinoma (Croft et al., 2002; Paulson et al., 2009). It is therefore clear that genomic instability arises early in BAC and increases over time, leading to accumulation of mutations and accompanied by progression of disease. Some of the mutations occurring in early stages of BAC affect antioncogenes, such as p16 and p53 (Koppert et al., 2005).
Mechanisms of genomic instability in cancer are not fully understood, and identification of these mechanisms could furnish novel targets for cancer prevention and treatment. It has been shown that tumorigenicity of cancer cell lines correlates with their karyotypic instability (Gee and Harris, 1979), suggesting that genomic rearrangements may have an important role in the development and/or progression of cancer. Although genetic recombination has been implicated in diverse aspects of maintaining the cell’s genetic integrity, including DNA repair (Resnick et al., 1989) and proper segregation of chromosomes in meiosis (Baker et al., 1976), aberrant or deregulated recombination activity can mediate genomic rearrangements and is likely involved in LOH (Honma et al., 1997), gene amplification (Windle et al., 1991) and chromosomal translocation (Cheng et al., 1997; Honma et al., 1997; Shammas et al., 1997) and therefore is a presumed mechanism for both oncogene activation and the LOH that inactivates the second copy of a mutated antioncogene. Consistent with this, recombination activity is altered by a variety of chemicals, carcinogens, radiation and oncogenic viruses (Rodarte-Ramon, 1972; Radman et al., 1982; Sengstag, 1994; Galli and Schiestl, 1995; Cheng et al., 1997; Li et al., 1997). Homologous recombination (HR) depends on extensive sequence homology between participating DNA molecules and is an important repair mechanism, which has a vital role in maintaining genomic integrity of a cell. Impairment or deregulation of HR can also be deleterious because it has the potential to cause unnecessary and ongoing genomic rearrangements. We have shown that HR activity is constitutively elevated in multiple myeloma and mediates genomic instability and development of drug resistance in myeloma cells (Shammas et al., 2009).
Repetitive sequences in the genome, such as Alu, which provide abundant, interspersed substrates for the HR machinery, may adversely affect the consequences of an overactive recombination pathway. The purpose of this study was to study genomic evolution in BAC cells and elucidate the roles of recombinase (hsRAD51), HR activity and repetitive DNA sequences in the genome. We show that recombinase (RAD51) expression is markedly elevated in BAC cell lines and tissue specimens, relative to normal cells. HR activity, as assessed by a plasmid-based assay, is also significantly elevated in BAC cells. Both RAD51 and HR are further upregulated by exposure of BAC cells to a carcinogen, known to be recombinogenic. Genome-wide analyses by both SNP (Affymetrix, Santa Clara, CA, USA) and CGH (Agilent Technologies, Santa Clara, CA, USA) arrays show that the suppression of RAD51 consistently reduces HR and significantly prevents the genomic evolution observed in BAC cells, in multiple independent experiments. We also show that mutational frequencies correlate well with Alu frequencies and their locations on individual chromosomes.
Results
Recombinase (hsRAD51) is overexpressed in Barrett’s esophageal adenocarcinoma cell lines and tissue specimens
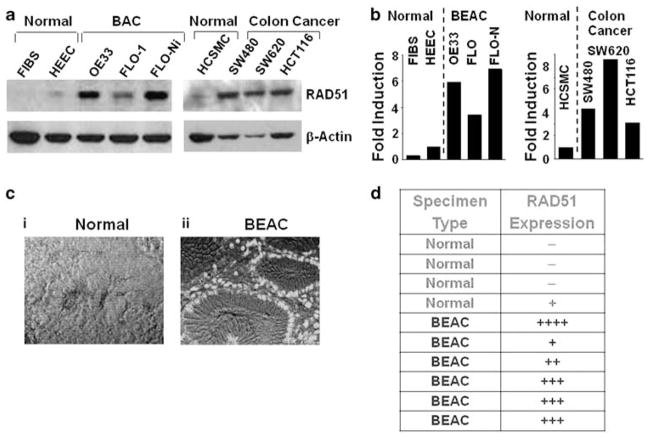
We analyzed RAD51 protein expression in BAC and other cancer cell lines, human BAC tissue specimens, and corresponding normal cells, using western blot and immunohistochemical analyses. RAD51 expression was near background level in normal fibroblasts, and very low or absent in normal human esophageal epithelial cells (HEEC), but was elevated 3.5- to 6-fold in two BAC cell lines, FLO-1 and OE33, relative to HEEC levels (Figures 1a and b). Treatment of FLO-1 cells with a known carcinogen and recombinogen, nickel chloride (Li et al., 1997; Shammas et al., 2009), led to a further twofold induction of RAD51 expression. To evaluate levels of RAD51 in tissue specimens, sections of paraffin-embedded BE and BAC tissues (from the Tissue Core at the Karmanos Cancer Institute, Detroit, MI, USA) were deparaffinized, blocked, and incubated with mouse monoclonal antibody against RAD51. The samples were then treated with a fluorescence- or horseradish peroxidase-labeled secondary antibody. Both immunofluorescence and immunohistochemical analyses indicated that RAD51 protein was either low or absent in four normal tissue sections of primary esophageal epithelium, but highly expressed in five of six BAC specimens tested (Figures 1c and d).
Figure 1.
Recombinase (hsRAD51) is overexpressed in Barrett’s esophageal adenocarcinoma (BAC) cell lines and tissue specimens. (a) RAD51 protein expression was analyzed by western blotting in normal human fibroblasts (FIBS), normal primary HEEC, BEAC cell lines (FLO-1, OE33), FLO-1 treated with recombinogen nickel chloride (0.6 mg/ml) for 3 h, normal human colonic smooth muscle cells (HCSMCs) and colonic adenocarcinoma cell lines (SW480, SW620, HCT116). (b) Bar graph showing fold elevation in RAD51 in the adenocarcinoma cell lines of panel (a), relative to control cells. (c) RAD51 immunostaining within BEAC, vs normal tissue. Sections of paraffin-embedded BAC tissues were deparaffinized and treated with anti-RAD51 mouse monoclonal antibody. The samples were then treated with Alexa-Fluor 488-labeled goat anti-mouse secondary antibody and viewed under a fluorescence microscope. The pictures show fluorescence merged with transmitted-light images. (d) Table showing relative expression of RAD51 in all human tissue specimens examined, including those of panel (c). BE, Barrett’s esophagus; BAC Barrett’s adenocarcinoma. A full colour version of this figure is available at the Oncogene journal online.
HR activity is elevated in Barrett’s esophageal adenocarcinoma cells, concordant with RAD51 expression
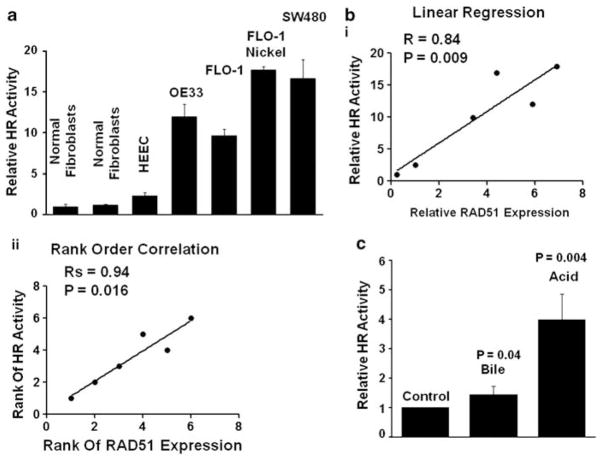
Using a plasmid-based assay, we have shown that HR activity is significantly elevated in hematologic cancer cells (Shammas et al., 2009). For this study, we used an improved HR substrate plasmid in which HR generates a functional firefly luciferase gene, Fluc, while Gaussia luciferase (Gluc) serves as an internal standard, which remains unaffected by HR. The plasmid is introduced into test cells, which are harvested after appropriate incubation, and HR is assessed by the ratio of FLuc to GLuc luciferase activities. Assessment of HR in various cell lines with the new substrate plasmid was consistent with our previously used plasmid substrate, R=0.993 (not shown). Consistent with recombinase protein assays, HR activity was near background level in two human diploid fibroblast strains, low in normal primary HEEC, but elevated 11-, 9- and 15- fold in OE33, FLO-1 and SW480 cells relative to normal fibroblasts (Figure 2a). Moreover, treatment of cells with nickel chloride, which induced the expression of recombinase protein in FLO-1 cells (Figures 1a and b), also led to a further twofold induction of HR activity in these cells (Figure 2a).
Figure 2.
HR activity is constitutively elevated in BEAC cells and correlates with RAD51 expression. (a) HR activity was assessed in normal diploid fibroblasts, primary HEECs (ScienCell), BEAC cell lines (OE33, FLO-1), FLO-1 treated with a known recombinogen, nickel chloride (0.6 mg/ml) for 3 h, and colonic adenocarcinoma cell line SW480, using a plasmid-based assay as described in Materials and methods section. Error bars represent s.e.m. of triplicate assays. (b) Correlation between HR activity and RAD51 expression levels was assessed using the data presented in Figures 1b and 2a. Pearson correlation coefficient (r) of 0.84 (P=0.009) for linear regression (panel I) and a Spearman coefficient, rs, of 0.94 (P=0.016) for rank-order regression (panel II) is shown. (c) Normal fibroblasts were transfected with HR substrate plasmid and exposed for 10 min to either bile (sodium glycochenodeoxycholate, 200 μM) or acid (by adjusting the pH of Dulbecco’s modified Eagle medium (DMEM) to 4 with HCl). Cells were washed twice with regular growth medium (RGM) and incubated in RGM for further 36 h at 37 °C, and evaluated for HR.
HR activity in various normal and BAC cells tested was highly concordant with the expression of RAD51 (Figure 2b), with a Pearson correlation coefficient (R) of 0.84 (P=0.009) for linear regression (Figure 4b, panel I), and a Spearman coefficient, RS, of 0.94 (P=0.016) for rank-order regression (panel II).
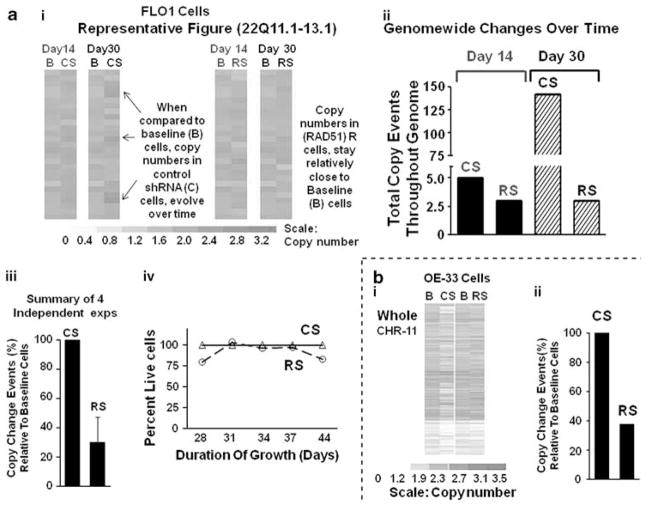
Figure 4.
Suppression of RAD51 prevents the acquisition of copy-number changes in BEAC cells. BEAC (FLO-1 and OE33) cells were transduced with lentivirus particles producing either control CS or RAD51-specific RS shRNAs. For each cell line, just before transduction, an aliquot of cells was harvested and stored at −150 °C to be used as baseline reference ‘B’. Transduced cells were allowed to recover, selected in puromycin for 3 days, and continued in cultured. Cells were harvested at various intervals, genomic DNA from these and baseline reference cells isolated, and evaluated for copy-number changes using 500K or 6.0 SNP arrays (Affymetrix) and software dChip (Kolomietz et al., 2002; Graves, 2004; Steinemann and Steinemann, 2005), as described in Materials and methods section. SNP hybridization intensities of baseline (day 0) cells were used to define the copy-number baseline, departures from which identified changes in transduced cells. A mutational event was then defined as a detectable change in copy number, in three or more consecutive SNPs. (a) Impact of RAD51 suppression on acquisition of copy-number changes in FLO-1 cells: panel (I) figure showing relative copy numbers in a region of chromosome 22, in baseline ‘B’ cells and those which were transduced with control CS or RAD51-specific RS shRNAs and cultured and harvested at two different time points, that is, day 14 or day 30. Color scale at the bottom shows relative copy numbers. Panel (II): for the experiment in panel (a), the number of copy-change events (that is, detectable change in three or more consecutive SNPs) detected throughout the genome, in transduced relative to baseline cells, is presented. Panel (III): bar graph shows prevention of copy-number changes in RAD51-suppressed RS cells, in four independent experiments; error bars represent s.e.m. CNCEs in transduced (CS and RS cells) relative to baseline cells are presented as percent of control shRNA-transduced CS cells. Panel (IV): line graph showing growth rates of control and RAD51-suppressed cells. (b) Impact of RAD51 suppression on acquisition of copy-number changes in OE33 cells: panel (I): OE33 cells were transduced with CS or RS shRNAs, cultured for 25 days and genomic changes evaluated. Relative copy numbers in chromosome 11 are shown as example. The color scale at the bottom shows relative copy numbers. Panel (II): for the experiment shown in panel (I), copy change events (that is, detectable change in three or more consecutive SNPs) detected throughout the genome, in treated relative to baseline cells, are presented as percent of control shRNA-treated CS cells. A full colour version of this figure is available at the Oncogene journal online.
As BAC is associated with acid reflux, we assessed whether HR in normal cells is affected by short-term exposure to acid or bile. Normal fibroblasts (described above) were transfected with HR substrate plasmid and exposed for 10 min to either bile (sodium glycochenodeoxycholate, 200 μM) or acid (by adjusting the pH of Dulbecco’s modified Eagle medium to 4 with HCl). Cells were washed twice with regular growth medium and incubated in regular growth medium for further 36 h at 37 °C, and evaluated for HR. Although short-term exposure to bile led to only 1.4-fold induction of HR, the exposure to acid caused a 4-fold induction of HR in normal human cells (P=0.004) (Figure 2c).
Suppression of HsRAD51 in BAC cells and impact on HR and gene expression
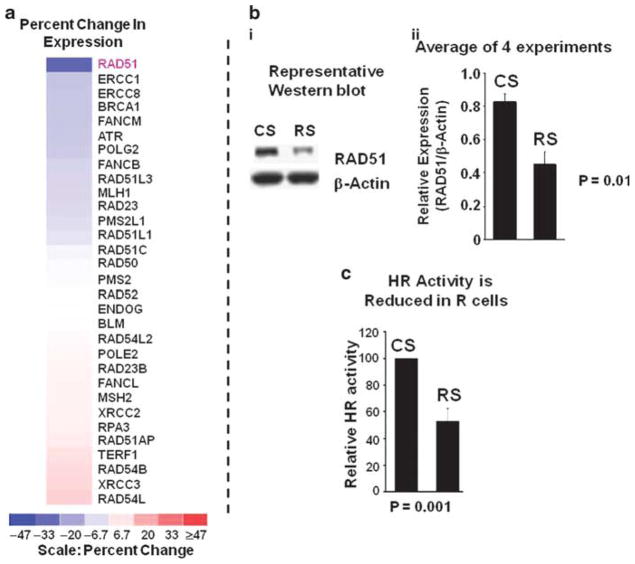
To evaluate the role of elevated recombinase and increased HR activity in evolving genomic changes in BAC cells, FLO-1 cells were transduced with lentiviruses, producing either control short hairpin RNA (shRNA) ‘CS’ or HsRAD51-targeting shRNA ‘RS’. Following recovery and selection in puromycin, at day 6 after transduction, the cells were evaluated for gene transcript levels using Human Gene 1.0 ST Arrays (Affymetrix). The color scale in Figure 3a represents percent change in expression of 31 genes associated with HR and genomic instability, in cells treated with RAD51 shRNA ‘RS’ relative to control cells expressing shRNA ‘CS’. Only HsRAD51, of all the HR and genomic-instability genes examined, had expression reduced by ≥40% in R-transduced cells, consistent with direct and specific silencing of the intended target.
Figure 3.
Impact of RAD51 suppression on expression profile and HR activity in BAC cells. (a) FLO-1 cells were transduced with lentiviruses, expressing control shRNA CS or RAD51-targeting shRNA RS. Following recovery and selection in puromycin, the cells were evaluated for gene expression, using Human Gene 1.0 ST Arrays. The color scale at the bottom of the figure represents % change in expression of HR and genomic instability associated genes in cells transduced with RAD51 shRNA ‘RS’, relative to control cells transduced with shRNA CS. (b) Panel (I): a representative western blot, showing Rad51 protein in FLO-1 cells transduced with control CS or RAD51 shRNA RS, at day 6 after transduction. Panel (II): bar graph showing relative expression of RAD51 following normalization with β-actin; error bars indicate s.e.m. of four independent experiments, each involving a new transduction. (c) HR activity is reduced in RAD51-suppressed cells. HR activity was assessed at day six after transduction, using a plasmid-based assay as described for Figure 2. Relative HR activity in RAD51-suppressed RS cells is shown as percentage of activity in control shRNA transduced CS cells: error bar indicates s.e.m. of four independent experiments.
In four independent experiments, transduction with RAD51-specific shRNA (RS) was associated with a 47% mean reduction in HsRAD51 protein in FLO-1 cells (P<0.01) (Figure 3b). Commensurate with decreased recombinase expression, HR activity was also reduced by 47% (P=0.001), in four independent experiments, each involving a new lentiviral transduction (Figure 3c).
Suppression of hsRAD51 and HR slows the acquisition of new genomic changes in BAC cells
We showed previously that cancer cell lines with constitutively elevated HR activity acquire new genomic changes over time; suppression of HR in these cells slows the acquisition of genomic changes whereas further induction by recombinogen treatment significantly increased the mutation rate (Shammas et al., 2009). We have consistently observed that cancer cells with higher HR levels acquire more genomic changes over time than do those with lower HR activity (Shammas et al., 2009 and unpublished data). To evaluate the role of elevated hsRAD51 and increased HR in evolving genomic changes in BAC cells, FLO-1 and OE33 cells transduced with control (CS) or RAD51-specific (RS) shRNAs were cultured for varying intervals before harvesting. An aliquot of cells was harvested and frozen at the beginning of each experiment (day 0), providing a baseline control. Accrual of genomic changes, in cells transduced with control vs RAD51 shRNA, was monitored by genome-wide microarray screens for heterozygosity using SNP arrays (Affymetrix), and for copy-number variation using both SNP arrays and CGH arrays (Agilent).
Figure 4a shows representative copy-number experiment based on relative signal intensity in SNP arrays probed with genomic DNA fragments. FLO-1 cells, transduced with control (CS) or RAD51 (RS) shRNAs and then maintained by serial subculture, were harvested at two time points: day 14 and day 30. Copy-number changes in cells transduced with CS or RS shRNA, are compared with baseline (B) cells for a region of chromosome 22 (Figure 4a, panel I) and summarized for the entire genome (Figure 4a, panel II). Copy-number change events (CNCEs, where one event is here defined as an observed change in three or more consecutive SNPs) increased over time in cells transduced with control shRNA, but remained constant in RAD51-suppressed cells (Figure 4a). Combining results from four independent experiments, RAD51 suppression reduced the accumulation of CNCE in FLO-1 cells by an average of 70±17%(Figure 4a, panel III). Similar results were obtained with another BAC cell line, OE33, in which RAD51 suppression reduced the time-dependent accrual of CNCE by 62% (Figure 4b). When results are compiled for both BAC lines (FLO-1 and OE33), shRNA suppression of RAD51 reduced the accumulation of genome-wide CNCE by an average of 68±14% (mean±s.d., P<0.0007).
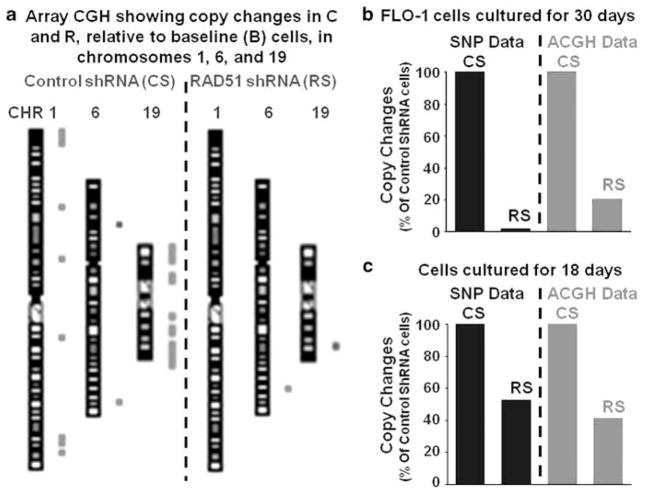
To confirm the prevention of copy-number changes following RAD51 suppression in BAC cells, the DNA samples from two independent experiments, which were analyzed by SNP, were also analyzed by CGH using Agilent’s newest high-density (1×1M) CGH arrays. These arrays have more evenly distributed and longer (60-mer) probes, with a median overall spacing of 2.1 kb, and therefore provide better coverage and more accurate measurement of copy-number changes than afforded by SNP arrays (Maciejewski and Mufti, 2008). Figure 5a is a representative array CGH figure showing chromosomes 1, 6 and 19 in ‘CS’ and ‘RS’ cells relative to baseline cells, as an example. Copy-number changes (deletions shown as green and amplifications as red dots) seen frequently in the chromosomes of control shRNA-transduced CS cells, are less frequent in RAD51-suppressed RS cells. Figures 5b and c represent two independent experiments in which DNA samples were analyzed with both the SNP and array CGH analyses. In both experiments, evaluation by CGH showed essentially the same suppression of copy-number changes in RS-treated cells, as were indicated by SNP data (R=0.99). These data confirm that suppression of RAD51 slows the acquisition of copy-number changes in BAC cells.
Figure 5.
Prevention of copy-number changes in RAD51-suppressed cells is confirmed by array CGH. To confirm the prevention of copy-number changes following RAD51 suppression in BAC cells, DNA samples from two independent experiments, which were analyzed by SNP arrays, were also analyzed by CGH assay using Agilent’s high density (1M) CGH arrays. (a) Representative CGH figure showing copy-number changes on chromosomes 1, 6 and 19, in control CS and RAD51-suppressed RS cells. With reference to the baseline genome, the amplifications (as red dots) and deletions (as green dots) acquired by CS and RS cells, are shown next to each chromosome. (b) For the experiment shown in panel (a), total numbers of copy-change events identified by SNP and CGH are presented as bar graphs. (c) Copy-change events identified by SNP and CGH in another independent experiment are shown. The number of copy-change events in control CS and RAD51-suppressed RS cells identified by SNP is consistent with those detected by CGH (R=0.99). A full colour version of this figure is available at the Oncogene journal online.
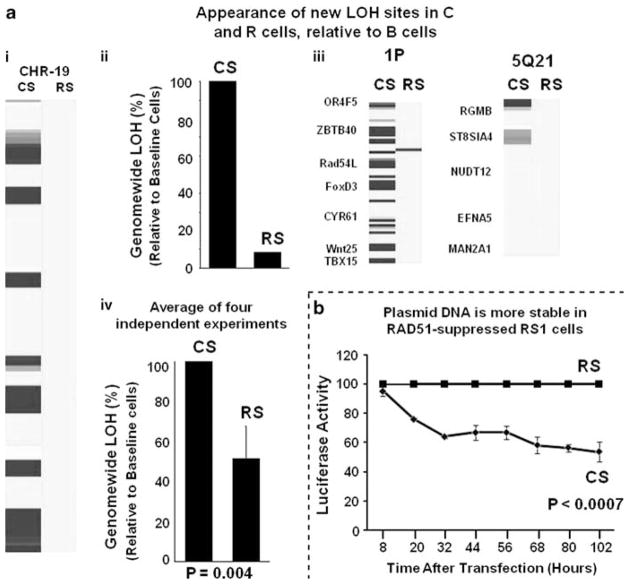
SNP data, analyzed for copy number in Figures 4 and 5, were also evaluated for evidence of LOH. Genotype calls from cells harvested and frozen at the beginning of each experiment (day 0) were used as baselines to identify new LOH loci in control and RAD51-suppressed cells. Figure 6a shows a representative experiment in which FLO-1 cells transduced with control (CS) and RAD51 (RS) shRNAs evaluated for LOH following 33 days of continuous subculture. Suppression of RAD51 in this experiment reduced the total accumulation of LOH by 92% in FLO-1 cells (Figure 6a, panels I and II). Some genomic regions (for example, 1P and 5Q21), previously reported to acquire LOH in esophageal adenocarcinomas (Romagnoli et al., 2001; van Dekken et al., 2001) are shown in Figure 6a, panel III. Whereas control (CS) cells indeed accrued LOH in these regions, RAD51-suppressed cells gained far fewer LOH sites. Combined data from four independent experiments indicate that RAD51 suppression reduces the appearance of new LOH loci by 50±15% in FLO-1 cell genomes (Figure 6a, panel IV). Thus, suppression of RAD51 appears to prevent both copy-number and LOH changes in BAC cells.
Figure 6.
Suppression of RAD51 prevents acquisition of LOH in BAC cells. SNP data analyzed for copy number (presented in Figures 4 and 5) was also evaluated for LOH. Genotype calls from ‘day 0’ baseline cells were used to define the allele type baseline, departures from which identified new LOH loci in cells transduced with control or Rad51-shRNA cells. If LOH was detected in three or more consecutive SNPs, it was called one LOH event. (a) Impact of RAD51 suppression on acquisition of LOH loci in FLO-1 cells. Panel (I): blue bars show LOH loci acquired in chromosome 19 in one of the four experiments, in cells transduced with control CS or RAD51-shRNA RS and cultured for 33 days. Panel (II): for the experiment of panel (a), the total number of LOH events detected throughout the genome, in transduced relative to baseline cells, is presented as a percentage of control–shRNA-treated CS cells. Panel (III): for the same experiment, LOH loci are shown (indicated by blue bars) in the regions of the genome known to acquire LOH in esophageal adenocarcinoma. Panel (IV): bar graph shows genome-wide incidence of LOH events (that is, detectable change in three or more consecutive SNPs), as a percent of control shRNA-treated cells, in four independent experiments; error bars represent s.e.m. (b) Plasmid DNA is more stable in RAD51-suppressed cells. Control RS and RAD51-suppressed RS cells were transfected with a plasmid carrying Gaussia secretory luciferase and plated at equal cell density, in triplicate dishes. Starting 8 h after transfection, the Gluc activity in the supernatants was measured every 12 h. Gradual loss of Gluc activity in control cells is presented as percent of activity in RAD51-suppressed RS cells. A full colour version of this figure is available at the Oncogene journal online.
The suppression of RAD51, under our experimental condition, did not affect the growth of BAC cells in culture (Figure 4, panel IV).
Plasmid DNA is more stably expressed in RAD51-suppressed cells
We next asked whether transfected plasmid DNA, like genomic DNA, is more stable in RAD51-suppressed (RS) cells than in control-treated cells. RS1 cells, transduced with either CS (control) or RS (RAD51-targeted) shRNAs, were then transfected with identical amounts of plasmid carrying the Gluc gene, encoding Gaussia secretory luciferase. Cells were plated at equal density in triplicate dishes, and beginning 8 h later, Gluc activity was measured in the supernatants at 12-h intervals. In Figure 6b, the gradual loss of Gluc activity in control cells is shown as a fraction of activity in RAD51-suppressed RS cells. Although there was no difference in observed Gluc activity between control and RAD51-suppressed cells at 8 h after transfection, indicating similar transfection efficiency and plasmid input, the relative amount of Gluc activity declined thereafter. This indicates that plasmid DNA continues to be expressed at substantially higher levels in RAD51-suppressed cells than in control cells (P<0.0007). Although the Gluc decline in control cells could reflect either plasmid degradation or silencing (for example, by DNA methylation) or both, these results suggest that suppression of RAD51 in BAC cells may stabilize DNA.
Alu frequency of individual chromosomes significantly correlates with mutational frequency
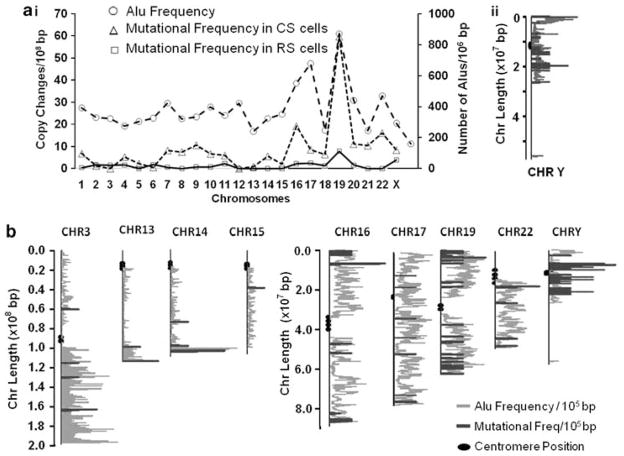
Alu elements in the human genome comprise a family of conserved short (300-bp) interspersed DNA repeat elements (Economou-Pachnis et al., 1985; Brookfield, 1994). As they can potentially serve as substrates for overactive HR machinery in cancer cells, we examined whether the frequency of Alu repeats (Alu elements per 106-bp) correlates with the frequency of copy-number variation across the chromosome complement. CGH arrays were used for this analysis, because (unlike SNP arrays) their sequence samples are evenly distributed along the genome. Data from two independent experiments, used for this analysis, produced similar results. Figure 7a (panel I) summarizes data from one such experiment, illustrating the parallel between Alu density (excluding the Y chromosome, as explained below) and the frequency of copy-number changes (R=0.75; P<0.00001) across the genome of FLO-1 cells transduced with control shRNA ‘CS’. A similar correlation between Alu density and copy-number alterations was also observed in a second, independent experiment (R=0.86; not shown). It is noteworthy that Alu frequency and copy-number changes remained correlated in RAD51-suppressed (‘RS’) cells, in which copy-number variation had been markedly reduced. This suggests that Alu repeat elements may be involved in chromosomal rearrangements even in pre-malignant cells, which have far lower levels of chromosomal instability than high-recombination cancer cells.
Figure 7.
Mutational frequency of chromosomes correlates with frequency of Alu elements. (a) (I) Data from a representative experiment showing that Alu frequency (excluding the Y chromosome) correlates strongly with copy-number variation. In FLO-1 cells treated with the control CS shRNAs, the correlation coefficient R=0.75 (P<0.00001); and for cells treated with Rad51-targeting RS shRNA. (II) Line plot showing the locations of acquired mutations with reference to Alu element on chromosome Y. The ends of each mutation and ALU frequencies were plotted at their corresponding locations, using the GraphPad Prism software. (b) Line plot showing locations of acquired mutations with reference to Alu elements, on various chromosomes, in another independent experiment. A full colour version of this figure is available at the Oncogene journal online.
Of all the human chromosomes, Y has the lowest overall Alu frequency and has an asymmetric distribution in which Alu elements are largely restricted to half of the chromosome. To indicate locations of mutations with reference to Alu elements in this and other chromosomes, the ends of each mutation and ALU frequencies were plotted along the base pair length on each chromosome. As depicted in Figures 7a (panel II) and 7b, combining data from two independent experiments, copy-number changes on the Y chromosome arose only in the Alu-rich area. The other half of this chromosome, lacking Alu elements, acquired no CNCE in either experiment. Similarly, for the majority of other chromosomes, most mutations arose in Alu-dense regions (Figure 7b). These data indicate a strong association of HR-mediated genomic instability in BAC cells, with areas rich in Alu repetitive DNA elements.
Discussion
We have demonstrated that: (1) recombinase (hsRAD51) is overexpressed in BAC and colon cancer cell lines, and in BAC tissue specimens, relative to corresponding normal cells. (2) Expression of recombinase is further induced by exposure of BAC cells to a carcinogen, nickel chloride, which is also recombinogenic; (Li et al., 1997; Shammas et al., 2009). (3) In concordance with RAD51 expression, HR activity is also elevated in BAC and colon cancer cells. (4) RAD51-specific shRNA (RS)-suppressed RAD51 without altering the expression of other genes associated with HR or genomic instability. (5) Suppression of hsRAD51 with RS was commensurate with reduced HR activity. (6) Whereas BAC cells transduced with control shRNA acquired genomic changes, increasing in number with time, the acquisition of such changes was significantly reduced following suppression of RAD51. (7) BAC cells transduced with control or RAD51-specific shRNAs had similar growth rate. (8) Both the high mutational frequency of cells transduced with control shRNA and the substantially lower frequency in cells expressing RAD51 shRNA, were strongly correlated with the Alu frequency in individual chromosome arms.
This is the first report to identify elevated HR activity as a potential target for minimizing genomic variegation in BAC cells, which accompanies (and may facilitate) tumor progression. Genomic instability in BAC is believed to arise early during oncogenesis and to be associated with cancer progression (Spechler and Goyal, 1986; Rabinovitch et al., 1989; Croft et al., 2002; Koppert et al., 2005; Finley et al., 2006; Cai et al., 2008; Akagi et al., 2009; Paulson et al., 2009). However, the mechanisms underlying the accumulation of genomic rearrangements in cancer remain to be identified. Here, we show that the human recombinase, hsRAD51, is overexpressed in BAC cell lines and tissue specimens, accompanied by significant elevation of HR in BAC cells (R=0.84, P=0.009; RS=0.94; P=0.016). HR is essential for DNA repair in cells exposed to DNA-damaging agents. In the normal cellular environment, the HR process is tightly regulated, but induction or dysregulation of HR can have harmful consequences if genomic integrity is compromised. Elevated/dysregulated recombination has been implicated in the generation of large DNA deletions (Cheng et al., 1997; Shammas et al., 1997), amplifications (Windle et al., 1991) and LOH (Bishop and Schiestl, 2003), and may lead to activation of oncogenes, inactivation of antioncogenes, telomere maintenance and ultimately the development and progression of cancer (Sengstag, 1994; Li et al., 1997; Xia et al., 1997; Sturgis et al., 1999; Barlund et al., 2000; Maacke et al., 2000; Bishop and Schiestl, 2003; Bastos et al., 2009; Shammas et al., 2009; Tal et al., 2009; Frank et al., 2010; Jara et al., 2010; Silva et al., 2010). Altered expression or mutation of recombinase (RAD51) and/or related HR genes have been associated with increased risk of several cancers including colorectal (Frank et al., 2010), breast (Jara et al., 2010; Silva et al., 2010) and thyroid (Bastos et al., 2009). Dysregulated expression or mutations of HR genes have been detected in a number of cancers including breast (Barlund et al., 2000; Tal et al., 2009), pancreatic (Maacke et al., 2000), head and neck (Sturgis et al., 1999), and multiple myeloma (Shammas et al., 2009). We have also reported that recombinase (RAD51) and HR activity are significantly elevated in immortal and cancer cell lines (Xia et al., 1997). Consistent with this study, in multiple myeloma we have shown that cancer cells with elevated HR acquire genomic changes over time; the incidence of these changes can be significantly prevented by suppression of HR, or increased by further induction of this activity (Shammas et al., 2009).
In this study, we also noted that the acquisition of copy-number changes on individual chromosomes strongly correlates with the density of Alu elements (Alu number per million base pairs). Alu elements are the most abundant family of repetitive DNA sequences, known collectively as short interspersed repetitive elements (Singer, 1982; Economou-Pachnis and Tsichlis, 1985; Brookfield, 1994). Alu elements comprise ~10% of the human genome, totaling over 106 copies (Abdurashitov et al., 2008). Long interspersed repetitive elements, present at 20 000–40 000 copies per haploid genome, are also widely distributed across the chromosome set. Owing to their greater length (1500–6000 bp), long interspersed repetitive element make up an even larger proportion, over 15%, of the human genome. In normal circumstances and on a very long time-scale, the interspersed repetitive sequences have an important role in the evolution of genes (Hess et al., 1983; Schimenti and Duncan, 1984; Brunner et al., 1986). However, retrotransposal insertion of these elements at vulnerable sites in the genome has also been implicated in the etiology of diverse diseases including cancer (Miki et al., 1992). Changes at repetitive DNA sequences, roughly indicative of microsatellite instability, as well as LOH, have been reported in all stages of BAC including premalignancy and BE (Cai et al., 2008). Our data suggest that Alu repetitive DNA elements, whether because of their size, number or distribution throughout the genome, provide an abundant source of homologous-sequence targets for an overactive HR machinery, and thus may adversely affect the short-term genomic evolution associated with progression from benign neoplasia to invasive, drug resistant and metastatic cancers.
Unlike somatic chromosomes, the Y chromosome does not have a homologue, and it is also unable to recombine with other chromosomes (Steinemann and Steinemann, 2005; Graves, 2006), presumably because of deleterious evolutionary consequences. Although Alu and long interspersed repetitive element frequencies are lowest on the Y chromosome, 97–99% of its repeat elements are confined to a span of just 30 million base pairs, the half of the Y chromosome-containing male-specific euchromatin. Interestingly, all copy-number changes also occurred within this region only. Repeat sequence families are preferred substrates for chromosomal aberrations in cancer, presumably mediated by unequal HR during mitoses (Kolomietz et al., 2002). Interestingly, under our experimental conditions, the Y chromosome consistently demonstrated the highest copy-number instability, despite having the lowest Alu density. Moreover, in both independent experiments, the evaluation by array CGH showed that copy-number changes in RAD51-suppressed cells were substantially reduced in all chromosomes except Y, which was unaffected or only slightly affected by RAD51-suppression. Thus, Y chromosome mutations represent a larger fraction of total mutations observed after RAD51-suppression. It is not clear how the copy-number changes on Y chromosome remain less responsive to HR suppression. However, the data indicate that the high mutational instability at Y chromosome may involve RAD51-independent mechanisms, in addition to Rad51-mediated HR. The genetic instability of the Y chromosome is not entirely unexpected. On an evolutionary timeframe, there is precedent for regarding the Y chromosome as genetically unstable. Although thought to have once contained more than 1000 genes, only about 45 active genes now reside on this chromosome, and it is estimated to lose 4.6 genes per million years (Graves, 2004). The low selection pressure, in the absence of any essential gene for survival, may be one of the reasons for the observed high mutation rate of Y DNA.
In summary, our data convincingly establish a direct correlation between elevated/dysregulated recombinase (RAD51) expression and ongoing genomic instability, studied in BAC cells in vitro as a model system for cancer progression. Significant stabilization of mutations on suppression of upregulated RAD51 convincingly shows the importance of this gene as a therapeutic target in cancer. Thus, inhibitors of RAD51 or other HR components/complexes have the potential to block or slow progression from benign to malignant disease and in particular to delay the acquisition of drug resistance in a wide variety of cancers. To date, no specific hsRAD51 inhibitors have been validated and approved for therapeutic use, although our work strongly implies that drug screening and identification of RAD51 inhibitors would be a valuable addition to our cancer-treatment armorarium. Additionally, our data suggest that repetitive DNA elements in the genome, in particular Alu elements, may contribute to the well-known genomic instability of cancer cells.
Materials and methods
Tissue specimens and cell lines
Specimens of BE and adenocarcinoma (from the Tissue Core Facility at Karmanos Cancer Institute) were used under a protocol approved by the IRB of Wayne State University, Detroit, MI, USA. The BAC cell line FLO-I was described previously (Aggarwal et al., 2000). The BAC cell line OE33, from the European Collection of Cell Cultures (purchased from Sigma-Aldrich, St Louis, MO, USA) has been described previously (Tselepis et al., 2003; Ogunwobi and Beales, 2008). Normal primary HEEC (ScienCell Research Laboratories, Carlsbad, CA, USA) were described previously (Yoshida et al., 2007; Shammas et al., 2008). Normal human diploid fibroblast strain GM01662 (from the Genetic Mutant Cell Repository funded by the National Institute for General Medical Sciences and maintained by the Coriell Institute for Medical Research, Camden, NJ, USA) have been described previously (Shammas et al., 2003). Normal fibroblasts and FLO-1 cells (Shammas et al., 1997, 1999) were cultured in Dulbecco’s modified Eagle medium (Sigma Chemical, St Louis, MO, USA) supplemented with 10% fetal bovine serum (HyClone, South Logan, UT, USA). OE33 cells were cultured in RPMI-1640 supplemented with 2mM L-glutamine and 10% fetal bovine serum. Normal HEEC cells were cultured in epithelial cell medium-2 (ScienCell Research Laboratories). Colon cancer cell lines SW480, SW620, HCT116 were obtained from the American Type Culture Collection (Manassas, VA, USA) and normal human colonic smooth muscle cells from ScienCell Research Laboratories. Cells were maintained in a state of logarithmic growth at 37 °C in humidified air with 5% CO2. For RNA and protein analyses, cultures were harvested at the same final cell density (5×105/ml), and immediately processed.
Recombination assay
We have previously used a plasmid substrate ‘DR1’ to assess HR frequency in human cells (Xia et al., 1997; Shammas et al., 2009). For this study, we used an improved HR substrate plasmid containing two incomplete but overlapping fragments of firefly luciferase (Fluc), separated by an AmpR gene. HR between identical sequences of two fragments generates a functional Fluc gene, resulting in removal of the AmpR gene. Gaussia luciferase (Gluc) serves as internal control and remains unaffected by HR. The plasmid is introduced into test cells, the cells are harvested after appropriate incubation, and HR is assessed by the ratio of firefly and Gaussia luciferase activities. Assessment of HR in various cell lines with this new HRreporter plasmid is consistent with our previously used plasmid substrate (R=0.993).
Immunohistochemical and western blot analyses
RAD51 mouse monoclonal and horseradish peroxidase- or Alexa-Fluor 488-labeled secondary antibodies were purchased and provided to the Tissue Core Facility of the Karmanos Cancer Institute. The tissue core facility used these antibodies to evaluate the expression of RAD51 in the specimens of human esophagi, under approved Institutional Review Board protocol. Briefly, the sections of paraffin-embedded BE and BAC tissues were deparaffinized, blocked, and sequentially treated with anti-RAD51 mouse monoclonal and horseradish peroxidase- or Alexa-Fluor 488-labeled secondary antibodies, and viewed under a fluorescence microscope (Nikon, Melville, NY, USA). For western blot analyses, extracts equivalent of 50 μg protein were suspended in Laemmli’s sample buffer (0.1M Tris–HCl buffer pH 6.8, 1% sodium dodecyl sulfate, 0.05% β-mercaptoethanol, 10% glycerol and 0.001% bromophenol blue), boiled for 2 min, and electrophoresed on 4–20% glycerol gradient sodium dodecyl sulfate–polyacrylamide gel for 4 h at 120 V. Gels were electroblotted onto Trans-Blot nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA, USA) at 40V for 3 h in a Tris-glycine buffer system.
Membranes were incubated with the indicated antibodies, with constant rocking for 2 h at room temperature in phosphate-buffered saline-Tween 20 containing 1% bovine serum albumin. Blots were washed with phosphate-buffered saline-Tween 20 and incubated in either anti-rabbit or antimouse antibody conjugated to horseradish peroxidase for 2 h in phosphate-buffered saline-Tween 20 containing 3% nonfat dry milk. After washing, specific proteins were detected using an enhanced chemiluminescence detection kit, according to the instructions provided by the manufacturer (Amersham Life Sciences Inc., Arlington Heights, IL, USA).
Lentiviruses and transductions
Lentivirus particles producing either non-targeted control (C) shRNAs or those targeting human RAD51 (R) were purchased from Sigma Chemical Co., St Louis, MO, USA. Cells (2×105 per well) were seeded into 24-well plates one day before transduction. On the day of transduction, medium was replaced with fresh medium containing hexadimethrine bromide at a final concentration of 8 μg/ml. Lentiviral particles (25 μl) were added to each well, mixed and incubated at 37 °C in a humidified incubator with 5% CO2. After 16 h, the virus-containing medium was replaced with fresh medium and cells maintained in culture. On the next day, cells were trypsinized and transferred to 25 cm2 flasks. After another 48 h, puromycin was added to the medium at a final concentration of 1 μg/ml. Suppression of HsRAD51 protein expression was confirmed by western blotting after 7 days of puromycin selection.
Gene expression analysis and biostatistics
Total RNA was isolated utilizing an ‘RNeasy’ kit (Qiagen Inc., Valencia, CA, USA) and the gene-expression profile was evaluated using Gene 1.0 ST Arrays (Affymetrix), as described previously (Munshi et al., 2004, 2004; Shammas et al., 2005, 2004, 2006). GeneChip arrays were scanned on a GeneArray Scanner (Affymetrix, Inc.), and normalized expression levels were calculated by the dChip Analyzer using the ‘Invariant Set’ normalization method at probe level to make arrays comparable, and the model-based method for probe selection and to compute adjusted expression values (Li and Hong, 2001; Li and Wong, 2001).
Evaluation of genomic evolution using SNP and CGH arrays
To evaluate the roles of RAD51 and HR in the progressive accrual (‘cellular evolution’) of genomic changes, BAC cells were transduced with non-targeted control (C) or RAD51-specific (R) shRNAs. For each experiment, an aliquot of cells was harvested and frozen at the beginning of the experiment (day 0), to be used as a reference. Transduced cells were cultured for various durations and the acquisition of changes, relative to reference cells, was monitored by use of genome-wide microarray screens for LOH and copy-number changes, based on SNP arrays (Affymetrix) and CGH arrays (Agilent Technologies), respectively.
For SNP analyses, genomic DNA was extracted using DNeasy Tissue Kit (Qiagen) as per the manufacturer’s instructions, and analyzed for copy-number and heterozygosity changes using 500k SNP arrays (Affymetrix, Inc.). Briefly, two aliquots of DNA (250 ng each) were digested with NspI and StyI restriction enzymes (New England Biolabs, Boston, MA, USA), ligated to an oligonucleotides adaptor, and amplified by PCR using Titanium Taq DNA Polymerase (Clontech, Mountain View, CA, USA). Three 100-μl PCR reactions were then set up for each StyI or NspI adaptor-ligated DNA sample. The PCR products from three reactions were pooled, concentrated, and fragmented with DNase I to a 25–200 bp size range. Fragmented PCR products were then labeled, denatured and hybridized to the array. After hybridization, the arrays were washed on an Affymetrix fluidics workstation and scanned using the Gene Chip Scanner 3000 and genotyping software, GTYPE 4.0. DChip software (http://www.dchip.org) was used for data analysis and visualization (Li and Hung Wong, 2001). Arrays were normalized to a baseline array set for moderate median probe intensity and 90% call rate. Genotype calls from cell samples harvested and frozen at the beginning of experiment (day 0) were used as baselines to identify new LOH loci in the transduced cells. For copy-number analyses, first the raw copy numbers were derived by trimmed mean analysis of signal values of each SNP in all the samples, and then inferred copy number at each SNP locus was derived by median smoothing of raw copy numbers in the neighboring five-marker window. The inferred signal values of treated samples were compared with the inferred signal values of the baseline reference samples (day 0) to estimate copy change events, defined as change in copy number in three or more consecutive SNPs, using Matlab (Natick, MA, USA). Copy-number changes were confirmed by array CGH as described below. Heterozygosity and copy-number changes throughout genome were represented as events, where an event was defined as a change in three or more consecutive SNPs.
For CGH array processing, 1 μg of each genomic DNA sample was digested with AluI and RsaI restriction endonucleases. Digested DNA was fluorescently labeled using a Genomic DNA Enzymatic Labeling Kit (Agilent). Unincorporated dye molecules were removed by desalting in TE buffer (pH8.0) using Microcon YM-30 filters (Millipore, Billerica, MA, USA). Sample concentrations (ng/ul) and dye intensities (pmol dye/μg of labeled DNA) were determined using a Nanodrop-1000 (Thermo Fisher, Waltham, MA, USA) UV-VIS spectrophotometer. Equal amounts of Cy3- and Cy5-labeled material were combined and hybridized to human CGH (1×1M) microarrays (Agilent Technologies), in accordance with the Agilent CGH protocol (v. 6.0). The array assemblies were contained within Agilent Sure-Hyb hybridization chambers during a 40-h incubation at 65 °C. At the end of hybridization, the arrays were washed using Agilent a-CGH wash buffers and scanned using Agilent DNA Microarray C-Scanner with Surescan High-Resolution Technology at the recommended settings. Agilent Feature Extraction software v. 10.7.1.1 was used to extract features, and data analysis was performed by Agilent CGH analytics. An aberration filter was created to include only those regions that contain three or more consecutive features.
Evaluating plasmid DNA stability in RAD51-suppressed cells
Control (C) and RAD51-suppressed (R) cells were transfected with a plasmid carrying the Gaussia secretory luciferase gene, Gluc, and plated at equal cell density, in triplicate dishes. Gluc activity in the supernatants was measured every 12 h, beginning 8 h after transfection. For each Gluc assay, medium was removed and replaced with fresh medium. Gradual loss of Gluc activity in control cells was represented as percent of activity in RAD51-suppressed (R) cells.
Evaluating correlation of HR with ALU frequency in the genome
FLO-1 cells transduced with non-targeting (C) or RAD51-specific (R) shRNAs, from two independent experiments, were cultured and copy-number changes were evaluated using CGH arrays and ‘day 0’ cells as the baseline control, as described above. Mutation frequency, defined as copy-number changes/108-bp, was calculated for each chromosome. The number of Alu elements in each chromosome was calculated based on the human genome database (Genome/Assembly: Human—March 2006—hg18) and Repeatmasker, version 3.2.7 (http://www.repeatmasker.org/cgi-bin/AnnotationRequest). The Alu frequency was expressed as Alu number/106-bp of a chromosome. To indicate locations of mutations with reference to Alu elements, point mutations or the ends of each deletion or insertion, and Alu positions, were plotted at their corresponding locations on each chromosome, using GraphPad Prism software (La Jolla, CA, USA), with centromeric position localized using the UCSC Genome Browser.
Acknowledgments
We are grateful to Dr Cheng Li, Department of Bioinformatics and Dr Samir Amin, Departments of Medical Oncology and Bioinformatics, Dana Farber Cancer Institute, Boston, MA, USA, for their critical review of paper and guidance in data analyses. This work was supported in part by grants from National Cancer Institute (R01CA125711 to MAS), from the Department of Veterans Affairs (Merit Review Awards to NCM and RJSR and a Research Career Scientist Award to RJSR) and from the National Institutes of Health (RO1-1375555, P50-100007 and PO1-78378 to NCM).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Abdurashitov MA, Tomilov VN, Chernukhin VA, Degtyarev S. A physical map of human Alu repeats cleavage by restriction endonucleases. BMC Genomics. 2008;9:305. doi: 10.1186/1471-2164-9-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal S, Taneja N, Lin L, Orringer MB, Rehemtulla A, Beer DG. Indomethacin induced apoptosis in esophageal adenocarcinoma cells involves upregulation of Bax and translocation of mitochondrial cytochrome C independent of COX-2 expression. Neoplasia. 2000;2:346–356. doi: 10.1038/sj.neo.7900097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Ito T, Kato M, Jin Z, Cheng Y, Kan T, et al. Chromosomal abnormalities and novel disease-related regions in progression from Barrett’s esophagus to esophageal adenocarcinoma. Int J Cancer. 2009;125:2349–2359. doi: 10.1002/ijc.24620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Carpenter AT, Esposito MS, Esposito RE, Sandler L. The genetic control of meiosis. Annu Rev Genet. 1976;10:53–134. doi: 10.1146/annurev.ge.10.120176.000413. [DOI] [PubMed] [Google Scholar]
- Barlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, et al. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 2000;60:5340–5344. [PubMed] [Google Scholar]
- Bastos HN, Antao MR, Silva SN, Azevedo AP, Manita I, Teixeira V, et al. Association of polymorphisms in genes of the homologous recombination DNA repair pathway and thyroid cancer risk. Thyroid. 2009;19:1067–1075. doi: 10.1089/thy.2009.0099. [DOI] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AJ, Schiestl RH. Role of homologous recombination in carcinogenesis. Exp Mol Pathol. 2003;74:94–105. doi: 10.1016/s0014-4800(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Brookfield JF. The human Alu SINE sequences–is there a role for selection in their evolution? Bioessays. 1994;16:793–795. doi: 10.1002/bies.950161104. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Schimenti JC, Duncan CH. Dual evolutionary modes in the bovine globin locus. Biochemistry. 1986;25:5028–5035. doi: 10.1021/bi00366a009. [DOI] [PubMed] [Google Scholar]
- Cai JC, Liu D, Liu KH, Zhang HP, Zhong S, Xia NS. Microsatellite alterations in phenotypically normal esophageal squamous epithelium and metaplasia-dysplasia-adenocarcinoma sequence. World J Gastroenterol. 2008;14:4070–4076. doi: 10.3748/wjg.14.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RZ, Shammas MA, Li J, Shmookler Reis RJ. Expression of SV40 large T antigen stimulates reversion of a chromosomal gene duplication in human cells. Exp Cell Res. 1997;234:300–312. doi: 10.1006/excr.1997.3649. [DOI] [PubMed] [Google Scholar]
- Croft J, Parry EM, Jenkins GJ, Doak SH, Baxter JN, Griffiths AP, et al. Analysis of the premalignant stages of Barrett’s oesophagus through to adenocarcinoma by comparative genomic hybridization. Eur J Gastroenterol Hepatol. 2002;14:1179–1186. doi: 10.1097/00042737-200211000-00004. [DOI] [PubMed] [Google Scholar]
- Economou-Pachnis A, Tsichlis PN. Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Res. 1985;13:8379–8387. doi: 10.1093/nar/13.23.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou-Pachnis A, Lohse MA, Furano AV, Tsichlis PN. Insertion of long interspersed repeated elements at the Igh (immunoglobulin heavy chain) and Mlvi-2 (Moloney leukemia virus integration 2) loci of rats. Proc Natl Acad Sci USA. 1985;82:2857–2861. doi: 10.1073/pnas.82.9.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber E. The multistep nature of cancer development. Cancer Res. 1984;44:4217–4223. [PubMed] [Google Scholar]
- Finley JC, Reid BJ, Odze RD, Sanchez CA, Galipeau P, Li X, et al. Chromosomal instability in Barrett’s esophagus is related to telomere shortening. Cancer Epidemiol Biomarkers Prev. 2006;15:1451–1457. doi: 10.1158/1055-9965.EPI-05-0837. [DOI] [PubMed] [Google Scholar]
- Frank B, Hoffmeister M, Klopp N, Illig T, Chang-Claude J, Brenner H. Colorectal cancer and polymorphisms in DNA repair genes WRN, RMI1 and BLM. Carcinogenesis. 2010;31:442–445. doi: 10.1093/carcin/bgp293. [DOI] [PubMed] [Google Scholar]
- Galli A, Schiestl RH. On the mechanism of UV and gamma-ray-induced intrachromosomal recombination in yeast cells synchronized in different stages of the cell cycle. Mol Gen Genet. 1995;248:301310. doi: 10.1007/BF02191597. [DOI] [PubMed] [Google Scholar]
- Gee CJ, Harris H. Tumorigenicity of cells transformed by Simian virus 40 and of hybrids between such cells and normal diploid cells. J Cell Sci. 1979;36:223–240. doi: 10.1242/jcs.36.1.223. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The degenerate Y chromosome-can conversion save it? Reprod Fertility Dev. 2004;16:527–534. doi: 10.10371/RD03096. [DOI] [PubMed] [Google Scholar]
- Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Hansen LT, Lundin C, Spang-Thomsen M, Petersen LN, Helleday T. The role of RAD51 in etoposide (VP16) resistance in small cell lung cancer. Int J Cancer. 2003;105:472–479. doi: 10.1002/ijc.11106. [DOI] [PubMed] [Google Scholar]
- Hess JF, Fox M, Schmid C, Shen CK. Molecular evolution of the human adult alpha-globin-like gene region: insertion and deletion of Alu family repeats and non-Alu DNA sequences. Proc Natl Acad Sci USA. 1983;80:5970–5974. doi: 10.1073/pnas.80.19.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M, Zhang LS, Hayashi M, Takeshita K, Nakagawa Y, Tanaka N, et al. Illegitimate recombination leading to allelic loss and unbalanced translocation in p53-mutated human lymphoblastoid cells. Mol Cell Biol. 1997;17:4774–4781. doi: 10.1128/mcb.17.8.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii A, Nakatsuru S, Miyoshi Y, Ichii S, Nagase H, Kato Y, et al. The APC gene, responsible for familial adenomatous polyposis, is mutated in human gastric cancer. Cancer Res. 1992;52:3231–3233. [PubMed] [Google Scholar]
- Jara L, Dubois K, Gaete D, de Mayo T, Ratkevicius N, Bravo T, et al. Variants in DNA double-strand break repair genes and risk of familial breast cancer in a South American population. Breast Cancer Res Treat. 2010;122:813–822. doi: 10.1007/s10549-009-0709-2. [DOI] [PubMed] [Google Scholar]
- Jones S, Chen WDm, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- Koppert LB, Wijnhoven BP, van Dekken H, Tilanus HW, Dinjens WN. The molecular biology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:169–190. doi: 10.1002/jso.20359. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hong F. Cluster-Rasch models for microarray gene expression data. Genome Biol. 2001;2(8):Research0031.1–0031.13. doi: 10.1186/gb-2001-2-8-research0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ayyadevera R, Shmookler Reis RJ. Carcinogens stimulate intrachromosomal homologous recombination at an endogenous locus in human diploid fibroblasts. Mutat Res. 1997;385:173–193. doi: 10.1016/s0921-8777(97)00054-2. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Lynch JF. Genetics of colonic cancer. Digestion. 1998;59:481–492. doi: 10.1159/000007525. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Smyrk T, Lynch JF. Molecular genetics and clinical-pathology features of hereditary nonpolyposis colorectal carcinoma (Lynch syndrome): historical journey from pedigree anecdote to molecular genetic confirmation. Oncology. 1998;55:103–108. doi: 10.1159/000011843. [DOI] [PubMed] [Google Scholar]
- Maacke H, Jost K, Opitz S, Miska S, Yuan Y, Hasselbach L, et al. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- Maciejewski JP, Mufti GJ. Whole genome scanning as a cytogenetic tool in hematologic malignancies. Blood. 2008;112:965–974. doi: 10.1182/blood-2008-02-130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Ohmori M, Perantoni AO, Enomoto T. K-ras activation in gastric epithelial tumors in Japanese. Cancer Lett. 1991;58:107–113. doi: 10.1016/0304-3835(91)90031-c. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Munshi NC, Hideshima T, Carrasco D, Shammas M, Auclair D, Davies F, et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2004;103:1799–1806. doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]
- Munshi NC, Hideshima T, Carrasco D, Shammas M, Auclair D, Davies F, et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2004;103:1799–1806. doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]
- Neiman PE, Hartwell LH. Malignant instability. New Biol; Workshop on genetic instability and its role in carcinogenesis sponsored by the Programs in Molecular Medicine of the Fred Hutchinson Cancer Research Center and the University of Washington; Seattle, WA, USA. January 11–12, 1991; 1991. pp. 347–351. [PubMed] [Google Scholar]
- Ogunwobi OO, Beales IL. Statins inhibit proliferation and induce apoptosis in Barrett’s esophageal adenocarcinoma cells. Am J Gastroenterol. 2008;103:825–837. doi: 10.1111/j.1572-0241.2007.01773.x. [DOI] [PubMed] [Google Scholar]
- Pathak S, Dave BJ, Gagos S. Chromosome alterations in cancer development and apoptosis. In vivo. 1994;8:843–850. [PubMed] [Google Scholar]
- Paulson TG, Maley CC, Li X, Li H, Sanchez CA, Chao DL, et al. Chromosomal instability and copy number alterations in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:3305–3314. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch PS, Reid BJ, Haggitt RC, Norwood TH, Rubin CE. Progression to cancer in Barrett’s esophagus is associated with genomic instability. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- Radman M, Jeggo P, Wagner R. Chromosomal rearrangement and carcinogenesis. Mutat Res. 1982;98:249–264. doi: 10.1016/0165-1110(82)90035-5. [DOI] [PubMed] [Google Scholar]
- Resnick MA, Skaanild M, Nilsson-Tillgren T. Lack of DNA homology in a pair of divergent chromosomes greatly sensitizes them to loss by DNA damage. Proc Natl Acad Sci USA. 1989;86:2276–2280. doi: 10.1073/pnas.86.7.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodarte-Ramon US. Radiation-induced recombination in Saccharomyces: the genetic control of recombination in mitosis and meiosis. Radiat Res. 1972;49:148–154. [PubMed] [Google Scholar]
- Romagnoli S, Roncalli M, Graziani D, Cassani B, Roz E, Bonavina L, et al. Molecular alterations of Barrett’s esophagus on microdissected endoscopic biopsies. Lab Invest. 2001;81:241–247. doi: 10.1038/labinvest.3780232. [DOI] [PubMed] [Google Scholar]
- Schimenti JC, Duncan CH. Ruminant globin gene structures suggest an evolutionary role for Alu-type repeats. Nucleic Acids Res. 1984;12:1641–1655. doi: 10.1093/nar/12.3.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C. The role of mitotic recombination in carcinogenesis. Crit Rev Toxicol. 1994;24:323–353. doi: 10.3109/10408449409017922. [DOI] [PubMed] [Google Scholar]
- Shammas MA, Neri P, Koley H, Batchu RB, Bertheau RC, Munshi V, et al. Specific killing of multiple myeloma cells by (−)-epigallocatechin-3gallate extracted from green tea: biologic activity and therapeutic implications. Blood. 2006;108:2804–2810. doi: 10.1182/blood-2006-05-022814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA, Xia SJ, Shmookler Reis RJ. Induction of duplication reversion in human fibroblasts, by wild-type and mutated SV40T antigen, covaries with the ability to induce host DNA synthesis. Genetics. 1997;146:1417–1428. doi: 10.1093/genetics/146.4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA, Simmons CG, Corey DR, Reis RJ. Telomerase inhibition by peptide nucleic acids reverses ‘immortality’ of transformed human cells. Oncogene. 1999;18:6191–6200. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
- Shammas MA, Shmookler Reis RJ, Akiyama M, Koley H, Chauhan D, Hideshima T, et al. Telomerase inhibition and cell growth arrest by Gquadruplex interactive agent in multiple myeloma. Mol Cancer Therapeut. 2003;2:825–833. [PubMed] [Google Scholar]
- Shammas MA, Koley H, Beer DG, Li C, Goyal RK, Munshi NC. Growth arrest, apoptosis, and telomere shortening of Barrett’s-associated adenocarcinoma cells by a telomerase inhibitor. Gastroenterology. 2004;126:1337–1346. doi: 10.1053/j.gastro.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Shammas MA, Koley H, Batchu RB, Bertheau RC, Protopopov A, Munshi NC, et al. Telomerase inhibition by siRNA causes senescence and apoptosis in Barrett’s adenocarcinoma cells: mechanism and therapeutic potential. Mol Cancer. 2005;4:24. doi: 10.1186/1476-4598-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA, Qazi A, Batchu RB, Bertheau RC, Wong JY, Rao MY, et al. Telomere maintenance in laser capture microdissection purified Barrett’s adenocarcinoma cells and effect of telomerase inhibition in vivo. Clin Cancer Res. 2008;14:4971–4980. doi: 10.1158/1078-0432.CCR-08-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA, Shmookler Reis RJ, Koley H, Batchu RB, Li C, Munshi NC. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009;113:2290–2297. doi: 10.1182/blood-2007-05-089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SN, Tomar M, Paulo C, Gomes BC, Azevedo AP, Teixeira V, et al. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol. 2010;34:85–92. doi: 10.1016/j.canep.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Singer MF. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982;28:433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Spechler SJ, Goyal RK. Barrett’s esophagus. N Engl J Med. 1986;315:362–371. doi: 10.1056/NEJM198608073150605. [DOI] [PubMed] [Google Scholar]
- Steinemann S, Steinemann M. Y chromosomes: born to be destroyed. Bioessays. 2005;27:10761083. doi: 10.1002/bies.20288. [DOI] [PubMed] [Google Scholar]
- Stilgenbauer S, Sander S, Bullinger L, Benner A, Leupolt E, Winkler D, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–1245. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- Stoler DL, Chen N, Basik M, Kahlenberg MS, Rodriguez-Bigas MA, Petrelli NJ, et al. The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc Natl Acad Sci USA. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgis EM, Clayman GL, Guan Y, Guo Z, Wei Q. DNA repair in lymphoblastoid cell lines from patients with head and neck cancer. Arch Otolaryngol Head Neck Surg. 1999;125:185–190. doi: 10.1001/archotol.125.2.185. [DOI] [PubMed] [Google Scholar]
- Tal A, Arbel-Goren R, Stavans J. Cancer-associated mutations in BRC domains of BRCA2 affect homologous recombination induced by Rad51. J Mol Biol. 2009;393:1007–1012. doi: 10.1016/j.jmb.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Tsancheva M. The molecular biology and genetics of colorectal carcinoma. Khirurgiia (Sofiia) 1997;50:40–44. [PubMed] [Google Scholar]
- Tselepis C, Morris CD, Wakelin D, Hardy R, Perry I, Luong QT, et al. Upregulation of the oncogene c-myc in Barrett’s adenocarcinoma: induction of c-myc by acidified bile acid in vitro. Gut. 2003;52:174–180. doi: 10.1136/gut.52.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dekken H, Alers JC, Riegman PH, Rosenberg C, Tilanus HW, Vissers K. Molecular cytogenetic evaluation of gastric cardia adenocarcinoma and precursor lesions. Am J Pathol. 2001;158:1961–1967. doi: 10.1016/S0002-9440(10)64666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle B, Draper BW, Yin YX, O’Gorman S, Wahl GM. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- Xia SJ, Shammas MA, Shmookler Reis RJ. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol Cell Biol. 1997;17:7151–7158. doi: 10.1128/mcb.17.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Yoshida T, Hayashi K, Sekiya T, Yokota J, Hirohashi S, et al. p53 gene mutations in gastric cancer metastases and in gastric cancer cell lines derived from metastases. Cancer Res. 1991;51:5800–5805. [PubMed] [Google Scholar]
- Ye CJ, Liu G, Bremer SW, Heng HH. The dynamics of cancer chromosomes and genomes. Cytogenet Genome Res. 2007;118:237–246. doi: 10.1159/000108306. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Katada K, Handa O, Takagi T, Kokura S, Naito Y, et al. Interleukin-8 production via protease-activated receptor 2 in human esophageal epithelial cells. Int J Mol Med. 2007;19:335–340. doi: 10.3892/ijmm.19.2.335. [DOI] [PubMed] [Google Scholar]