Abstract
This study aimed to screen the pathogenic genes and pathways that relate to the transformation of hamster buccal mucosa from precancerous lesions to squamous cell carcinoma by whole genome microarray and bioinformatics analysis. A DMBA (7,12-dimethylbenz[a]anthracene)-induced hamster model of a precancerous lesion and squamous cell carcinoma was established. The differentially expressed genes were detected using an Agilent whole rat genome microarray, which contains 41,000 genes/ESTs. Gene ontology (GO) functional classification and pathway analyses were performed, and a subset of differentially expressed genes were validated using RT-PCR. The results showed that during the transformation of hamster buccal mucosa from the precancerous lesion to squamous cell carcinoma, a total of 1,981 genes were differentially expressed, of which 1,037 were up-regulated and 944 were down-regulated. GO analysis revealed that the differentially expressed genes are mainly involved in 14 functional groups including those of metabolism and cell structure. Additionally, 9 significantly altered pathways were identified. Among the 1,861 known differentially expressed genes, 14 genes including Casp3, CCL5 and CXCL12 were enriched in the 9 altered pathways. The up-regulation of SPARC and down-regulation of Casp3 were confirmed by RT-PCR. In conclusion, a total of 1,981 differentially expressed genes and 9 significantly altered pathways were identified in the transformation of hamster buccal mucosa from precancerous lesions to squamous cell carcinoma. A total of 14 pathway-enriched genes including Casp3, CCL5 and CXCL12 may play critical roles in the alteration of cellular pathways leading to the transformation of buccal mucosa from precancerous lesions to squamous cell carcinoma. Future studies focusing on these genes and pathways are required in order to gain a better understanding and provide effective prevention and treatment of oral squamous cell carcinoma.
Keywords: oral, precancerous lesion, squamous cell, carcinoma, gene, pathway
Introduction
Oral squamous cell carcinoma (OSCC) accounts for approximately 90% of cases of oral cancers and is currently ranked sixth among malignant human cancers (1). Although advances have been made in various treatment methods for oral cancer, the five-year survival rate of OSCC is only 55–60% after standard therapy (1,2). Long-term clinical observations have shown that a number of OSCCs are derived from various precancerous lesions (1–3). However, no effective treatment currently exists to prevent or block the development of malignant precancerous lesions. Therefore, reversal of the precancerous lesions or protection from the malignant transformation through effective intervention is likely to have a great impact on the prevention and treatment of oral cancer.
Recent studies (3–5) have shown that the initiation of OSCC is a multi-staged complex pathological process involving the changes of numerous genes. The toxic effect of chemical substances is one of the major causes of OSCC. Due to its similarity to the malignant transformation process of human oral mucosal epithelia, the DMBA (7,12-dimethylbenz[a]anthracene)-induced squamous cell carcinoma model with buccal mucosa from a hamster cheek pouch is currently considered to be the most ideal animal model of oral mucosal cancer (6,7). However, no studies are available at the whole genome level regarding the changes in genes and pathways during the DMBA-induced malignant transformation of oral mucosa from precancerous lesions to OSCC. Subsequently, a DMBA-induced Syrian hamster model of malignant transformation of cheek pouch mucosa from precancerous lesions to squamous cell carcinoma was established. Additionally, the differentially expressed gene profile at the whole genome level was analyzed during this transformation process using an Agilent rat whole genome microarray, which contains 41,000 genes/ESTs. Oral mucosa malignant transformation-related genes and pathways were examined at the whole genome level using bioinformatics methods such as gene ontology (GO) functional classification and pathway analysis. These methods are crucial to the study of molecular mechanisms involved in the initiation and progress of the malignant transformation of oral mucosa precancerous lesions, reversal of the precancerous lesions, and intervention methods which block the malignant transformation.
Materials and methods
Animals and reagents
Twelve Syrian hamsters were purchased from the Chengdu Institute of Biological Products (Chengdu, China). The reagents used were: DMBA (Sigma, St. Louis, MO, USA), PCR instrument (PTC-100, MJ Company, USA), hybridization oven (G2545A, Agilent Technologies, Santa Clara, CA, USA), Agilent scanner (G2565AA, Agilent Technologies), a spectrophotometer (ND1000, NanoDrop Technologies Inc., Wilmington, DE USA), an RT-PCR kit (Bioflux, Japan), and an Agilent rat genome cDNA microarray (Agilent Technologies).
Experimental methods
To establish the hamster model of cheek pouch precancerous lesions and squamous cell carcinoma, a total of 12, 6–8-week old Syrian hamsters, weighing 90–120 g, were randomly divided into groups I and II, with six animals in each group. The cheek pouch was opened, and 5% DMBA in acetone solution was applied to the bilateral cheek pouch every Monday, Wednesday and Friday. The animals were sacrificed after 6 weeks (precancerous lesion group) and 12 weeks (squamous cell carcinoma group), respectively, following the DMBA treatment. Samples of the cheek pouch tissues were removed. Half of the sample was stored immediately in liquid nitrogen, and the remaining half was fixed with 10% formalin, paraffin-embedded, sectioned, stained with hematoxylin and eosin (H&E), and observed under a light microscope. The experimental procedures were approved by the Laboratory Animal Management Committee of Chongqing Medical University.
Extraction and purification of total RNA
Total RNA from the two groups was isolated from frozen tissues using a TRIzol RNA Extraction kit (Invitrogen, Carlsbad, CA, USA) and further purified using a RNeasy mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. The purified RNA was dissolved in RNase-free water and stored in a freezer at -80°C until required.
cRNA labeling and synthesis
Extracted total RNA in the two groups was fluorescently labeled with Cy3 through reverse transcription according to the kit's instructions. Following the synthesis, cRNAs of each group were labeled with aaUTP and purified with a RNeasy mini kit (Qiagen). The cRNAs were then quantified with a spectrophotometer and purified again.
cRNA fragmentation and microarray chip hybridization
Cy3-labeled cRNA (875 ng) from each group was used in the experiment. Fragmentation was carried out following the manufacturer's instructions. The microarray chips were hybridized with 100 μl of fragmented cRNAs for 17 h (65°C, 10 r/min) and then washed subsequently in solution 1 and 2 at 37°C for 1 min each time.
Chip scanning and data processing
Hybridized microarray chips were scanned with an Agilent G2565AA Microarray Scanner (Agilent Technologies) and the results were normalized with feature extraction software. Ratio ≥2 and ≤0.5 criteria were used to select differentially expressed genes.
Bioinformatics analysis
Abnormally expressed genes were functionally classified based on the GO standard (http://www.ncbi.nlm.nih.gov/). The abnormally changed pathways were identified by searching pathway databases KECG, GENMAPP and BIOCARTA (www.biorag.com). P<0.05 was used to select the pathways that were significantly altered.
Validation of microarray results by RT-PCR
Up-regulated gene SPARC and down-regulated gene Casp3 were randomly selected for RT-PCR validation. PCR primers (Table I) were designed using Primer3 software.
Table I.
RT-PCR primers.
| Gene name | GenBank accession | Primer sequence | Product length (bp) |
|---|---|---|---|
| SPARC | NM_012656 | F: 5′-GTGCCAGGACCCCACCAG-3′ R: 5′-ATGGGAATGAGGGGAGCG-3′ |
504 |
| Casp3 | NM_012922 | F: 5′-ACCCTGAAATGGGCTTGT-3′ R: 5′-GTTTCGGCTTTCCAGTCA-3′ |
348 |
| β-actin | NM_001101.2 | F: 5′-GTAAAGACCTCTATGCCAACA-3′ R: 5′-GGACTCATCGTACTCCTGCT-3′ |
227 |
Results
Pathological examination of precancerous lesion and squamous cell carcinoma tissues
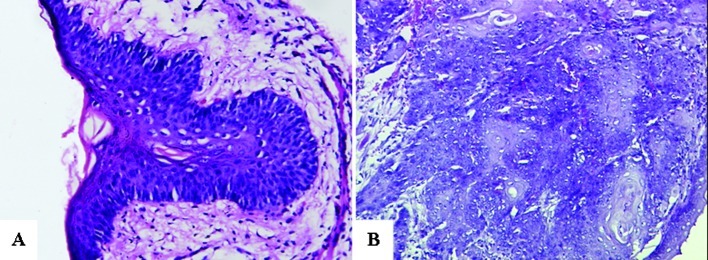
Routine H&E staining showed that among the six cases of precancerous lesions in group I, there were five cases of moderate dysplasia and one case of mild dysplasia. An experiment was performed with the five moderate dysplasia cases to detect precancerous lesions. The six cases in group II were all squamous cell carcinoma tissues (Fig. 1).
Figure 1.
Pathological examination under a light microscope (H&E; magnification, ×200). (A) Precancerous lesion (moderate dysplasia). (B) Squamous cell carcinoma.
Screening the genes that are differentially expressed in precancerous lesions and squamous cell carcinoma tissues
A total of 1,981 genes were differentially expressed during the transformation of hamster cheek mucosa from the precancerous lesions to OSCC. Of these genes, 1,861 are known, 120 are unknown, 1,033 are up-regulated and 944 are down-regulated.
Functional classification of differentially expressed genes
The 1,861 differentially expressed known genes were classified into 14 functional gene categories (Table II) based on the GO functional classification.
Table II.
Gene ontology (GO) analysis of the differentially expressed genes.
| GO functions | Up-regulated genes | Down-regulated genes | Total | % |
|---|---|---|---|---|
| Metabolism | 298 | 185 | 483 | 24.38 |
| Development | 179 | 212 | 391 | 19.74 |
| Cell structure | 133 | 178 | 311 | 15.70 |
| Stress | 72 | 107 | 179 | 9.04 |
| Signal transduction | 83 | 42 | 125 | 6.31 |
| Immune | 41 | 52 | 93 | 4.69 |
| Transportation | 47 | 15 | 62 | 3.13 |
| Cellular localization | 19 | 32 | 51 | 2.57 |
| Cell adhesion | 17 | 24 | 41 | 2.07 |
| Transcription | 25 | 9 | 34 | 1.72 |
| Cell growth | 13 | 19 | 32 | 1.62 |
| Cell differentiation | 23 | 8 | 31 | 1.56 |
| Apoptosis | 6 | 12 | 18 | 0.91 |
| Cell cycle | 7 | 3 | 10 | 0.50 |
| Unknown function | 74 | 46 | 120 | 6.06 |
Pathway analysis
By checking the 1,861 known differentially expressed genes against three major pathway databases, 9 pathways were determined to have been significantly alered during the transformation from precancerous lesions to squamous cell carcinomas (P<0.05). However, only 14 out of the 1,861 known genes were enriched in these 9 pathways (Table III).
Table III.
Pathway analysis.
| Pathway | P-value | Gene changes |
|---|---|---|
| Activation of caspase-3 | 0.0366 | Up: 0 Down: Casp3 |
| Chemokine receptors bind to chemokines | 0.0406 | Up: CCL5, CXCL12 and CCL24 Down: 0 |
| E2F-mediated regulation of DNA replication | 0.0471 | Up: 0 Down: Dhfr and Tyms |
| G1/S transition | 0.0370 | Up: 0 Down: Dhfr and Tyms |
| Integrin cell surface interactions | 0.0070 | Up: Vcam1, Vtn and Fn1 Down: 0 |
| Lipoprotein metabolism | 0.0082 | Up: Apoe and Alb Down: 0 |
| Response to elevated platelet cytosolic Ca | 0.0366 | Up: Alb, Fn1, Serping1 and Sparc Down: 0 |
| Arachidonic acid metabolism | 0.012 | Up: 0 Down: Cyp2b13 |
| Metabolism of xenobiotics by cytochrome P450 | 0.040 | Up: 0 Down: Cyp2b13 |
RT-PCR validation
The results from the RT-PCR validation of the up-regulated gene SPARC and the down-regulated gene Casp3 are shown in Fig. 2. The gene expression of Sparc and Casp3, which is shown as integral optical densities of the PCR products, has been normalized with the expression of β-actin and compared (Table IV). The PCR results confirmed the trend of gene expression determined in the microarray analysis.
Figure 2.
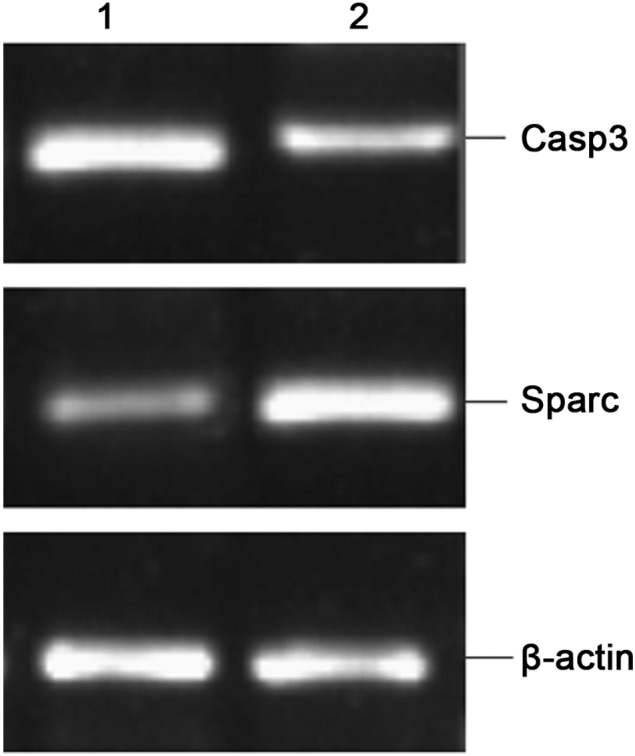
RT-PCR validation of differentially expressed genes SPARC and Casp3. Lane 1, precancerous lesions and lane 2, squamous cell carcinoma.
Table IV.
Validation of microarray results by RT-PCR.
| Gene name | Target gene mRNA/β-actin mRNA (mean ± SD) | B/A | Microarray | |
|---|---|---|---|---|
|
|
||||
| A | B | |||
| SPARC | 0.1157±0.0031 | 0.5023±0.0131 | 4.34 | 4.70 |
| Casp3 | 0.4401±0.0024 | 0.1467±0.0008 | 0.32 | 0.3a |
A, precancerous lesion; B, squamous cell carcinoma;
absolute value of the reciprocal of the original microarray fold change −3.2.
Discussion
This study has established a DMBA-induced hamster model of transformation of buccal mucosa from precancerous lesions to squamous cell carcinoma. A total of 1,981 differentially expressed genes were identified through microarray analysis of whole mouse genome. Of these, 1,861 are known genes. These differentially expressed genes were grouped into 14 functional categories: metabolism, development, cell structure, stress, signal transduction, immune, chemical transportation, cellular localization, cell adhesion, transcription, cell growth, cell differentiation, apoptosis and cell cycle. This classification indicates that the malignant transformation of the oral mucosa precancerous lesion is a complex pathological process involving changes in numerous genes.
Further pathway analysis shows that there were 9 significantly altered pathways in the DMBA-induced malignant transformation of oral mucosa precancerous lesions. However, out of the 1,861 differentially expressed known genes, only 14 genes were found to be enriched in the 9 abnormally altered pathways. These genes were: Casp3, CCL5, CXCL12, CCL24, Dhfr, Tyms, Vcam1, Vtn, Fn1, Apoe, Alb, Serping1, Sparc and Cyp2b13. Thus, we consider these 14 genes to play significant pathogenic roles in the malignant transformation of oral mucosa precancerous lesions. Studies have shown that Casp3 is an apoptosis regulatory gene; and that its down- regulation promotes tumor cell proliferation and inhibits tumor cell apoptosis (8). CCL5, CXCL12 and CCL24 are all chemokines, whose high expression promotes cell proliferation and tumor metastasis (9–11). Dhfr and Tyms are key enzymes of intracellular folate metabolism (12–14), both of which are the target of the folic acid antagonist class of anti-cancer drugs and are responsible for drug resistance. Their expression levels correlate to the prognosis and drug resistance of the cancer, and a low expression level indicates a favorable prognosis. Vcam1 belongs to the immunoglobulin superfamily, and its high expression enhances the migration of endothelial cells and angiogenesis, thereby promoting tumor invasion and metastasis (15). A high expression of either Vtn or Fn1 promotes tumor formation and metastasis (16–18). Apoe is an arginine-rich basic protein, whose high expression also promotes tumor cell proliferation and metastasis (19,20). Alb expression correlates to cancer prognosis; the prognosis of tumors expressing high levels of Alb protein is better than that of tumors with a low Alb expression (21). Serping1 gene encodes the C1 inhibitor, which inhibits the activity of coagulation factors XIIa and XIa, kallikrein and plasmin, thereby playing significant roles in the anticoagulation system of the body. Although studies have shown that the expression levels of Serping1 are variable among different types of tumors (22,23), Serping1 expression is up-regulated in the present study. Therefore, it is hypothesized that Serping1 exerts diverse effects on different types of tumors; however, the underlying mechanisms should be investigated. A high expression of SPARC enhances the mobility of tumor cells and promotes tumor invasion and metastasis (24–26). Cyp2b13 belongs to the second family of cytochrome P450. It participates in the metabolism of a number of carcinogens and toxins, induces mutation in a number of genes, and inhibits certain gene expressions (27). The down-regulation of Cyp2b13 causes a decrease in carcinogen metabolism and detoxification. The present study has shown a similar expression pattern of the above-mentioned genes during the malignant transformation process of oral mucosa precancerous lesions. The low expression of Dhfr and Tyms and the high expression of Alb indicate a positive prognosis of the tumor, which may be related to the fact that the animal model of OSCC in this study is currently at an early stage.
The present study is a preliminary one, and the gene expression results should be validated and verified. However, an overview is provided of the gene and pathway changes occurring during the transformation of oral buccal mucosa from precancerous lesions to squamous cell carcinoma at a whole genome level. The identified abnormally altered genes and pathways in this study potentially shed light on further in-depth investigations.
Acknowledgements
The authors would like to express their appreciation to Shanghai Biochip Co., Ltd., for the technical support in this study. This study was supported by the Medical Foundation of Chongqing City Health Bureau (no. 03-2-073).
References
- 1.Kademani D, Bell RB, Schmidt BL, Blanchaert R, Fernandes R, Lambert P, Tucker WM. Oral and maxillofacial surgeons treating oral cancer: a preliminary report from the American Association of Oral and Maxillofacial Surgeons Task Force on Oral Cancer. J Oral Maxillofac Surg. 2008;66:2151–2157. doi: 10.1016/j.joms.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Lumerman H, Freedman P, Kerpel S. Oral epithelial dysplasia and the development of invasive squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1995;79:321–329. doi: 10.1016/s1079-2104(05)80226-4. [DOI] [PubMed] [Google Scholar]
- 3.Yang K, Zhang G, Mei J, Chen D, Wu M. Screening and analysis of pathogenic genes during DMBA-induced buccal mucosa carcinogenesis in golden hamsters. Oncol Rep. 2010;23:1619–1624. doi: 10.3892/or_00000803. [DOI] [PubMed] [Google Scholar]
- 4.Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87:14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 5.Carinci F, Lo Muzio L, Piattelli A, et al. Genetic portrait of mild and severe lingual dysplasia. Oral Oncol. 2005;41:365–374. doi: 10.1016/j.oraloncology.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Feng L, Wang Z. Chemopreventive effect of celecoxib in oral precancers and cancers. Laryngoscope. 2006;116:1842–1845. doi: 10.1097/01.mlg.0000233778.41927.c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimenez-Conti IB, Slaga TJ. The hamster cheek pouch carcinogenesis model. J Cell Biochem Suppl. 1993;9:83–90. doi: 10.1002/jcb.240531012. [DOI] [PubMed] [Google Scholar]
- 8.Meggiato T, Calabrese F, De Cesare CM, Baliello E, Valente M, Del Favero G. C-JUN and CPP32 (CASPASE 3) in human pancreatic cancer: relation to cell proliferation and death. Pancreas. 2003;26:65–70. doi: 10.1097/00006676-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki K, Natsugoe S, Ishigami S, et al. Expression of CXCL12 and its receptor CXCR4 in esophageal squamous cell carcinoma. Oncol Rep. 2009;21:65–71. [PubMed] [Google Scholar]
- 10.Vaday GG, Peehl DM, Kadam PA, Lawrence DM. Expression of CCL5 (RANTES) and CCR5 in prostate cancer. Prostate. 2006;66:124–134. doi: 10.1002/pros.20306. [DOI] [PubMed] [Google Scholar]
- 11.Cheadle EJ, Riyad K, Subar D, et al. Eotaxin-2 and colorectal cancer: a potential target for immune therapy. Clin Cancer Res. 2007;13:5719–5728. doi: 10.1158/1078-0432.CCR-07-1145. [DOI] [PubMed] [Google Scholar]
- 12.Joerger M, Omlin A, Cerny T, Früh M. The role of pemetrexed in advanced non small-cell lung cancer: special focus on pharmacology and mechanism of action. Curr Drug Targets. 2010;11:37–47. doi: 10.2174/138945010790030974. [DOI] [PubMed] [Google Scholar]
- 13.Jensen SA, Vainer B, Witton CJ, Jørgensen JT, Sørensen JB. Prognostic significance of numeric aberrations of genes for thymidylate synthase, thymidine phosphorylase and dihydrofolate reductase in colorectal cancer. Acta Oncol. 2008;47:1054–1061. doi: 10.1080/02841860801942158. [DOI] [PubMed] [Google Scholar]
- 14.Shimokawa H, Uramoto H, Onitsuka T, Iwata T, Nakagawa M, Ono K, Hanagiri T. TS expression predicts postoperative recurrence in adenocarcinoma of the lung. Lung Cancer. 2010 Oct 21; doi: 10.1016/j.lungcan.2010.08.024. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 15.Nakao S, Kuwano T, Ishibashi T, Kuwano M, Ono M. Synergistic effect of TNF-alpha in soluble VCAM-1-induced angiogenesis through alpha 4 integrins. J Immunol. 2003;l70:5704–5711. doi: 10.4049/jimmunol.170.11.5704. [DOI] [PubMed] [Google Scholar]
- 16.Gubała E, Wiench M, Oczko-Wojciechowska M, et al. Gene expression analysis by DNA microarray in papillary thyroid cancer. Endokrynol Pol. 2005;56:752–757. [PubMed] [Google Scholar]
- 17.Heyman L, Kellouche S, Fernandes J, Dutoit S, Poulain L, Carreiras F. Vitronectin and its receptors partly mediate adhesion of ovarian cancer cells to peritoneal mesothelium in vitro. Tumour Biol. 2008;29:231–244. doi: 10.1159/000152941. [DOI] [PubMed] [Google Scholar]
- 18.Heyman L, Leroy-Dudal J, Fernandes J, Seyer D, Dutoit S, Carreiras F. Mesothelial vitronectin stimulates migration of ovarian cancer cells. Cell Biol Int. 2010;34:493–502. doi: 10.1042/CBI20090331. [DOI] [PubMed] [Google Scholar]
- 19.Huvila J, Brandt A, Rojas CR, et al. Gene expression profiling of endometrial adenocarcinomas reveals increased apolipoprotein E expression in poorly differentiated tumors. Int J Gynecol Cancer. 2009;19:1226–1231. doi: 10.1111/IGC.0b013e3181b33be0. [DOI] [PubMed] [Google Scholar]
- 20.Su WP, Chen YT, Lai WW, Lin CC, Yan JJ, Su WC. Apolipo-protein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer. 2010 Apr 27; doi: 10.1016/j.lungcan.2010.04.009. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 21.Chen SS, Chen KK, Lin AT, Chang YH, Wu HH, Chang LS. Correlation between pre-treatment serum biochemical markers and treatment outcome for prostatic cancer with bony metastasis. J Chin Med Assoc. 2009;72:301–306. doi: 10.1016/S1726-4901(09)70376-4. [DOI] [PubMed] [Google Scholar]
- 22.Santin AD, Zhan F, Bignotti E, et al. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology. 2005;331:269–291. doi: 10.1016/j.virol.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Ranvier GG, Weng J, Yeh RF, et al. Candidate diagnostic markers and tumor suppressor genes for adrenocortical carcinoma by expression profile of genes on chromosome 11q13. World J Surg. 2008;32:873–881. doi: 10.1007/s00268-008-9521-0. [DOI] [PubMed] [Google Scholar]
- 24.Podhajcer OL, Benedetti LG, Girotti MR, Prada F, Salvatierra E, Llera AS. The role of the matricellular protein SPARC in the dynamic interaction between the tumor and the host. Cancer Metastasis Rev. 2008;27:691–705. doi: 10.1007/s10555-008-9146-7. [DOI] [PubMed] [Google Scholar]
- 25.Alonso SR, Tracey L, Ortiz P, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 26.Briggs J, Chamboredon S, Castellazzi M, Kerry JA, Bos TJ. Transcriptional up-regulation of SPARC, in response to c-Jun overexpression, contributes to increased motility and invasion of MCF7 breast cancer cells. Oncogene. 2002;21:7077–7091. doi: 10.1038/sj.onc.1205857. [DOI] [PubMed] [Google Scholar]
- 27.Brookman-Amissah N, Mackay AG, Swann PF. Isolation and sequencing of the cDNA of a novel cytochrome P450 from rat oesophagus. Carcinogenesis. 2001;22:155–160. doi: 10.1093/carcin/22.1.155. [DOI] [PubMed] [Google Scholar]