Abstract
Two-photon imaging of fluorescence in brain enables analysis of the structure and dynamic activity of neurons and glial cells in living animals. However, vital functions such as beating of the heart cause pulsations in brain tissue, leading to image distortion and loss of resolution. We find that synchronizing imaging scans to the cardiac cycle reduces motion artifacts, significantly improving the resolution of cellular structures. By interlacing multiple heartbeat triggered imaging scans, it was possible to image large brain volumes with negligible distortion. This approach can be readily incorporated into conventional microscopes to achieve substantial reductions in motion artifacts during two-photon imaging.
Key points
Two-photon fluorescence microscopy enables analysis of the structure and dynamic activity of different cell types in the brains of living animals at high temporal and spatial resolution.
Our ability to accurately resolve small structures, including dendrites and organelles in vivo, is hampered by pulsations in brain tissue that are caused by vital functions such as breathing and beating of the heart.
Here we show that beating of the heart is the major cause of brain pulsations in the cerebral cortex and find that synchronizing imaging scans to cardiac cycles significantly reduces motion artifacts.
We introduce a strategy (interlaced scanning) that enables electrocardiogram (ECG)-triggered scanning of large brain volumes, and provide software for implementation of ECG-triggered scanning on conventional two-photon microscopes.
Introduction
In vivo two-photon imaging has become an indispensable approach for monitoring changes in the morphology and physiological activity of cells in the central nervous system (Kerr & Denk, 2008). To accurately follow cellular dynamics, such as Ca2+ changes in subcellular compartments, the growth of dendritic spines, and organelle movements, it is essential to maximize spatial resolution. However, rapid movements of the brain due to beating of the heart and breathing present an inherent challenge for in vivo studies. The complex motions produced by these vital functions reduce image stability, compromise the resolving power of the microscope, and limit our ability to study both physiological responses and pathological changes in the intact CNS.
In vivo imaging of fluorescent structures in the brain requires removal or thinning of the overlying skull. Although it is possible to reduce brain movement in the open skull configuration by pressing a glass coverslip or other transparent material against the surface of the brain, such manipulations are not possible when imaging is combined with drug application, or when imaging is performed using less traumatic thinned skull preparations (Dombeck et al. 2007; Greenberg & Kerr, 2009; Hofer et al. 2009; Holtmaat et al. 2009; Schultz et al. 2009; Drew et al. 2010; O’Connor et al. 2010; Yang et al. 2010), leaving the challenge of brain motion unresolved. Post hoc application of whole-frame registration algorithms based on cross-correlation (Rosenfeld & Kak, 1982) is effective at reducing the impact of both translational movements of the brain and thermal drift arising from instrumentation. However, it is more difficult to compensate motion artifacts that occur during the acquisition of individual image frames (in-frame motion artifacts). The impact of these frequent movements can be reduced using line-by-line correction algorithms based on the Lucas–Kanade framework or hidden Markov models (Dombeck et al. 2007; Greenberg & Kerr, 2009; Chen et al. 2010). Nevertheless, these post hoc in-frame correction approaches rely on statistical assumptions that are difficult to validate, and data lost during image acquisition cannot be recovered. Here, we show that the impact of brain motion on images collected through in vivo two-photon microscopy can be substantially reduced by synchronizing image acquisition to the cardiac cycle.
Methods
Ethical approval
All experiments were performed in strict accordance with protocols approved by the Animal Care and Use Committee at Johns Hopkins University. We used six adult Thy1-enhanced green fluorescent protein (EGFP)M mice (Feng et al. 2000) for cranial window experiments in up to three imaging sessions and three adult mice for polished and reinforced thinned skull (PoRTS) window experiments.
Animal surgery
For craniotomies, mice were anaesthetized by i.p. injection of ketamine (100 mg kg−1) and xylazine (10 mg kg−1). As soon as animals were unconscious, dexamethasone (Vedco; 0.1 mg per mouse) was injected i.p. and petroleum jelly was applied to the eyes. The scalp was incised, resected, and a mixture of lidocaine (lignocaine) HCl and adrenaline (epinephrine) (Hospira; 10 mg ml−1 and 10 μg ml−1, respectively) were applied topically to the exposed skull. The periostium was then shaved off and approximately 3 mm of muscle surrounding the exposed skull was covered with a thin layer of cyanoacrylate cement. After drying, an 8 mm wide aluminum head plate with a 4.5 mm diameter circular opening centred 1.5 mm posterior from bregma and 1.5 mm left of midline was attached to the skull using dental cement (C&B Metabond; Parkell Bio-Materials Div.). A 2 mm × 2 mm area of skull in the centre of the opening was removed using a dental drill (Osada; XL-230) fitted with a conical drill bit (Meisinger; HM23SR007) under forced air cooling. A No.1 cover glass was placed on the dura mater and the edges sealed with dental cement (Caulk Division, Dentsply International; Grip Cement). Imaging was initiated at least 2 weeks after surgery. For PoRTS window experiments, a head plate was attached using a similar procedure, and then a 2 mm × 2 mm region of skull was thinned to a thickness of approximately 15 μm and processed further, as described (Drew et al. 2010). Animals with this configuration were imaged 1–2 days after surgery.
In vivo two-photon imaging
Fluorescence images were collected using a Movable Objective Microscope (MOM) (Sutter Instrument) with a Zeiss ×20, 1.0 NA or an Olympus ×40, 0.8 NA objective. The microscope was controlled by a personal computer equipped with an Intel Core i7 CPU 950 @ is part of the technical description 3.07 GHz and 3 GB of RAM running ScanImage (v3.6) (Pologruto et al. 2003) software and custom scripts written in MATLAB 7 (see TrImager below) on Windows XP Professional (Microsoft). Two-photon excitation was achieved using a titanium:sapphire laser (Chameleon Ultra II, Coherent) tuned to 900 nm and attenuated, so that 15 mW or less average power entered the brain. The head of the animal was immobilized by attaching the head plate to a custom machined stage mounted on the microscope table. All experiments except for those shown in Fig. 2J (right) were conducted under isoflurane (1.5–2% vol./vol. in O2) anaesthesia. The experiments for Fig. 2J (right) were conducted under ketamine/xylazine (100 mg kg−1/10 mg kg−1, respectively) anaesthesia. The depth of anaesthesia was monitored by hind paw retraction in response to toe pinch, and by the rate of heart beats; anaesthesia level was increased when mice retracted their hind paw in response to toe pinch. The body temperature of the mice was maintained at 37°C using a feedback-controlled heating pad, and mice were kept on the stage for a maximum of 2 h.
Figure 2. ECG-triggered scanning allows real-time motion correction during in vivo two-photon imaging.
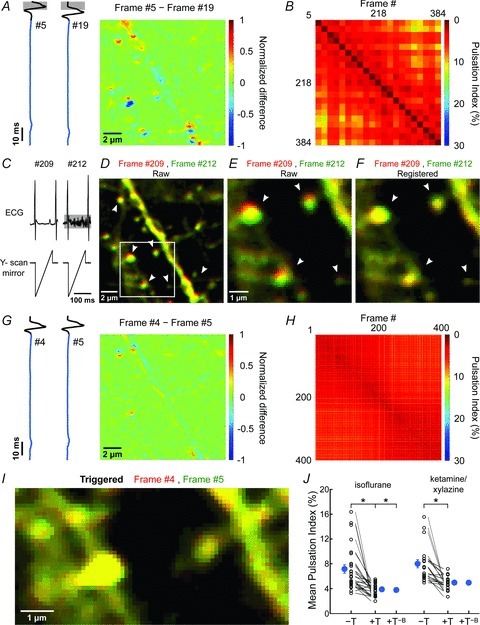
A, pseudocoloured difference image obtained by subtracting two frames serendipitously collected during the same phase of the cardiac R-wave. Grey boxes highlight the search window for the cardiac R-wave. B, plot of PIs of image pairs for which acquisition began within 15 ms after the cardiac R-wave. This condition was met by 21 of 400 frames. C, ECG trace (top traces) and simultaneously acquired Y-scan mirror position signal (bottom traces) for two images shown in D, for which scanning serendipitously occurred in between heartbeats. Note that during acquisition of frame #212 the surface electrodes also recorded the electromyogram (EMG) signal caused by breathing (highlighted by grey horizontal box). D, overlay of two images collected during the scans shown in C. Frame #209 is shown in red and #212 in green. Arrowheads highlight regions where vertical displacement of the dendrite is prominent. White box highlights region shown in panels E and F. E and F, images shown in highlighted region in D before (E) and after (F) performing whole-frame cross correlation registration (see Methods for details). G, pseudocoloured difference image obtained by subtracting two consecutive images collected with ECG-triggered acquisition. Traces at left show the simultaneously collected ECG signals. H, plot of PIs of image pairs during this recording. I, overlay of two consecutive pseudocoloured images (red and green) (region highlighted in Fig. 1A) collected with ECG-triggering. J, plot of the average PI measured with different anaesthetic drugs. PIs calculated from triggered (+T) and untriggered (–T) time series collected sequentially in the same field are connected by lines. ‘+T−B’ represent PIs calculated from triggered time series with frames acquired during breathing removed. Note that this does not lower PI further. Blue symbols, mean ± SEM; *P < 0.001 (paired Student's t test).
Electrocardiogram recording
The electrocardiogram (ECG) was recorded using an ECG amplifier (Sigmann Elektronik) and an instrumentation amplifier (Brownlee Precision; Model 440), as the potential difference between silver electrodes inserted subcutaneously in the right shoulder and the left thigh. The signal was amplified ×3000, low-pass filtered at 3 kHz, and applied to the digital input channel; the baseline was adjusted so that only the peak of the R-wave exceeded the threshold (1.6 V) necessary to trigger a low to high transition. This transition was monitored by TrImager to initiate individual frame scans. The analog ECG signal also was digitized at 50 kHz (Digidata 1440A, Molecular Devices) for post hoc analysis.
TrImager MATLAB scripts
For triggering individual frame scans and implementing interlaced frame scanning, we developed a graphical user interface in MATLAB 7 to control individual scripts. TrImager interfaces with the microscope hardware configuration used in combination with ScanImage (see ‘https://openwiki.janelia.org/wiki/display/ephus/ScanImage+r3.6.x+Documentation#ScanImager3.6.xDocumentation-WiringYou’– Wiring Your System) and collects information from ScanImage about the most recent power attenuation setting (Pockels cell command voltage), but otherwise operates autonomously. For a flow chart of the operation of TrImager, see Supplemental Fig. 1 (available online only). Independent from TrImager, it should be possible to implement ECG-triggered imaging at commercial laser scanning microscopes if they accept TTL triggers for individual image scans and maintain a fixed delay between trigger and initiation of scanning. Additional modifications to their proprietary control software would be required to implement interlaced scanning. Three scripts were used for all presented data. For Figs 1–3 and Supplemental Figs 2–4 (online), a 15 μm × 15 μm field of view was scanned at a resolution of 128 by 128 picture elements (pixels) within 99 ms. Each scan was triggered when the ECG signal connected to a digital input port (NI-6110, National Instruments), exceeded ∼1.6 V, which only occurred during the R-wave of the ECG signal. This strategy was chosen to avoid any delay due to signal processing. For untriggered scanning, a constant offset of 2–3 V was added to the ECG signal, lifting the ECG baseline above triggering level. Upon completion of one scan, 12 consecutive data samples were averaged to yield one pixel value and the linear stream of pixel values was transformed into a 2-dimensional (2-D) matrix yielding one image frame. The averaging and framing steps were completed by functions written in C and compiled to MATLAB extension (MEX) files to improve speed. The minimum time interval between scans was 63 ms, reflecting the time required to process data and initialize the next acquisition in the MATLAB Data Acquisition Toolbox. This step of TrImager could be further optimized to allow a larger fraction of the time between two heart beats to be used for scanning. For Fig. 4, a 100 μm × 100 μm field of view was scanned at 500 × 500 pixels resolution. In the untriggered mode, one frame was scanned within 751 ms and processed as described above, while in triggered mode, ten individually triggered subframes were scanned with an identical pixel dwell time, as described in the main text. Data were processed after completion of all ten subframes. After completion of one z-stack there was a break of 2 s (triggered) or 3 s (untriggered) before the next z-stack was initiated. All MATLAB scripts for TrImager, MEX files, their source codes and wiring instructions are available for download at http://www.bergleslab.com/Publications.htm.
Figure 1. Complex in-frame motion artifacts induced during in vivo two-photon imaging of the brain.
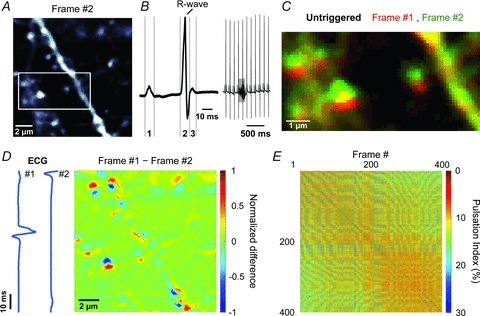
A, representative fluorescence image (#2) from a series of 400 images of a 14 μm × 14 μm region of cortex from a Thy1-EGFPM mouse. White box highlights region shown in panels C and Fig. 2I. B, left, representative ECG waveform showing the P-wave (1), representing depolarization of the atrial myocardium; the QRS-complex (2), representing depolarization of the ventricular myocardium; and the T-wave (3), representing repolarization of the ventricular myocardium. B, right, continuous ECG recording showing 11 heartbeats and 1 breath (highlighted by the grey box). C, overlay of two consecutive pseudocoloured images (red and green) (region highlighted in A). D, pseudocoloured difference image obtained by subtracting two consecutive images. Values were normalized to the largest absolute difference. Blue traces represent ECG signal recorded during each frame. E, plot of pulsation indices (PIs) of image pairs among 400 consecutive frames. Warmer colours indicate smaller pulsations (higher correlation).
Figure 3. ECG-triggered motion correction reduces artifacts when imaging through the thinned skull.
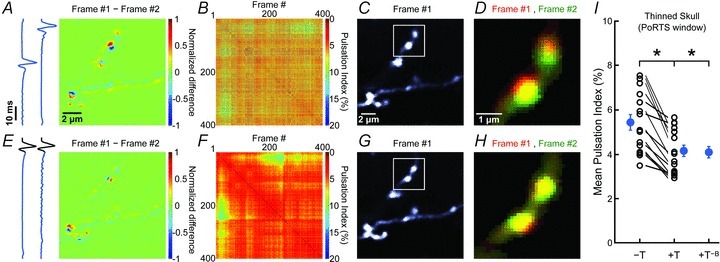
A, pseudocoloured difference image obtained by subtracting two consecutive images of a 14 μm × 14 μm region of cortex from a Thy1-EGFPM mouse. Values were normalized to the largest absolute difference. Blue traces at left show the ECG signals recorded during each frame. B, plot of PIs for image pairs among 400 consecutive frames. Warmer colours indicate smaller pulsations (higher correlation). C, representative fluorescence image (#1) from the 400 frames collected. White box highlights region shown in panel D. D, overlay of two consecutive pseudocoloured images (red and green) (region highlighted in C) collected without ECG-triggering. E–H, same analysis as in A–D for a subsequent imaging session in which ECG-triggered image acquisition was applied. I, plot of the average PI. PIs calculated from triggered (+T) and untriggered (–T) time series collected sequentially in the same field are connected by lines. ‘+T−B’ represent PIs calculated from triggered sessions in which frames acquired during breathing were removed. Note that this does not lower PI further. Blue symbols, mean ± SEM; *P < 0.001 (paired Student's t test).
Figure 4. ECG-triggered interlaced scanning reduces motion artifacts during in vivo 3-D imaging.
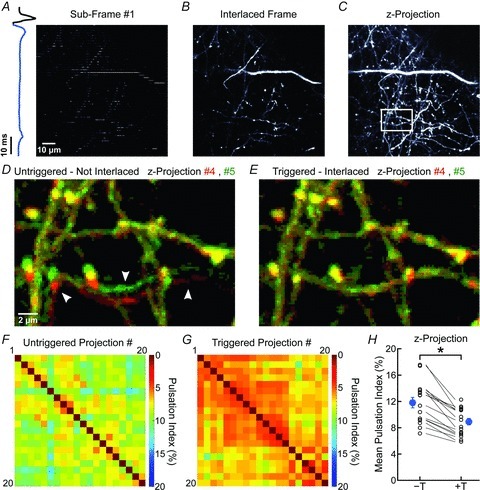
A, representative fluorescence image from a 100 μm × 100 μm region of the cortex in a Thy1-EGFPM mouse showing one sub-frame (50 lines in the 500 × 500 pixels area) collected between two heartbeats. Blue trace at left represents the simultaneously recorded ECG. B, fluorescence image of a single optical section reconstructed from 10 sub-frames. C, maximum intensity projection fluorescence image created through reconstruction of 14 interlaced frames covering 19.5 μm. White box highlights region shown at higher magnification in D and E. D and E, overlay of two consecutive pseudocoloured images (red and green) (region highlighted in C) collected without (D) or with (E) ECG-triggering. In D, arrowheads highlight movement artifacts. F and G, plot of PIs for pairs of images for 20 consecutive maximum intensity z-projections collected during untriggered, non-interlaced (F) or triggered, interlaced (G) z-stack time series. Warmer colours indicate smaller pulsations (higher correlation). H, plot of average PI measured for z-projections collected through untriggered (–T) and triggered (+T) acquisition. PIs calculated from triggered and untriggered time series collected sequentially in the same field are connected by lines. Blue symbols, mean ± SEM; *P= 2 × 10−6 (paired Student's t test).
Data analysis
Data were processed and analysed in MATLAB using built-in functions integrated into custom routines. Images were first processed with a 3 × 3 pixel median filter (built-in function ‘medfilt2’) to reduce stochastic noise of the detector. To compensate for motion artifacts and thermal drift during image series without in-frame manipulations, a whole-frame normalized 2-D cross-correlation (built-in function ‘normxcorr2’) was determined and individual frames were registered to maximize correlation. This procedure was applied to all data except that shown in Fig. 2D and E. For Fig. 4, maximum intensity projections were obtained before cross-correlation was applied. To determine correlation matrices of time-series data, each frame was linearized by concatenating all columns of a frame. Then, all linearized frames were concatenated in the second dimension, and the Pearson's linear correlation coefficient for each pair of columns determined (built-in function ‘corr’). Since identical images would yield a correlation coefficient of 1, we defined a pulsation index (PI; %) as PI = (1 – average correlation coefficient (excluding main diagonal)) × 100. It should be noted that the reproducibility of pixel values of low-intensity fluorescence signals is limited by the accuracy of scan mirror movements and instrumentation noise. As a result, even a sample that is stationary will not obtain a PI of 0%. Moreover, PI is influenced by the complexity of image content. An image series in which a large fraction of pixels are devoid of fluorescence will carry less instrumentation-caused image variability (resulting in a lower achievable PI) than an image series in which most pixels represent fluorescent structures (Supplemental Fig. 2). Nevertheless, PI provides a quantitative measure of reproducibility of images acquired under comparable conditions.
Results
To determine which components of brain motion are caused by heart beating or breathing, we monitored vital functions while collecting full-field images (128 × 128 pixels) of neuronal dendrites in the upper layers of mouse cortex. Unless otherwise stated, experiments were performed using anaesthetized adult Thy1-EGFPM mice at least 2 weeks after replacing a portion of the skull with a coverslip (for details see Methods), and neuronal processes were imaged in the somatosensory cortex 40–100 μm below the pial surface (Fig. 1A). The simultaneously recorded electrocardiogram (ECG) revealed that mice in these conditions had a heart rate of 8.7 ± 0.1 Hz and a respiration rate of 0.9 ± 0.1 Hz (n= 6 mice) (Fig. 1B). Although post hoc image registration was performed using whole-frame cross-correlation, motion artifacts were still apparent when consecutive frames were viewed (Supplemental Movie 1, available online). Image distortions consisted of a complex combination of stretches, compressions and torsions leading to displacement of entire dendritic spines by up to ∼1.5 μm (Fig. 1C and D).
To quantify image stability, we calculated the Pearson's linear correlation coefficient among each pair of frames collected during a series of 400 consecutive images (Fig. 1E). A ‘pulsation index’ (PI; %) equal to (1 – average correlation coefficient) × 100 was then determined to assess changes in stability of a given image series and compare movements between experiments. The consistent appearance of within-frame movements during high frame acquisition rates (6.2 Hz) suggested that the predominant source of movement was due to beating of the heart rather than breathing. If such changes in intravascular pressure cause reproducible cardiac-induced image distortions (CIDs), aligning each frame scan with the onset of the heartbeat should minimize these pressure-induced distortions. Indeed, comparisons among frames in which the rising phase of the associated cardiac R-wave occurred within 15 ms preceding the start of the scan significantly reduced PI from 7.19 ± 0.70% to 3.86 ± 0.20% (n= 26 fields of view (FOVs), 6 mice; P= 3.0 × 10−5, paired Student's t test) (Fig. 2A and B; Supplemental Movie 2). PI was comparably reduced when frames were selected in which the rising phase of the cardiac R-wave fell within a 6 ms time window in the middle of the frame scan (PI = 3.85 ± 0.19%; P= 2.8 × 10−5, paired Student's t test) (Supplemental Fig. 3), providing further evidence of the reproducibility of CIDs. In contrast to CIDs, breathing was associated with lateral shifts of whole image frames by 0.7 ± 0.1 μm (n= 26 FOVs, 6 mice), which could be reliably compensated using post hoc image registration (Fig. 2C–F and J).
Heartbeat induced motion artifacts have been described in a variety of other biomedical imaging contexts. In ophthalmology, investigation of the retina using optical coherence tomography (OCT) is hampered by axial motions correlated with the heartbeat, which can be compensated through feedback control of the OCT probe using estimates of displacement obtained from spectral domain partial coherence interferometry (Pircher et al. 2007; de Kinkelder et al. 2011). In addition, ECG-triggered image acquisition has been applied to both medical imaging of the cardiovascular system and intrinsic optical imaging of brain activity (van Dijk, 1984; Frostig et al. 1990). However, this approach has, to our knowledge, not been applied to in vivo two-photon microscopy of CNS tissues. To facilitate application of this method, we developed software (see Supplemental Fig. 1 and Methods for details) that allows acquisition of individual frame scans synchronized to the cardiac cycle. Triggering individual frame scans at the onset of cardiac cycles (Fig. 2G) significantly improved image stability, leading to a reduction of PI to 3.91 ± 0.19% (P= 8.6 × 10−5, paired Student's t test) (Fig. 2H–J; Supplemental Movie 3). Moreover, this approach reduced motion artifacts in a variety of conditions, independent of anaesthetic drug or surgical procedure. When ketamine/xylazine anaesthesia was used rather than isoflurane, PI was reduced from 8.04 ± 0.67% to 4.99 ± 0.24% (P= 1.3 × 10−4, paired Student's t test; n= 20 FOVs, 4 mice) with triggering (Fig. 2J), and with the recently described PoRTS window preparation (Drew et al. 2010), representing one variant of a thinned skull preparation, ECG triggering lowered PI from 5.45 ± 0.35% to 4.17 ± 0.26% (P= 4.3 × 10−6, paired Student's t test; n= 15 FOVs, 3 mice) (Fig. 3; Supplemental Movie 4), indicating that this approach improves image stability in both open skull and intact skull configurations. It is often desirable to average multiple consecutive frames during in vivo imaging to minimize the contribution of instrument noise, particularly when the fluorescence intensity is low. Synchronizing image acquisition to the cardiac cycle also minimized blurring of structures following frame averaging, due to tissue movement between frames (Supplemental Fig. 4).
In the simplest configuration, one frame scan should be completed within the time between two heartbeats (∼100 ms) to ensure that the trajectory taken by the tissue during each scan is consistent. However, several hundred milliseconds are typically required to acquire high resolution images from larger FOVs, due to limitations of laser scanning with conventional galvanometers. To overcome this limitation, we developed an interlacing strategy, in which one FOV was reconstructed from 10 sequentially triggered scans, each covering every tenth line (Fig. 4A and B). This approach was then repeated in different focal planes to obtain heartbeat triggered image stacks (Fig. 4C). When compared to conventionally acquired image stacks of identical volume (same pixel dwell time), triggered acquisition reduced the PI among 20 consecutive stacks from 11.78 ± 0.78% to 8.88 ± 0.52% (P= 2.0 × 10−6, paired Student's t test; n= 16 FOVs, 5 mice) (Fig. 4D–H; Supplemental Movie 5). PI values were higher for z-projections than single frames, as instrumentation noise has a greater impact on the correlation coefficient when the complexity of the images increases (Supplemental Fig. 2) (see Methods).
Discussion
Vital functions such as breathing and contraction of the heart induce movements of brain tissue that impair the ability to resolve small structures and follow the dynamics of cells in vivo using two-photon microscopy. While global, translational movements of the brain within the focal plane can be compensated by image registration, in-frame motion artifacts are more difficult to correct. To date, the most efficient approaches to eliminate in-frame artifacts involve post hoc manipulation of imaging data; however, these manipulations are difficult to validate and often result in loss of data. We found that the majority of in-frame distortions are caused by heartbeat-induced pulsations. By triggering acquisition of individual imaging scans to the cardiac cycle it was possible to substantially reduce frame-to-frame variability and improve resolution of cell structure in vivo.
In-frame tissue movements arising from CIDs were the most prominent cause of motion artifacts in the cerebral cortex, independent of whether imaging was performed through a chronic cranial window or through thinned skull. It is likely that the relative contribution of breathing and heartbeat to tissue motion may vary among CNS regions. In particular, to reduce motion artifacts during in vivo two-photon imaging of the spinal cord, previous studies have found it necessary to temporarily halt animal respiration during scanning (Kerschensteiner et al. 2005). By combining animal ventilation with ECG-triggered scanning, it may be possible to improve image stability further (Frostig et al. 1990), particularly when imaging the spinal cord, where breathing may induce larger tissue displacements.
An inherent limitation of this approach is that single interleaved scans can be acquired at a maximum rate of 8–10 Hz, as determined by the mouse heart rate. This acquisition rate is suitable for measurements of cell and organelle movement and whole-frame in vivo Ca2+ imaging (Dombeck et al. 2007; Ozden et al. 2009; Schultz et al. 2009; Komiyama et al. 2010; O’Connor et al. 2010). It may be possible to accelerate image acquisition further by completing several scans during each cycle, limited only by the duration of CIDs and the speed with which the laser beam can be displaced. Laser beam displacement can be strongly accelerated with acousto-optic deflector (AOD) based approaches, which enable line scan rates of up to 30 kHz (Reddy et al. 2008; Kirkby et al. 2010).
By interlacing multiple heartbeat triggered subframes for image reconstruction, it is possible to image large fields of view at high pixel resolution. However, individual scans that comprise a subframe are not registered with the approach described here. For preparations with more pronounced breathing induced brain motion, the TrImager interface offers the option to reject heartbeats that occur during breathing for triggering (Supplemental Fig. 1). Heartbeat triggered imaging is not limited to frame scanning; because of the reproducibility of CIDs, it is likely that 3-D line scans also would greatly benefit from this approach (Göbel et al. 2007; Reddy et al. 2008; Kirkby et al. 2010). Moreover, the application of super-resolution imaging techniques such as stimulated emission depletion (STED) microscopy to in vivo settings, will place higher demands on image stability to achieve improvements in resolution (Ding et al. 2009). Together, these studies show that ECG triggered scanning can be implemented with minimal hardware modifications to reduce the impact of complex tissue motion on in vivo two-photon imaging of fluorescent structures.
Acknowledgments
We thank Dr Ethan Hughes for insightful discussions during the project, Dr Robert Cudmore for valuable advice regarding chronic cranial window surgery and Terry Shelley for his machining expertise. The study was supported by grants from the Brain Science Institute (BSI) at Johns Hopkins University and the NIH (MH084020 and NS050274).
Glossary
- CID
cardiac-induced image distortio/n
- EGFP
enhanced green fluorescent protein
- FOV
field of view
- PI
pulsation index
- pixel
picture element
- PoRTS
polished and reinforced thinned skull
Author contributions
M.P. and D.E.B. designed the experiments. M.P. developed the software and performed the experiments. M.P. and D.E.B. wrote the manuscript.
Supplementary material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
References
- Chen T, Xue Z, Wang C, Qu Z, Wong KK, Wong STC. Motion artifact correction of multi-photon imaging of awake mice models using speed embedded HMM. Med Image Comput Assist Interv. 2010;13:473–480. doi: 10.1007/978-3-642-15711-0_59. [DOI] [PubMed] [Google Scholar]
- de Kinkelder R, Kalkman J, Faber DJ, Schraa O, Kok PHB, Verbraak FD, van Leeuwen TG. Heartbeat-induced axial motion artifacts in optical coherence tomography measurements of the retina. Invest Ophthalmol Vis Sci. 2011;52:3908–3913. doi: 10.1167/iovs.10-6738. [DOI] [PubMed] [Google Scholar]
- Ding JB, Takasaki KT, Sabatini BL. Supraresolution imaging in brain slices using stimulated-emission depletion two-photon laser scanning microscopy. Neuron. 2009;63:429–437. doi: 10.1016/j.neuron.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew PJ, Shih AY, Driscoll JD, Knutsen PM, Blinder P, Davalos D, Akassoglou K, Tsai PS, Kleinfeld D. Chronic optical access through a polished and reinforced thinned skull. Nat Methods. 2010;7:981–984. doi: 10.1038/nmeth.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Frostig RD, Lieke EE, Ts’o DY, Grinvald A. Cortical functional architecture and local coupling between neuronal activity and the microcirculation revealed by in vivo high-resolution optical imaging of intrinsic signals. Proc Natl Acad Sci U S A. 1990;87:6082–6086. doi: 10.1073/pnas.87.16.6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel W, Kampa BM, Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat Methods. 2007;4:73–79. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- Greenberg DS, Kerr JND. Automated correction of fast motion artifacts for two photon imaging of awake animals. J Neurosci Methods. 2009;176:1–15. doi: 10.1016/j.jneumeth.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hübener M. Experience leaves a lasting structural trace in cortical circuits. Nature. 2009;457:313–317. doi: 10.1038/nature07487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hübener M, Keck T, Knott G, Lee WA, Mostany R, Mrsic-Flogel TD, Nedivi E, Portera-Cailliau C, Svoboda K, Trachtenberg JT, Wilbrecht L. Long-term high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JND, Denk W. Imaging in vivo: Watching the brain in action. Nat Rev Neurosci. 2008;9:195–205. doi: 10.1038/nrn2338. [DOI] [PubMed] [Google Scholar]
- Kerschensteiner M, Schwab ME, Lichtman JW, Misgeld T. In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat Med. 2005;11:572–577. doi: 10.1038/nm1229. [DOI] [PubMed] [Google Scholar]
- Kirkby PA, Srinivas NKMN, Silver RA. A compact acusto-optic lens for 2D and 3D femtosecond based 2-photon microscopy. Opt Express. 2010;18:13721–13745. doi: 10.1364/OE.18.013720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Sato TR, O’Connor DH, Zhang Y, Huber D, Hooks BM, Gabitto M, Svoboda K. Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature. 2010;464:1182–1186. doi: 10.1038/nature08897. [DOI] [PubMed] [Google Scholar]
- O’Connor DH, Peron SP, Huber D, Svoboda K. Neural activity in barrel cortex underlying vibrissa-based object localization in mice. Neuron. 2010;67:1048–1061. doi: 10.1016/j.neuron.2010.08.026. [DOI] [PubMed] [Google Scholar]
- Ozden I, Sullivan MR, Lee HM, Wang SS. Reliable coding emerges from coactivation of climbing fibers in microbands of cerebellar Purkinje neurons. J Neurosci. 2009;29:10463–10473. doi: 10.1523/JNEUROSCI.0967-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher M, Baumann B, Götzinger E, Sattmann H, Hitzenberger CK. Simultaneous SLO/OCT imaging of the human retina with axial eye motion correction. Opt Express. 2007;15:16922–16932. doi: 10.1364/oe.15.016922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GD, Kelleher K, Fink R, Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat Neurosci. 2008;11:713–720. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld A, Kak AC. Digital Picture Processing. Vol. I and II. Orlando, FL: Academic Press; 1982. [Google Scholar]
- Schultz SR, Kitamura K, Post-Uiterweer A, Krupic J, Häusser M. Spatial pattern coding of sensory information by climbing fiber-evoked calcium signals in networks of neighboring cerebellar Purkinje cells. J Neurosci. 2009;29:8005–8015. doi: 10.1523/JNEUROSCI.4919-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk P. ECG-triggered NMR imaging of the heart. Diagn Imaging Clin Med. 1984;53:29–37. [PubMed] [Google Scholar]
- Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan W. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat Protoc. 2010;5:201–208. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


