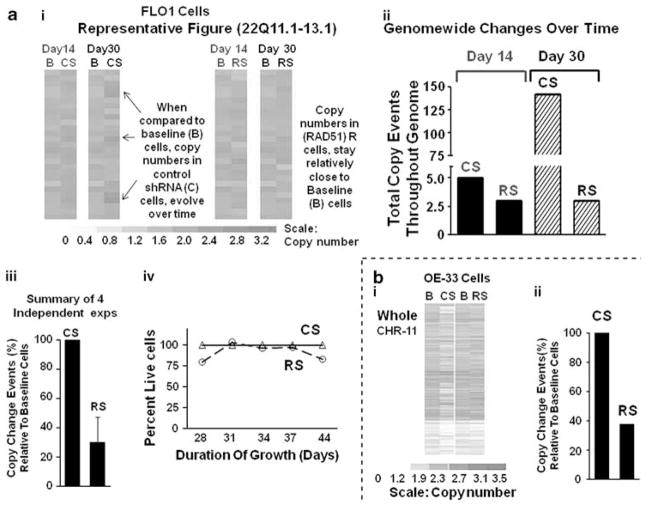
Figure 4.
Suppression of RAD51 prevents the acquisition of copy-number changes in BEAC cells. BEAC (FLO-1 and OE33) cells were transduced with lentivirus particles producing either control CS or RAD51-specific RS shRNAs. For each cell line, just before transduction, an aliquot of cells was harvested and stored at −150 °C to be used as baseline reference ‘B’. Transduced cells were allowed to recover, selected in puromycin for 3 days, and continued in cultured. Cells were harvested at various intervals, genomic DNA from these and baseline reference cells isolated, and evaluated for copy-number changes using 500K or 6.0 SNP arrays (Affymetrix) and software dChip (Kolomietz et al., 2002; Graves, 2004; Steinemann and Steinemann, 2005), as described in Materials and methods section. SNP hybridization intensities of baseline (day 0) cells were used to define the copy-number baseline, departures from which identified changes in transduced cells. A mutational event was then defined as a detectable change in copy number, in three or more consecutive SNPs. (a) Impact of RAD51 suppression on acquisition of copy-number changes in FLO-1 cells: panel (I) figure showing relative copy numbers in a region of chromosome 22, in baseline ‘B’ cells and those which were transduced with control CS or RAD51-specific RS shRNAs and cultured and harvested at two different time points, that is, day 14 or day 30. Color scale at the bottom shows relative copy numbers. Panel (II): for the experiment in panel (a), the number of copy-change events (that is, detectable change in three or more consecutive SNPs) detected throughout the genome, in transduced relative to baseline cells, is presented. Panel (III): bar graph shows prevention of copy-number changes in RAD51-suppressed RS cells, in four independent experiments; error bars represent s.e.m. CNCEs in transduced (CS and RS cells) relative to baseline cells are presented as percent of control shRNA-transduced CS cells. Panel (IV): line graph showing growth rates of control and RAD51-suppressed cells. (b) Impact of RAD51 suppression on acquisition of copy-number changes in OE33 cells: panel (I): OE33 cells were transduced with CS or RS shRNAs, cultured for 25 days and genomic changes evaluated. Relative copy numbers in chromosome 11 are shown as example. The color scale at the bottom shows relative copy numbers. Panel (II): for the experiment shown in panel (I), copy change events (that is, detectable change in three or more consecutive SNPs) detected throughout the genome, in treated relative to baseline cells, are presented as percent of control shRNA-treated CS cells. A full colour version of this figure is available at the Oncogene journal online.