Summary
Non-aggregative multicellularity requires strict control of cell number. The Hippo signaling pathway coordinates cell proliferation and apoptosis and is a central regulator of organ size in animals. Recent studies have shown the presence of key members of the Hippo pathway in non-bilaterian animals, but failed to identify this pathway outside Metazoa. Through comparative analyses of recently sequenced holozoan genomes, we show that Hippo pathway components, such as the kinases Hippo and Warts, the co-activator Yorkie and the transcription factor Scalloped, were already present in the unicellular ancestors of animals. Remarkably, functional analysis of Hippo components of the amoeboid holozoan Capsaspora owczarzaki, performed in Drosophila, demonstrate that the growth-regulatory activity of the Hippo pathway is conserved in this unicellular lineage. Our findings show that the Hippo pathway evolved well before the origin of Metazoa and highlight the importance of Hippo signaling as a key developmental mechanism pre-dating the origin of Metazoa.
Introduction
The emergence of multicellularity represents one of the most important transitions in animal evolution. Non-aggregative multicellularity requires strict control over cell differentiation, proliferation and survival and this is attained by sophisticated cell-cell communication systems. A startling revelation from decades of developmental genetic studies is that these cell communications are largely mediated by just a handful of signaling pathways in metazoans such as Notch, Wnt, TGF-β, Hedghog, and receptor tyrosine kinase (RTK) (Pires-daSilva and Sommer, 2003). A longstanding question in evolutionary developmental biology concerns the genetic mechanisms underlying the transition from unicellular eukaryotes to metazoa, in particular the relationship between multicellularity and the evolutionary origin of the major metazoan signaling pathways (King et al., 2003; Adamska et al., 2007; King et al., 2008; Srivastava et al., 2010; Sebe-Pedros et al., 2010). Comparative genomic analyses have so far confirmed the absence of Notch, Wnt, TGF-β, and Hedghog signaling in any unicellular organisms, supporting the view that these signaling pathways are metazoan synapomophies (Pires-daSilva and Sommer, 2003). In contrast, the discovery of RTKs in the closest unicellular relatives of animals (choanoflagellates) suggests that RTKs may serve as preadaptations in the metazoans’ unicellular ancestors for co-option into the multicellular lifestyle (King et al., 2003; Manning et al., 2008).
The most recent addition to the metazoan signaling “toolkit” is the Hippo signaling pathway. The Hippo pathway was first discovered in Drosophila as a critical regulator of imaginal disc growth, and more recent studies have implicated a conserved function of this pathway in organ size control in mammals (Zhao et al., 2010; Pan, 2010; Halder and Johnson, 2011; Zeng and Hong, 2008; Harvey and Tapon, 2007; Reddy and Irvine, 2008; Badouel et al., 2009). The core of the Hippo pathway is a functionally conserved kinase cascade leading from the Ste20-like kinase Hippo (Hpo) (Mst1/2 in mammals) and the NDR family kinase Warts (Wts) (Lats1/2 in mammals) to the transcription factor complex formed by the coactivator Yorkie (Yki) (YAP/TAZ in mammals) and its major DNA-binding partner Scalloped (Sd) (TEAD1/2/3/4 in mammals) (Figure 1A). The Sd-Yki transcription factor complex, in turn, regulates an array of target genes involved in cell proliferation and cell survival, such as the cell death inhibitor diap1. Diverse upstream inputs into the core kinase cascade have been identified in Drosophila. These include an apical protein complex composed of the WW- and C2-domain-containing protein Kibra, and two FERM-domain containing proteins Expanded (Ex) and Merlin (Mer); the Fat signaling module composed of the atypical cadherins Fat and its effectors such as Four-jointed and Dachs; the apical-basal polarity regulators Crumbs (Crb), atypical Protein Kinase C (aPKC) and the WD40 scaffold protein Lethal giant larvae (Lgl) (Figure 1A). With the exception of Kibra and Mer, these upstream inputs have not been functionally linked to Hippo signaling in mammals (Zhao et al., 2010; Pan, 2010; Halder and Johnson, 2011; Zeng and Hong, 2008; Harvey and Tapon, 2007; Reddy and Irvine, 2008; Badouel et al., 2009).
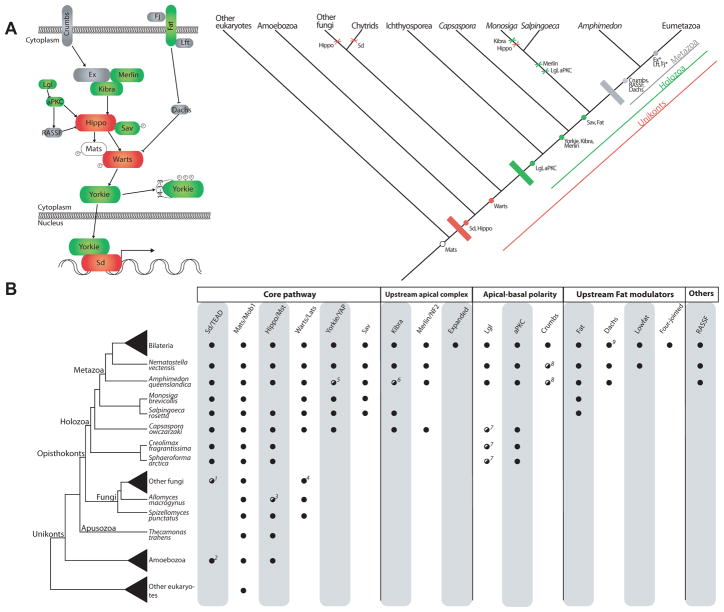
Figure 1. Evolution of the Hippo signaling pathway.
(A) Schematic representation of the Hippo pathway evolution. The canonical metazoan Hippo pathway is shown on the left. The colours correspond to the three main steps in the evolution of the pathway, as shown in the cladogram (white=eukaryotes, red=unikonts, green=Holozoa, grey=Metazoa). Dots indicate origin and crosses indicate losses. Asterisks in Expanded (Ex) and Four-jointed (Fj) indicate these genes are exclusive to Bilateria.
(B) Schematic representation of the eukaryotic tree of life showing the distribution of the different components of the Hippo pathway. A black dot indicates the presence of clear homologs, while a striped white-black dot indicates the presence of putative or degenerate homologs. Absence of a dot indicates that a homolog is lacking in that taxon. The taxon sampling for Bilateria includes Homo sapiens, Drosophila melanogaster, Daphnia pulex and Capitella teleta; other fungi includes the Ascomytoca Neurospora crassa and the Basidiomycota Ustilago maydis; Amoebozoa includes Acanthamoeba castellanii and Dictyostelium discoideum; other eukaryotes includes Arabidopsis thaliana, Chlamydomonas reinhardtii, Naegleria gruberi, Trichomonas vaginalis, Thalassiosira pseudonana, and Tetrahymena thermophila. Notes: 1 Fungi Sd homologs do not have the C-terminal Y460 residue. 2 Sd/TEAD is present in the amoebozoan A. castellanii (whose homolog includes the C-terminal Y460 residue), but not in D. discoideum. 3 A. macrogynus Hippo homolog does not contain the SARAH domain. 4 N. crassa does not encode any homolog of Warts/Lats, although other Ascomycota such as Schizosaccharomyces pombe and Aspagillus niger do encode this gene. 5 Putative A. queenslandica Yorkie homolog contains just one, instead of two, WW protein domains. 6 Putative A. queenslandica Kibra homolog contains an extra N-terminal PDZ domain. 7 C. owczarzaki, C. fragrantissima and S. arctica have proteins with the LLGL protein domain that in phylogenetic analysis appear as sister-group to a clade of the LLGL-containing Tomosyn and Lgl proteins. 8 Protein domain architecture is aberrant compared to bilaterian homologs. 9 Absent in H. sapiens.
Despite its essential role in animal development, the evolutionary history of the Hippo signaling pathway has been unresolved. Two recent comparative analyses reported key components of the pathway in non-bilaterian animals, but failed to identify any of these components outside Metazoa (Srivastava et al., 2010; Hilman and Gat, 2011). These two analyses concluded that key components of the pathway were metazoan innovations and that the Hippo pathway originated in the last common ancestor of cnidarians and bilaterians (Hilman and Gat, 2011) or sometime within the early metazoan evolution (Srivastava et al., 2010). Through comparative genomic analysis of several recently sequenced holozoan genomes coupled with functional genetic characterization, we provide compelling evidence that an active Hippo signaling pathway was already present in the unicellular ancestors of Metazoa, thus significantly pushing back the origin of this important cell signaling mechanism.
Results
Comparative genomic analysis reveals a premetazoan origin of the Hippo signaling pathway
To trace the evolutionary origin of Hippo signaling, we performed an extensive search of pathway components in several recently sequenced holozoan genomes (Ruiz-Trillo et al., 2007), including ichthyosporeans, filastereans and choanoflagellates, the closest unicellular relatives of Metazoa (Torruella et al., 2011), as well as in other eukaryotes. Our comparative genomic analysis reconstructed with unprecedented detail the evolutionary history of the Hippo signaling pathway and allowed us to trace the birth of Hippo signaling well before the origin of Metazoa (Figure 1A).
We have identified clear Yki homologs in two independent non-metazoan lineages, the filastereans (Capsaspora owczarzaki, from herein called “Capsaspora”) and the choanoflagellates (Monosiga brevicollis and Salpingoeca rosetta) (Figure 1B). Phylogenetic analysis clusters them unequivocally with metazoan Yki homologs with high nodal support and well differentiated from the WWP1 and other ubiquitin ligases that also contain WW domains (Supplemental Figure S1). Importantly, all these non-metazoan Yki homologs contain highly conserved functional sites like the Hippo pathway responsive phosphorylation site S168/127 and the N-terminal homology region that is critical for interaction with Sd/TEAD transcription factor (Figure 2A). Indeed, these holozoan species contain homologues of Sd/TEAD with the C-terminal Y460 residue known to be important for YAP-TEAD interaction (Li et al., 2010; Chen et al., 2010).
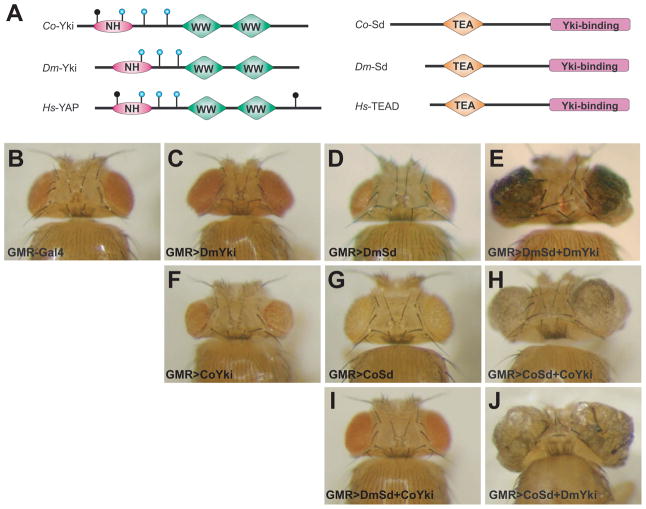
Figure 2. The Sd-Yki transcription factor complex from Capsaspora promotes tissue growth in Drosophila.
(A) Schematic structures of Yki (left) and Sd (right) orthologues from Capsaspora owczarzaki (Co), Drosophila melanogaster (Dm) and Homo sapiens (Hs). Wts phosphorylation motifs (HxRxxS/T) in each Yki homologue are indicated by vertical lines ending with circles, with the blue circles indicating the three conserved Wts phosphorylation motifs. “NH” refers to Yki’s N-terminal Homology domain which binds to Sd/TEAD. “TEA” refers to the DNA-binding domain of the Sd orthologues.
(B-J) Dorsal view of adult heads from the indicated genotypes. All images were taken under the same magnification.
(B) GMR-Gal4/+. Wildtype control.
(C) GMR-Gal4 UAS-DmYki/+. Overexpression of DmYki resulted in an increase in eye size (compare C to B).
(D) GMR-Gal4/UAS-DmSd. Overexpression of DmSd caused a decrease in eye size (compare D to B).
(E) GMR-Gal4 UAS-DmYki/UAS-DmSd. The eye tissue was massively overgrown and folded.
(F) GMR-Gal4/UAS-CoYki. Overexpression of CoYki resulted in small and rough eyes (compare F to B).
(G) GMR-Gal4 UAS-CoSd/+. The eye size was similar to wildtype control (compare G to B).
(H) GMR-Gal4 UAS-CoSd/UAS-CoYki. The eye tissue was massively overgrown and folded.
(I) GMR-Gal4 UAS-DmSd/UAS-CoYki. The eye size was similar to wildtype control (compare I to B).
(J) GMR-Gal4 UAS-CoSd/UAS-DmYki. The eye tissue was massively overgrown and folded.
Our searches further identified homologues of Hpo, defined by the presence of a Ste20-like kinase domain and a SARAH domain (Scheel and Hofmann, 2003), in amoebozoans, apusozoans and most opisthokonts, except for M. brevicollis and non-chytrid fungi, most likely due to secondary loses (Figures 1A–B). Likewise, Wts homologues are present along all opisthokonts, except maybe for the ichthyosporeans, where the incompleteness of genome data makes it difficult to ensure its absence. The adaptor protein Mats is present in all eukaryotes, whereas the other adaptor of the core pathway, Salvador, is only present in choanoflagellates and Metazoa. Besides these critical components of the core Hippo kinase cascade, the amoeboid Capsaspora encodes several upstream regulators of Hpo such as Kibra, Mer, aPKC and Lgl but not Ex, Crb or the Fat signaling module (Figures 1A–B).
Thus, our data has pinpointed with unprecedented detail the evolutionary history of all members of the Hippo pathway, and shows that a well-constituted Hippo pathway was present well before the origin of Metazoa, acting in a unicellular context. Moreover, given that Capsaspora encodes both Kibra and Mer and the apical-basal polarity proteins Lgl and aPKC, the level of upstream regulatory complexity of the Hippo/YAP pathway in Capsaspora is potentially very high. Although little is known about the receptors that lead to activation of the Hippo pathway in Metazoa, Hippo signaling is known to be activated in a cell-density dependent manner (Zhao et al., 2010). In this regard, Mer has indeed been shown to directly mediate contact inhibition of proliferation in cell cultures (Okada et al., 2007; McClatchey and Giovannini, 2005), where Mer is known to engage reciprocal signaling with key effectors of the integrin signaling and adhesion machinery (Pugacheva et al., 2006). Interestingly, Capsaspora is so far the only analyzed non-metazoan organism known to harbour all the components of the integrin-mediated adhesion and signaling system present in metazoans (Sebe-Pedros et al., 2010). Moreover, Capsaspora also encodes some genes known to be downstream of the Hippo pathway, such as Myc (Sebe-Pedros et al., 2011) and cyclin E (unpublished results). This suggests that the regulatory complexity of cell proliferation control in the closest unicellular relatives of animals is remarkably high. Thus a possible function of the Hippo pathway in this unicellular context could be the control of cell proliferation in a cell density- and/or cell adhesion-dependent manner.
The Sd-Yki transcription factor complex from the unicellular amoeboid Capsaspora owczarzaki promotes tissue growth and Hippo target gene expression in Drosophila
To test the functional relevance of our evolutionary analysis, we assayed the activities of Capsaspora owczarzaki (Co) Hippo pathway components in Drosophila (see Supplemental Information for sequence alignment of Yki, Sd, Hpo, Wts and Mats homologues among Capsaspora, Drosophila and humans). Given their critical roles in Drosophila and mammalian Hippo signaling, we first examined the Capsaspora homologues of Sd and Yki (Figure 2A). We have shown previously that overexpression of Drosophila melanogaster (Dm) Yki by the GMR-Gal4 driver (GMR>Dm-Yki) leads to increased eye size (Huang et al., 2005) (Figure 2C), whereas overexpression of DmSd by the same Gal4 driver (GMR>Dm-Sd) results in smaller eye size (Figure 2D), likely due to a dominant-negative effect whereby overexpressed Dm-Sd titrates (or squelches) certain endogenous Sd cofactor(s) (Wu et al., 2008). We found that overexpression of Co-Sd (GMR>Co-Sd) in Drosophila did not result in an appreciable change in eye size (Figure 2G), suggesting a reduced ability of Co-Sd to squelch endogenous Dm-Sd cofactors. Surprisingly, we found that unlike its Drosophila counterpart, overexpression of Co-Yki (GMR>Co-Yki) did not result in any tissue overgrowth, but rather caused a small and rough eye phenotype (Figure 2F). While the exact reason for this rough eye phenotype is unclear, the failure of Co-Yki overexpression to promote Drosophila eye growth suggests that Co-Yki has greatly diminished ability to interact productively with endogenous Dm-Sd to drive tissue overgrowth. Alternatively, Co-Yki may not possess intrinsic ability to drive tissue overgrowth (for example, due to its lack of general or specific co-activator activity to turn on growth-promoting genes), even if Co-Yki can form a transcription factor complex with Sd.
To distinguish between these models, we examined pair-wise combinatorial overexpression between Co-Sd/Co-Yki and Dm-Sd/Dm-Yki. As shown previously, co-expression of Dm-Sd and Dm-Yki (GMR>Dm-Sd+Dm-Yki) resulted in tremendous overgrowth of eye tissue (Figure 2E), consistent with the well-established role of the Sd-Yki complex in promoting tissue growth. In agreement with the inability of Co-Yki alone to drive tissue overgrowth, co-expression of Co-Yki and Dm-Sd (GMR>Dm-Sd+Co-Yki) failed to drive eye overgrowth (Figure 2I). We noted that the GMR>Dm-Sd+Co-Yki eyes were larger than GMR>Dm-Sd or GMR>Co-Yki eyes, suggesting that when both proteins were overexpressed at high levels, they may interact with each other, albeit in a greatly attenuated manner. Most strikingly, despite the inability of Co-Yki or Co-Yki+Dm-Sd to induce tissue overgrowth, co-expression of Co-Yki and Co-Sd (GMR>Co-Sd+Co-Yki) resulted in massive tissue overgrowth resembling that caused by co-expression of their Drosophila counterparts (compare Figures 2H and 2E). A similar and massive tissue overgrowth was also observed when Co-Sd was co-expressed with Dm-Yki (GMR>Co-Sd+Dm-Yki) (Figure 2J). Thus, despite the greatly attenuated cross-species interactions between Co-Yki and Dm-Sd, the Sd-Yki complex evolves as a functional entity – it is the function of the Sd-Yki complex rather than the individual subunit that is pivotal to growth control.
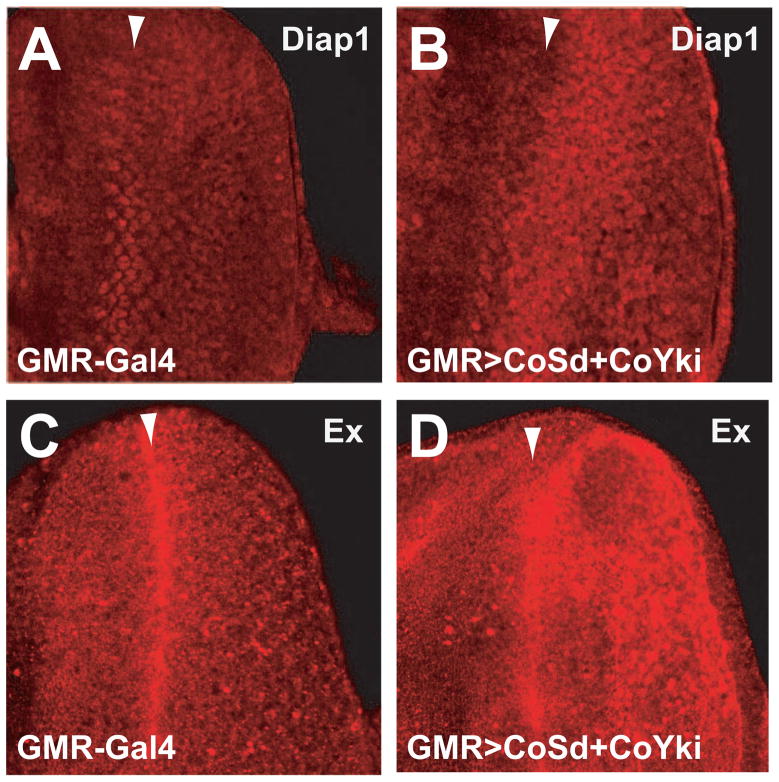
To understand the molecular mechanism by which co-expression of Co-Sd and Co-Yki induces tissue overgrowth, we examined the expression of Diap1 and Ex, two well characterized Hippo/Yki target genes. Third instar eye imaginal discs of GMR>Co-Sd+Co-Yki animals showed a marked upregulation of Diap1 and Ex staining posterior to the morphogenetic furrow (where the GMR-Gal4 driver is active) (Figure 3). Thus, despite their enormous evolutionary distance from each other, the Sd-Yki complex from a unicellular holozoan still retains the ability to promote tissue growth and to activate similar transcriptional targets as its Drosophila counterpart.
Figure 3. The Sd-Yki transcription factor complex from Capsaspora activates Hippo target genes in Drosophila.
Confocal images of 3rd instar eye imaginal discs from wildtype control (GMR-Gal4) (A and C) and animals with GMR-Gal4-meidated co-overexpression of Co-Sd and Co-Yki (GMR>CoSd+CoYki) (B and D). Arrowheads mark the position of the morphogenetic furrow (MF), and all eye discs are oriented anterior to the left.
(A–B) eye imaginal discs showing Diap1 immunostaining (red). Note the elevated Diap1 expression posterior to the MF in GMR>CoSd+CoYki eye discs (compare B to A).
(C–D) eye imaginal discs showing Ex immunostaining (red). Note the elevated Ex expression posterior to the MF in GMR>CoSd+CoYki eye discs (compare D to C).
The unicellular amoeboid Capsaspora owczarzaki contains an active Hippo kinase cascade leading from Hpo to the Sd-Yki complex
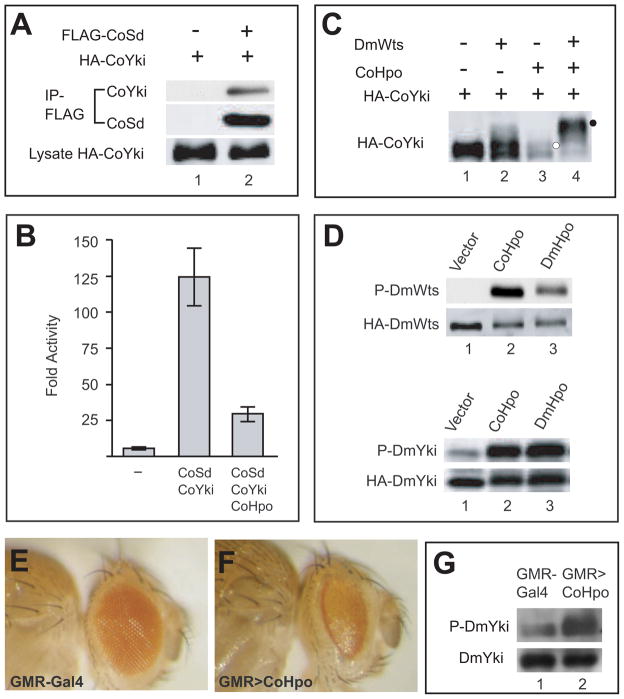
Our transgenic experiment predicted that Co-Sd and Co-Yki should physically interact with each other. Indeed, epitope-tagged Co-Sd and Co-Yki immunoprecipitated with each other in Drosophila S2R+ cells (Figure 4A), demonstrating their ability to form a protein complex. Using a well-characterized luciferase reporter driven by the minimal Hippo-Responsive Element (HRE) derived from the Hippo target gene diap1 (Wu et al., 2008), we found that co-expression of Co-Sd and Co-Yki stimulated the transcription of the HRE-luciferase reporter in Drosophila S2R+ cells (Figure 4B). Together with the synergistic effect of Co-Sd and Co-Yki in inducing tissue overgrowth (Figure 2H) and Diap1 expression (Figures 3A–B) in vivo, these data demonstrate the ability of Co-Sd and Co-Yki to form a functional transcription factor complex with striking specificity to activate similar target genes as its Drosophila counterpart.
Figure 4. The unicellular amoeboid Capsaspora owczarzaki contains an active Hippo kinase cascade leading from Hpo to Yki phosphorylation.
(A) Physical association between Co-Sd and Co-Yki. S2R+ cell lysates expressing the indicated constructs were immunoprecipitated (IP) and probed with the indicated antibodies. HA-CoYki was detected in FLAG-IP in the presence (lane 2), but not the absence (lane 1), of FLAG-CoSd.
(B) Co-Hpo antagonized Co-Sd/Co-Yki-mediated activation of an HRE-luciferase reporter in S2R+ cells. S2R+ cells were transfected with HRE-luciferase reporter along with the indicated expression constructs for Co-Sd, Co-Yki and Co-Hpo. Luciferase activity was quantified in triplicates and plotted. Note the activation of the HRE-luciferase reporter by Co-Sd/Co-Yki, and the inhibition of Co-Sd/Co-Yki-stimulated HRE-luciferase activity by Co-Hpo.
(C) Co-Hpo induced Co-Yki phosphorylation in cultured Drosophila cells. S2R+ cell lysates expressing HA-CoYki together with the indicated constructs were probed with HA antibody. Note the mobility shift of HA-CoYki induced by Co-Hpo (retarded band indicated by white circle), and the supershift induced by Co-Hpo plus DmWts (supershifted band indicated by black circle).
(D) Co-Hpo stimulated Dm-Wts and Dm-Yki phosphorylation in cultured Drosophila cells. S2R+ cells expressing HA-DmWts (top two gels) or HA-DmYki (lower two gels) in combination with Co-Hpo or Dm-Hpo were probed with P-Wts-T1077 or P-Yki-S168, respectively. Note that both Co-Hpo and Dm-Hpo resulted in increased levels of P-DmWts-T1077 or P-DmYki-S168 (compare lanes 2 and 3 with lane 1 in both gels).
(E–F) Growth-suppressing activity of Co-Hpo in Drosophila. Side views of adult heads of control (GMR-Gal4/+) (E) and flies that overexpressed Co-Hpo in the eye (GMR-Gal4/UAS-CoHpo) (F). Note the reduced eye size of GMR>CoHpo flies (compare F to E).
(G) Overexpression of Co-Hpo stimulated phosphorylation of endogenous Yki in Drosophila. Protein extracts from control (GMR-Gal4) or GMR>CoHpo adult heads were probed with antibodies against endogenous DmYki and P-DmYki-S168. Note the increase in P-DmYki signal in GMR>CoHpo adult head extracts (compare lane 2 to lane 1).
Next, we tested the functionality of Co-Hpo in inducing the phosphorylation of Co-Yki or Dm-Yki by co-expression of the respective constructs in Drosophila S2R+ cells. We found that Co-Hpo significantly inhibited Co-Sd/Co-Yki-mediated activation of the diap1 HRE-luciferase reporter in S2R+ cells (Figure 4B), suggesting that Co-Hpo can negatively regulate the transcriptional activity of the Co-Yki/Co-Sd complex. Consistent with this finding, expression of Co-Hpo induced phosphorylation of Co-Yki in S2R+ cells, and this phosphorylation was further enhanced by co-expression of Dm-Wts (Figure 4C). Interestingly, Co-Hpo also stimulated the phosphorylation of Dm-Wts and Dm-Yki, as revealed by phospho-specific antibodies against P-Dm-Wts-T1077 and P-Dm-Yki-S168, respectively (Figure 4D). Thus, Co-Hpo can engage a canonical kinase cascade through the phosphorylation of the intermediary kinase Wts and the ultimate phosphorylation target Yki
To corroborate the cell-based assays described above in a more physiological setting, we used a transgenic overexpression assay to examine the activity of Co-Hpo in vivo. Overexpression of Co-Hpo by the GMR-Gal4 driver (GMR>Co-Hpo) resulted in a small eye phenotype (Figures 4E–F) reminiscent of that caused by overexpression of its Drosophila counterpart, suggesting that the growth-inhibitory activity of Hpo is conserved in the unicellular Capsaspora. The GMR>Co-Hpo animals also allowed us to examine the influence of Co-Hpo on endogenous Yki phosphorylation in vivo. Using a phospho-specific antibody against the critical Hippo-responsive Ser168 phosphorylation site (Dong et al., 2007), we found that protein extracts from GMR>Co-Hpo fly heads showed increased Yki-S168 phosphorylation compared to control extracts (Figure 4G). Thus, Co-Hpo not only possesses growth-suppressing activity but also functionally activates a signaling cascade leading to the phosphorylation of endogenous Yki in Drosophila.
Concluding Remarks
In conclusion, our study demonstrates that key components of the Hippo pathway are encoded in the genomes of unicellular relatives of metazoans. We provide compelling evidence that the amoeboid Capsaspora contains functional homologues of the core Hippo kinase cascade leading from the tumor suppressor protein Hpo to the transcriptional coactivator Yki, suggesting the existence of an active Hippo kinase cascade well before the origin of Metazoa. In particular, our data shows that a well-constituted Hippo pathway originated within the Holozoa, before the divergence of filastereans, choanoflagellates, and Metazoa. Most remarkably, we demonstrate that despite the enormous evolutionary divergence, the growth-promoting and gene-regulatory activity/specificity of the Sd-Yki complex, as well as the growth-inhibitory activity of Hpo, are still retained in the unicellular Capsaspora. Our findings further pinpoint the Sd-Yki complex, rather than each subunit of this transcription factor complex, as a critical functional entity in the evolution of growth control mechanisms.
The surprising conservation of biochemical functionality for different Hippo pathway elements in such phylogenetically divergent species (bilaterian metazoan vs amoeba) could probably be explained by strong functional constraints due to the varied network of interactions of these components. In theory, these different network elements could have as well independently co-evolved and, therefore, not be functional within another species context. The fact that the different Hippo pathway elements of Capsaspora are indeed functional within the multicellular Drosophila background, strongly supports a functional homology between the unicellular and multicellular Hippo pathway. This suggests that the current function of the Hippo signaling pathway might be somehow similar within these two biological contexts.
How the ancestral Hippo kinase cascade is used in a unicellular organism such as Capsaspora remains a mystery at present. We speculate that this pathway might be used to coordinate cell proliferation in response to cell density or cell polarity (e.g., upon substrate adhesion), given the established roles of Merlin, Kibra, aPKC and Lgl (all of which are encoded in Capsaspora genome) in these biological processes. The absence of other developmental signaling pathways, such as Notch, Hedgehog, Wnt or BMP, in Capsaspora or any other analyzed unicellular holozoan, emphasizes the relevance of Hippo signaling as a key developmental mechanism predating the origin of Metazoa. The exaptation of this pathway may have easily provided a mechanism for strict control of cell proliferation in early metazoans, an essential property for any integrated multicellular entity. The presence of a functional and highly conserved Hippo pathway in Capsaspora accentuates not only the importance of analyzing the unicellular prehistory of animals to understand their origin, but also the role that gene co-option may have played in the unicellular-to-multicellular transition.
Experimental Procedures
Gene searches and phylogenetic analysis
Genes were searched by using blast (blastp, blastn, and tblastn) and blast reverse with several sequences used as queries, as described previously (Sebe-Pedros et al., 2010). Some genes could only be identified by performing phylogenetic analyses. A full list of the genes and sequences used, including the newly annotated ones is shown in Table S1. The taxon sampling used is shown in Figure 1B and includes new genome sequences obtained by the UNICORN genome project (see http://www.broadinstitute.org/annotation/genome/multicellularity_project/MultiHome.html) (Ruiz-Trillo et al., 2007). Domain arrangements were confirmed by Pfam and SMART. Alignments were constructed using MAFFT online server (Katoh et al., 2002) and then manually inspected and edited in Geneious. Only those species and those positions that were unambiguously aligned were included in the final analysis. Maximum likelihood (ML) phylogenetic trees were estimated by Raxml (Stamatakis, 2006) using PROTGAMMAIWAG+Γ+I model of evolution. Nodal supports were assessed by performing 100-bootstrap replicates with the same evolutionary model. Bayesian analysis was performed with MrBayes 3.1 (Ronquist and Huelsenbeck, 2003) using WAG+Γ+I model of evolution, four chains and two parallel runs. Runs were stopped when the average standard deviation of split frequencies of the two parallel runs was < 0.01, usually around 1,000,000 generations, and burn-in length was established by checking the two LnL graphs, stationarity of the chain typically occurred after <15% of the generations.
The annotation of Capsaspora Yorkie (Co-Yki) was further checked by using 5′ and 3′ RACE PCR using standard conditions. An excel file with all the sequences of the genes used in this study can be downloaded from the webpage http://www.multicellgenome.com.
Drosophila genetics and cell culture
Capsaspora Hpo, Yki and Sd cDNAs were amplified by PCR and inserted into pUAST vector to generate pUAST-CoHpo, pUAST-CoYki and pUAST-CoSd constructs, respectively. Transgenic flies were made by P-element mediated germline transformation pUAST constructs. Flies were raised on standard cornmeal medium at 25°C. Eye imaginal discs of wandering third instar larvae were fixed and stained as described (Yu et al., 2010) using α-Ex (1:5000) (gift of R.G. Fehon) (Maitra et al., 2006) and α-Diap1 (1:600) (gift of B. A. Hay) (Yoo et al., 2002). For analysis of endogenous Yki phosphorylation in fly tissues, twenty fly heads from control (GMR-Gal4) or Co-Hpo-transgenic (GMR>CoHpo) animals were smashed in 50μL 2×SDS loading buffer and then boiled for 5 min. After centrifugation at 12000rpm for 5 min, 10μL supernatants were separated on 8% SDS-PAGE and transferred to an Immobilon-P PVDF membrane. The western blots were probed with rabbit α-P-S168-Yki and rabbit α-Yki antibodies (Dong et al., 2007).
Drosophila S2R+ cells were propagated in Schneider’s medium (GIBCO) supplemented with 10% FBS and antibiotics. Luciferase assay was carried out using the HRE (Hippo Responsive Element)-luciferase reporter as described (Wu et al., 2008), using pUAST-CoHpo, pUAST-CoYki and pUAST-CoSd constructs in combination with pAc-Gal4 construct. Expression constructs for HA-DmWts and HA-DmYki have been described previously (Ling et al., 2010; Huang et al., 2005). Yki and Wts phosphorylation was probed using rabbit α-P-S168-Yki (Dong et al., 2007) and rabbit α-P-Wts-T1077 (Yu et al., 2010).
For immunoprecipitation, HA-CoYki and FLAG-CoSd were constructed in the pAc5.1/V5-HisB vector by adding the respective epitope at the N-terminus of each protein. S2R+ cells transiently transfected with these constructs were lysed in lysis buffer (50mM Tris [Ph7.4], 150mM NaCl, 1mM EDTA, 0.5% Triton X-100) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (20uM Sodium fluoride, 4mM Sodium orthovanadate, 4mM Sodium Pyrophosphate, 12mM β-glycerophosphate). Lysate was cleared by centrifugation at 14000rpm for 5min. Supernatant was incubated with ANTI-FLAG® M2 Affinity Gel (Sigma-Aldrich) at 4°C for 2hrs, followed by centrifugation and washing as described (Yu et al., 2010).
Supplementary Material
Acknowledgments
The genome sequences of C. owczarzaki, S. rosetta, A. macrogynus, S. punctatus, and T. trahens are being determined by the Broad Institute of MIT/Harvard University under the auspices of the National Human Genome Research Institute (NHGRI) and within the UNICORN initiative. We thank JGI, BI, and BCM for making data publicly available. We also thank Dr. Kim Worley and her colleagues in the Human Genome Sequencing Center of Baylor College of Medicine for allowing us to use A. castellanii genome sequence. We are grateful to Drs. Rick Fehon and Bruce Hay for providing antibodies used in this study. This work was supported by an ICREA contract, a European Research Council Starting Grant (ERC-2007-StG-206883) and a grant (BFU2008-02839/BMC) from Ministerio de Ciencia e Innovación (MICINN) to I. R.-T., and National Institutes of Health (R01 EY015708) to D.P.. A.S-P. was supported by a pre-graduate FPU grant from MICINN. D. P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamska M, Matus DQ, Adamski M, Green K, Rokhsar DS, Martindale MQ, Degnan BM. The evolutionary origin of hedgehog proteins. Curr Biol. 2007;17:R836–R837. doi: 10.1016/j.cub.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Badouel C, Garg A, McNeill H. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol. 2009;21:837–843. doi: 10.1016/j.ceb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, Song H. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Hilman D, Gat U. The Evolutionary History of YAP and the Hippo/YAP Pathway. Mol Biol Evol. 2011;28:2403–2417. doi: 10.1093/molbev/msr065. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis--the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- Okada T, You L, Giancotti FG. Shedding light on Merlin’s wizardry. Trends Cell Biol. 2007;17:222–229. doi: 10.1016/j.tcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Roegiers F, Golemis EA. Interdependence of cell attachment and cell cycle signaling. Curr Opin Cell Biol. 2006;18:507–515. doi: 10.1016/j.ceb.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PW, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Scheel H, Hofmann K. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr Biol. 2003;13:R899–R900. doi: 10.1016/j.cub.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Sebe-Pedros A, de MA, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol. 2011;28:1241–1254. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci U S A. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single copy protein domains. Mol Biol Evol. 2011 Jul 28; doi: 10.1093/molbev/msr185. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates hippo signaling in conjunction with Merlin and expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.