Abstract
Little is known about the correlation between TGFBR2 G-875A and breast cancer risk. Moreover, the associations of the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2) in breast cancer tissues with the TGFB1 C-509T, T+29C and TGFBR2 G-875A polymorphisms remain to be determined. In this study, we genotyped for TGFB1 C-509T, T+29C and TGFBR2 G-875A in fresh surgically resected tissues (n=82) and archived paraffin-embedded specimens (n=88) from 170 patients with breast cancer, as well as peripheral blood samples from 178 cancer-free female individuals. Evaluation of ER, PR and HER2 expression was performed using immunohistochemical staining. Logistic regression analysis was carried out to determine the risk of breast cancer by calculating the odds ratios (ORs) and their 95% confidence intervals (CIs). As a result, no difference was observed in the TGFB1 C-509T, T+29C genotype and allele frequencies between patients and controls. However, the frequency of the TGFBR2 −875A allele was marginally higher in cancer-free female individuals than that of women with breast cancer (24.2 vs. 17.9%, P=0.05). Notably, when stratification was performed by ER, PR and HER2 expression, the TGFBR2 −875A allele was found to correlate significantly to a decreased risk of breast cancer with ER+ (OR=0.57, 95% CI 0.35–0.92), PR+ (OR=0.54, 95% CI 0.34–0.88), ER+PR+ (OR=0.55, 95% CI 0.33–0.92) and HER2− (OR=0.55, 95% CI 0.34–0.88) under a dominant genetic model. In conclusion, this is the first study to suggest that the TGFBR2 −875A allele modifies predisposition to breast cancer with an expression of ER+, PR+, ER+PR+ and HER2−.
Keywords: TGFBR2, TGFB1, polymorphism, breast cancer, risk
Introduction
Breast cancer is the leading cause of death from cancer in America, and its incidence is currently increasing in China. By 2021, it is anticipated to have increased substantially from the current rate, estimated at 10–60 cases per 100,000 women, to more than 100 new cases per 100,000 women aged 55–69 years (1,2). Breast cancer has been suggested to be caused by interactions between genetic and environmental factors. Of these factors, genetic factors, such as family history and gene-related variants, may play an important role in the development and progression of breast cancer.
Signaling from transforming growth factor-β (TGF-β) was conducted through serine-threonine kinases of transmembrane receptors, including TGFBR1 and TGFBR2. TGF-β1 is the most abundant form of TGF-β and regulates cellular processes by binding to TGFBR2, which then activates TGFBR1 through phosphorylation (3). Inactivation of the TGF-β signaling pathway may lead to acquisition of resistance to the anti-mitogenic effects of TGF-β and contribute to tumor development and progression (4,5). Therefore, defective expression or inactivation of TGF-β1 and its receptors, such as TGFBR2, may play a significant role in carcinogenesis. Compelling evidence shows that TGF-β signaling may play a crucial role in mammary development (6,7) and a complex role in breast cancer tumorigenesis via both tumor-suppressor and oncogenic activities (8).
Although a number of polymorphisms in TGFB1 and its receptor TGFBR2 were originally reported in the western population (9,10), no frequency of the polymorphisms G-800A, codon25 and codon263 of TGFB1 was detected in the Chinese population (11). Two polymorphisms (C-509T and T+29C) in TGFB1 have been associated with increased levels of TGF-β1 in serum from breast cancer patients (12). The TGFBR2 −875A allele was reported to enhance transcriptional activity in normal epithelial cells (10). Based on previous findings, we hypothesize that polymorphisms affecting the activity of genes involved in the TGF-β signaling pathway play a role in the development of breast cancer. However, little is known about the relationship between TGFBR2 G-875A and breast cancer risk in the Chinese population. Furthermore, the correlation of the expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2) in breast cancer tissue with TGFB1 C-509T, T+29C and TGFBR2 G-875A polymorphisms remains to be determined. In this study, three functional polymorphisms, TGFB1 C-509T, T+29C and TGFBR2 G-875A, were selected to evaluate whether they modify predisposition to breast cancer.
Materials and methods
Study participants
A total of 170 breast cancer patients and 178 cancer-free female controls were enrolled in this study. Fresh surgically resected tissues (n=82) and archived paraffin-embedded specimens (n=88) from patients diagnosed with breast cancer between 2006 and 2010 were selected from the Second Affiliated Hospital of Soochow University, China. In addition, blood samples from 178 cancer-free donors, frequency-matched to the patients by age (Table I), were collected for use as controls at the same hospital. With respect to immunohistochemistry, expression data on ER were available for 138, PR for 138 and HER2 for 103 patients among all of the breast cancer cases. Immunohistochemistry staining permitted the detection and localization of ER, PR and HER2 within sections of paraffin-embedded tissues. Positive expression is considered as >20% of tumor cell nuclei staining, borderline is 5–19% and negative is <5%. Both borderline and overtly positive results were considered as positive (13,14). HER2 expression was considered as positive (score 3+) and negative (score 0–1+). Samples with a score of 2+ were excluded from the analysis (15). This study was approved by the Academic Advisory Board of Soochow University.
Table I.
Characteristics of breast cancer cases and controls.
| Cases | Controls | P-valuea | |
|---|---|---|---|
| Age (mean ± SD) | 53.75±11.22 | 52.67±10.28 | 0.35 |
| Tumor stage | |||
| 0 | 3 | ||
| I | 26 | ||
| II | 34 | ||
| III | 60 | ||
| IV | 0 | ||
| Missing | 47 | ||
| Cancer type | |||
| Intraductal carcinoma | 5 | ||
| Infiltrative ductal carcinoma | 158 | ||
| Infiltrative lobular carcinoma | 1 | ||
| Mucinous adenocarcinoma | 4 | ||
| Adenocarcinoma infiltrating | 2 | ||
| Histologic grade | |||
| Well-differentiated | 5 | ||
| Moderately differentiated | 115 | ||
| Poorly differentiated | 12 | ||
| Missing | 26 | ||
| Tumor size | |||
| ≤2 cm | 38 | ||
| 2.1–5 cm | 81 | ||
| >5 cm | 4 | ||
| Missing | 47 | ||
| Lymph node status | |||
| Positive | 61 | ||
| Negative | 63 | ||
| Not examined | 46 | ||
| Tumor subtypesb | |||
| ER+ | 88 | ||
| ER− | 50 | ||
| PR+ | 88 | ||
| PR− | 50 | ||
| ER+ and PR+ | 74 | ||
| ER+ or PR+ | 28 | ||
| ER− and PR− | 36 | ||
| HER2+ | 9 | ||
| HER2− | 94 | ||
Independent t-test applied to age.
+, positive expression; −, negative expression.
DNA isolation and genotyping
Genomic DNA from fresh tissues and peripheral blood samples was extracted using the proteinase K digestion standard method. Isolation of DNA from paraffin-embedded tissues was performed using a microwave-based DNA extraction method, as previously described (16,17). Single nucleotide polymorphism (SNP) analysis was performed using a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay. In total, two SNPs in TGFB1 and one SNP in TGFBR2 were genotyped. C-509T and G-875A were located in the promoter region of TGFB1 and TGFBR2, respectively. Additionally, T+29C was located in exon 1 of TGFB1. The primer sequences, annealing temperatures, sizes of PCR products and restriction enzymes are shown in Table II. The PCR reaction was carried out in a total volume of 25 μl, containing 50–100 ng of genomic DNA, 1 unit of Ex Taq DNA polymerase (Takara, Japan), 0.2 μmol/l of each primer, 1X Ex Taq Buffer (Mg2+ Plus) and 0.25 mmol/l of each deoxynucleotide triphosphate. Briefly, PCR amplification was performed according to the following conditions: initial denaturation at 95°C for 5 min followed by 35 cycles of 94°C for 45 sec, 63 or 59°C for 45 sec and 72°C for 45 sec. PCR was completed by a final extension cycle at 72°C for 10 min.
Table II.
Primers and restriction enzymes used in PCR-RFLP assays for genotyping.
| Gene | Polymorphism | Primer sequence 5′→3′ | T (°C) | Size (bp) | Restriction enzyme |
|---|---|---|---|---|---|
| TGFB1 | C-509T | F: TTG AGT GAC AGG AGG CTG CTT A R: GCT GGG AAA CAA GGT AGG AGA A |
63 | 178 | Eco81I |
| T+29C | F: CCA CCA CAC CAG CCC TGT T R: TCC GCT TCA CCA GCT CCA T |
63 | 186 | MspA1I | |
| TGFBR2 | G-875A | F: GGA ATG TCT TGG GCA AAT CT R: ACC TGA ATG CTT GTG CTT TTA TT |
59 | 152 | TaaI |
F, forward primer sequence; R, reverse primer sequence.
Statistical analysis
The independent samples t-test was used to compare the difference in age between breast cancer patients and the controls. Differences in the distributions of genotypes and alleles of TGFB1 and TGFBR2 variants between patients and controls were evaluated using the χ2 test. The odds ratios (ORs), their 95% confidence intervals (CIs) and the P-value were assessed by logistic regression analyses, which were adjusted for age. Hardy-Weinberg equilibrium (HWE) was tested by a goodness-of-fit χ2 test. Statistical analysis was performed using SPSS 16.0.
Results
Association of polymorphisms in TGFB1 and TGFBR2 with risk of breast cancer
As summarized in Table III, the genotype and allele frequencies were obtained for TGFB1 C-509T, T+29C and TGFBR2 G-875A in breast cancer cases and controls. The genotype distributions for the three polymorphisms did not deviate from HWE in the patients (P=0.17, 0.22 and 1.00, respectively) or controls (P=0.55, 0.14 and 0.54, respectively). No statistical difference was found between the patients and controls for TGFB1 C-509T, T+29C genotype and allele frequencies. Additionally, no overall association was found between TGFBR2 G-875A genotype and breast cancer risk. However, the frequency of the TGFBR2 -875A allele was marginally higher in cancer-free individuals than that in breast cancer patients (24.2 vs. 17.9%, P=0.05), indicating that TGFBR2 −875A may predispose to breast cancer (adjusted OR=0.69, 95% CI 0.48–0.99).
Table III.
Genotype and allele distributions for the polymorphisms of TGFB1 and TGFBR2 in breast cancer patients and controls.
| Gene | Genotype | Case | Control | OR (95% CI) | P-value | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| n | % | n | % | ||||
| TGFB1 | C-509T | ||||||
| CC | 28 | 16.5 | 41 | 23.0 | 1 | ||
| CT | 93 | 54.7 | 84 | 47.2 | 1.35 (0.73–2.50) | 0.35 | |
| TT | 49 | 28.8 | 53 | 29.8 | 1.59 (0.91–2.81) | 0.11 | |
| CT/TT | 142 | 83.5 | 137 | 77.0 | 1.50 (0.88–2.56) | 0.14 | |
| C | 149 | 43.8 | 166 | 46.6 | 1 | ||
| T | 191 | 56.2 | 190 | 53.4 | 1.12 (0.83–1.51) | 0.46 | |
| T+29C | |||||||
| TT | 38 | 22.4 | 49 | 27.5 | 1 | ||
| TC | 76 | 44.7 | 79 | 44.4 | 1.24 (0.73–2.10) | 0.43 | |
| CC | 56 | 32.9 | 50 | 28.1 | 1.45 (0.82–2.57) | 0.20 | |
| TC/CC | 132 | 77.6 | 129 | 72.5 | 1.32 (0.81–2.15) | 0.27 | |
| T | 152 | 44.7 | 177 | 49.7 | 1 | ||
| C | 188 | 55.3 | 179 | 50.3 | 1.22 (0.91–1.65) | 0.19 | |
| TGFBR2 | G-875A | ||||||
| GG | 114 | 67.1 | 104 | 58.4 | 1 | ||
| GA | 51 | 30.0 | 62 | 34.8 | 0.74 (0.47–1.17) | 0.20 | |
| AA | 5 | 2.9 | 12 | 6.7 | 0.38 (0.13–1.12) | 0.08 | |
| GA/AA | 56 | 32.9 | 74 | 41.5 | 0.68 (0.44–1.06) | 0.09 | |
| G | 279 | 82.1 | 270 | 75.8 | 1 | ||
| A | 61 | 17.9 | 86 | 24.2 | 0.69 (0.48–0.99) | 0.05 | |
Odds ratios; 95% confidence intervals (CIs) and P-values were assessed by the logistic regression analysis, which was adjusted for age.
Relationship of TGFBR2 −875A allele to breast cancer with the expression of ER, PR and HER2
Based on the effects of hormones on breast carcinogenesis, the specimens were divided into various subgroups, including ER−, ER+, PR−, PR+, ER+PR+, ER+PR−/ER−PR+ and ER−PR− (Fig. 1A–D). The samples were additionally classified into subgroups of HER2− and HER2+ (Fig. 1E and F). Information on ER, PR and HER2 expression was available for 138, 138 and 103 patients with breast cancer, respectively. In detail, 88 ER+, 50 ER−, 88 PR+, 50 PR−, 74 ER+PR+, 28 ER+PR−/ER−PR+, 36 ER−PR−, 94 HER2− and 9 HER2+ breast cancer cases were included (Table I). When performing stratification by ER, PR and HER2 expression, we found no difference in the frequencies of TGFB1 C-509T, T+29C and TGFBR2 G-875A between ER+ and ER−, PR+ and PR−, and HER2+ and HER2− breast cancer cases (data available upon request). However, when comparing the ER+, ER−, PR+, PR−, ER+PR+, ER+PR−/ER−PR+, ER−PR−, HER2−, HER2+ breast cancer cases to the controls, our results showed that TGFBR2 G-875A was associated with a decreased risk of breast cancer with ER+ (OR=0.57, 95% CI 0.35–0.92), PR+ (OR=0.54, 95% CI 0.34–0.88), ER+PR+ (OR=0.55, 95% CI 0.33–0.92) and HER2− (OR=0.55, 95% CI 0.34–0.88) under the dominant genetic model (Table IV).
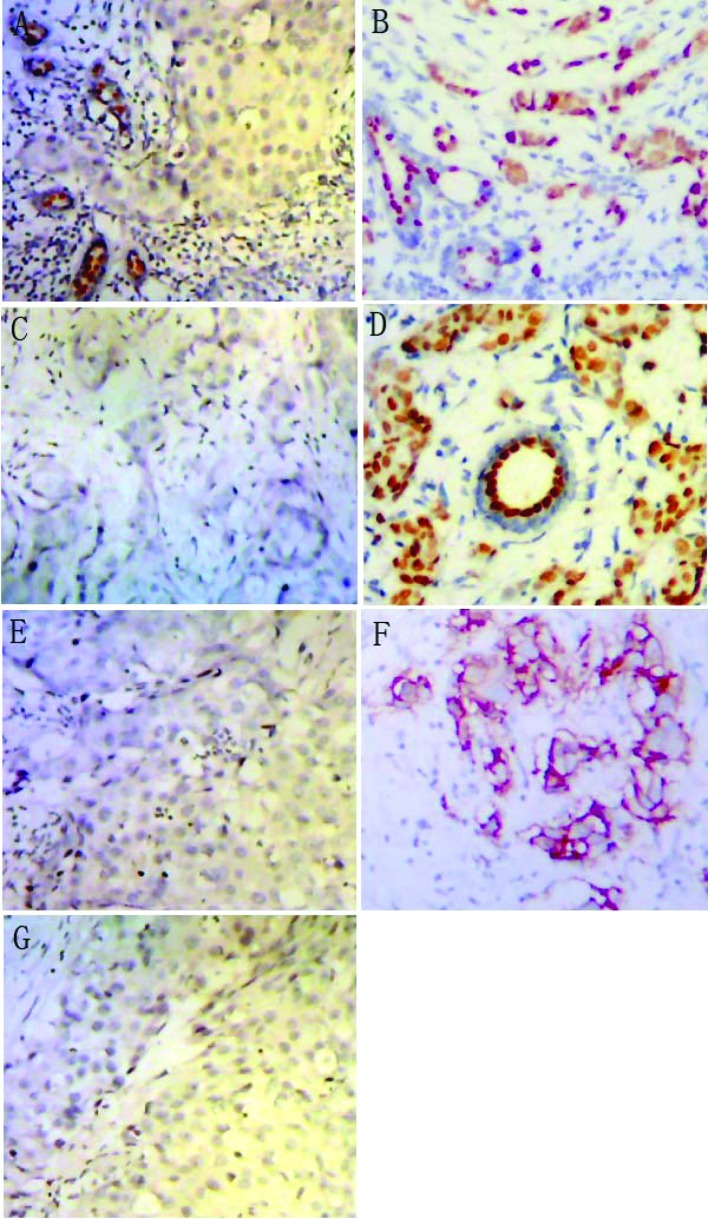
Figure 1.
Schematic representation of the expression of ER, PR and HER2 in breast cancer specimens by immunohistochemical staining (magnification, ×100). Breast cancer with (A) ER−, (B) ER+, (C) PR−, (D) PR+, (E) HER2− and (F) HER2+, (G) negative control.
Table IV.
Association of TGFBR2 G-875A genotypes and alleles with ER, PR and HER2 expression.
| Genotype | Cases (%)/Controls (%) | OR (95% CI) | P-value |
|---|---|---|---|
| ER+ breast cancer cases vs. controls | |||
| GG | 62 (70.5)/104 (58.4) | 1 | |
| GA | 25 (28.4)/62 (34.8) | 0.66 (0.37–1.16) | 0.15 |
| AA | 1 (1.1)/12 (6.7) | 0.14 (0.02–1.12) | 0.06 |
| GA+AA | 26 (29.5)/74 (41.6) | 0.58 (0.33–1.00) | 0.05 |
| G | 149 (84.7)/270 (75.8) | 1 | |
| A | 27 (15.3)/86 (24.2) | 0.57 (0.35–0.92) | 0.02 |
| PR+ breast cancer cases vs. controls | |||
| GG | 64 (72.7)/104 (58.4) | 1 | |
| GA | 22 (25)/62 (34.8) | 0.57 (0.32–1.01) | 0.06 |
| AA | 2 (2.3)/12 (6.7) | 0.27 (0.06–1.26) | 0.10 |
| GA+AA | 24 (27.3)/74 (41.5) | 0.52 (0.30–0.91) | 0.02 |
| G | 150 (85.2)/270 (75.8) | 1 | |
| A | 26 (14.8)/86 (24.2) | 0.54 (0.34–0.88) | 0.01 |
| ER+ and PR+ breast cancer cases vs. controls | |||
| GG | 53 (71.6)/104 (58.4) | 1 | |
| GA | 20 (27)/62 (34.8) | 0.62 (0.34–1.14) | 0.13 |
| AA | 1 (1.4)/12 (6.7) | 0.17 (0.02–1.31) | 0.09 |
| GA+AA | 21 (28.4)/74 (41.5) | 0.55 (0.31–0.99) | 0.05 |
| G | 126 (85.1)/270 (75.8) | 1 | |
| A | 22 (14.9)/86 (24.2) | 0.55 (0.33–0.92) | 0.02 |
| HER2− breast cancer cases vs. controls | |||
| GG | 67 (71.3)/104 (58.4) | 1 | |
| GA | 26 (27.7)/62 (34.8) | 0.62 (0.36–1.09) | 0.09 |
| AA | 1 (1.1)/12 (6.7) | 0.13 (0.02–1.01) | 0.05 |
| GA+AA | 27 (28.8)/74 (41.5) | 0.54 (0.32–0.93) | 0.03 |
| G | 160 (85.1)/270 (75.8) | 1 | |
| A | 28 (14.9)/86 (24.2) | 0.55 (0.34–0.88) | 0.01 |
Odds ratios (ORs), 95% confidence intervals (CIs) and P-values were assessed by the logistic regression analysis, which was adjusted for age.
Discussion
In this study, no difference was found in the genotype and allele frequencies of TGFB1 C-509T and T+29C between breast cancer patients and cancer-free female controls. This finding is consistent with a meta-analysis involving 10,633 cases and 13,648 controls for the C-509T polymorphism and 20,022 cases and 24,423 controls for T+29C (18). A previous study concerning T+29C showed decreased risk for Japanese pre-menopausal women with CC genotype, but not for post-menopausal women (19). Kaklamani et al found that breast cancer patients carrying the TGFB1*CC allele of the polymorphism T+29C were more likely to have ER− and PR− tumors (14). Cox et al found that an 18% decreased risk of ER+/PR+ breast cancer occurred among women heterozygous at C-509T and a 38% decrease occurred among women homozygous for the T allele as compared to those with CC genotype (20). In this study, breast cancer cases were stratified with ER, PR and HER2 expression. No significant association was found between the two polymorphisms (C-509T and T+29C) and breast cancer risk in the Chinese population.
Additionally, previous studies indicated that the TGFBR2 G-875A variant was not associated with breast cancer risk in the European population (21). However, results of the present study have shown that the −875A allele was marginally associated with a decreased risk of breast cancer. These findings are consistent with previous ones indicating that the TGFBR2 G-875A polymorphism significantly correlates with a decreased risk of gastric and esophageal squamous cell carcinomas in the Chinese population (22,23). In particular, we found that the TGFBR2 G-875A polymorphism may modify the risk of breast cancer with ER+, PR+, ER+PR+ and HER2−.
Activation of the TGF-β-mediated signal transduction is subject to hormonal regulation (24). TGF-β signal transduction may play a crucial role in the activation of various complex TGF-β receptors, including TGFBR1 and TGFBR2 (25). The expression of TGFBR2 is hormonally regulated and anti-estrogen may induce activation of the TGFBR2 promoter (26). For instance, breast cancer cell lines expressing ER+ are refractory to TGF-β effects, whereas ER− cells are sensitive (27). Loss or undetectable expression of TGFBR2 has been reported to contribute to TGF-β resistance in ER+ breast cancer cells (28). Paiva et al have suggested that the absence of TGFBR2 was associated with poorer prognosis in HER2− breast tumors (15). TGFBR2 −875A allele was proven to enhance the transcriptional activity of TGFBR2 in normal epithelial cells (10). In this study, breast cancer subgroups, comprising ER+, PR+, ER+PR+ and HER2−, had a higher frequency of TGFBR2 −875G than the controls, indicating that the TGFBR2 −875A allele correlated significantly to a decreased risk of breast cancer with ER+, PR+, ER+PR+ and HER2−.
In conclusion, the present study suggests that the functional polymorphism G-875A in TGFBR2, but neither of the common genetic variants (C-509T and T-29C) in TGFB1, modify the predisposition to breast cancer in Chinese females. Notably, the TGFBR2 −875A allele is significantly associated with a decreased risk of breast cancer with ER+, PR+ and HER2− expression.
Acknowledgements
The authors sincerely acknowledge the participation and cooperation of the patients with NSCLC and the individuals with no history of cancer. This study was supported by grants from the Program for New Century Excellent Talents in University (NCET-09-0165), the Science and Technology Committee of Jiangsu Province (BK2008162), the SRF for ROCS, State Education Ministry (2008890), the Qing-Lan Project of Education Bureau of Jiangsu Province, the ‘333’ Project of Jiangsu Province Government, and the Soochow Scholar Project of Soochow University (all to H.-T. Zhang).
References
- 1.Ziegler RG, Anderson WF, Mh G. Increasing breast cancer incidence in China: the numbers add up. JNCI. 2008;100:1339–1341. doi: 10.1093/jnci/djn330. [DOI] [PubMed] [Google Scholar]
- 2.Linos E, Rosner BA, Linos K. Effects of reproductive and demographic changes on breast cancer incidence in China: a modeling analysis. JNCI. 2008;100:1352–1359. doi: 10.1093/jnci/djn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 4.Gobbi H, Arteaga CL, Jensen RA, Simpson JF, Dupont WD, Olson SJ, Schuyler PA, Plummer WD, Jr, Page DL. Loss of expression of transforming growth factor beta type II receptor correlates with high tumour grade in human breast in situ and invasive carcinomas. Histopathology. 2000;36:168–177. doi: 10.1046/j.1365-2559.2000.00841.x. [DOI] [PubMed] [Google Scholar]
- 5.Gobbi H, Dupont WD, Simpson JF, Plummer WD, Jr, Schuyler PA, Olson SJ, Arteaga CL, Page DL. Transforming growth factor-beta and breast cancer risk in women with mammary epithelial hyperplasia. J Natl Cancer Inst. 1999;91:2096–2101. doi: 10.1093/jnci/91.24.2096. [DOI] [PubMed] [Google Scholar]
- 6.Daniel CW, Robinson SD. Regulation of mammary growth and function by TGF-beta. Mol Reprod Dev. 1992;32:145–151. doi: 10.1002/mrd.1080320210. [DOI] [PubMed] [Google Scholar]
- 7.Barcellos-Hoff MH, Ewan KB. Transforming growth factor-beta and breast cancer: mammary gland development. Breast Cancer Res. 2000;2:92–99. doi: 10.1186/bcr40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White RL. Tumor suppressing pathways. Cell. 1998;92:591–592. doi: 10.1016/s0092-8674(00)81124-1. [DOI] [PubMed] [Google Scholar]
- 9.Cambien F, Ricard S, Troesch A, Mallet C, Generenaz L, Evans A, Arveiler D, Luc G, Ruidavets JB, Poirier O. Polymorphisms of the transforming growth factor-beta 1 gene in relation to myocardial infarction and blood pressure. The Etude Cas-Temoin de l’Infarctus du Myocarde (ECTIM) Study. Hypertension. 1996;28:881–887. doi: 10.1161/01.hyp.28.5.881. [DOI] [PubMed] [Google Scholar]
- 10.Seijo ER, Song H, Lynch MA, Jennings R, Qong X, Lazaridis E, Muro-Cacho C, Weghorst CM, Munoz-Antonia T. Identification of genetic alterations in the TGFbeta type II receptor gene promoter. Mutat Res. 2001;483:19–26. doi: 10.1016/s0027-5107(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Liu B, Jin M, Ni Q, Liang X, Ma X, Yao K, Li Q, Chen K. Genetic polymorphisms of transforming growth factor-beta1 and its receptors and colorectal cancer susceptibility: a population-based case-control study in China. Cancer Lett. 2009;275:102–108. doi: 10.1016/j.canlet.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Dunning AM, Ellis PD, McBride S, et al. A transforming growth factorbeta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003;63:2610–2615. [PubMed] [Google Scholar]
- 13.Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4–13. doi: 10.3121/cmr.2009.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaklamani VG, Baddi L, Liu J, et al. Combined genetic assessment of transforming growth factor-beta signaling pathway variants may predict breast cancer risk. Cancer Res. 2005;65:3454–3461. doi: 10.1158/0008-5472.CAN-04-2961. [DOI] [PubMed] [Google Scholar]
- 15.Paiva CE, Drigo SA, Rosa FE, Moraes Neto FA, Caldeira JR, Soares FA, Domingues MA, Rogatto SR. Absence of transforming growth factor-beta type II receptor is associated with poorer prognosis in HER2-negative breast tumours. Ann Oncol. 2010;21:734–740. doi: 10.1093/annonc/mdp518. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee SK, Makdisi WF, Weston AP, Mitchell SM, Campbell DR. Microwave-based DNA extraction from paraffin-embedded tissue for PCR amplification. Biotechniques. 1995;18:768–770. 772–763. [PubMed] [Google Scholar]
- 17.Sato Y, Sugie R, Tsuchiya B, Kameya T, Natori M, Mukai K. Comparison of the DNA extraction methods for polymerase chain reaction amplification from formalin-fixed and paraffin-embedded tissues. Diagn Mol Pathol. 2001;10:265–271. doi: 10.1097/00019606-200112000-00009. [DOI] [PubMed] [Google Scholar]
- 18.Qi X, Zhang F, Yang X, Fan L, Zhang Y, Chen L, Zhou Y, Chen X, Zhong L, Jiang J. Transforming growth factor-beta1 polymorphisms and breast cancer risk: a meta-analysis based on 27 case-control studies. Breast Cancer Res Treat. 2010;122:273–279. doi: 10.1007/s10549-010-0847-6. [DOI] [PubMed] [Google Scholar]
- 19.Hishida A, Iwata H, Hamajima N, Matsuo K, Mizutani M, Iwase T, Miura S, Emi N, Hirose K, Tajima K. Transforming growth factor B1 T29C polymorphism and breast cancer risk in Japanese women. Breast Cancer. 2003;10:63–69. doi: 10.1007/BF02967627. [DOI] [PubMed] [Google Scholar]
- 20.Cox DG, Penney K, Guo Q, Hankinson SE, Hunter DJ. TGFB1 and TGFBR1 polymorphisms and breast cancer risk in the Nurses’ Health Study. BMC Cancer. 2007;7:175. doi: 10.1186/1471-2407-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Q, Hemminki K, Grzybowska E, Klaes R, Soderberg M, Zientek H, Rogozinska-Szczepka J, Utracka-Hutka B, Pamula J, Pekala W, Forsti A. Polymorphisms and haplotype structures in genes for transforming growth factor beta1 and its receptors in familial and unselected breast cancers. Int J Cancer. 2004;112:94–99. doi: 10.1002/ijc.20370. [DOI] [PubMed] [Google Scholar]
- 22.Jin G, Wang L, Chen W, Hu Z, Zhou Y, Tan Y, Wang J, Hua Z, Ding W, Shen J, Zhang Z, Wang X, Xu Y, Shen H. Variant alleles of TGFB1 and TGFBR2 are associated with a decreased risk of gastric cancer in a Chinese population. Int J Cancer. 2007;120:1330–1335. doi: 10.1002/ijc.22443. [DOI] [PubMed] [Google Scholar]
- 23.Jin G, Deng Y, Miao R, Hu Z, Zhou Y, Tan Y, Wang J, Hua Z, Ding W, Wang L, Chen W, Shen J, Wang X, Xu Y, Shen H. TGFB1 and TGFBR2 functional polymorphisms and risk of esophageal squamous cell carcinoma: a case-control analysis in a Chinese population. J Cancer Res Clin Oncol. 2008;134:345–351. doi: 10.1007/s00432-007-0290-1. [DOI] [PubMed] [Google Scholar]
- 24.Knabbe C, Lippman ME, Wakefield LM, Flanders KC, Kasid A, Derynck R, Dickson RB. Evidence that transforming growth factor-beta is a hormonally regulated negative growth factor in human breast cancer cells. Cell. 1987;48:417–428. doi: 10.1016/0092-8674(87)90193-0. [DOI] [PubMed] [Google Scholar]
- 25.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 26.Buck M, von der Fecht J, Knabbe C. Antiestrogenic regulation of transforming growth factor beta receptors I and II in human breast cancer cells. Ann NY Acad Sci. 2002;963:140–143. doi: 10.1111/j.1749-6632.2002.tb04104.x. [DOI] [PubMed] [Google Scholar]
- 27.Arteaga CL, Tandon AK, von Hoff DD, Osborne CK. Transforming growth factor beta: potential autocrine growth inhibitor of estrogen receptor-negative human breast cancer cells. Cancer Res. 1988;48:3898–3904. [PubMed] [Google Scholar]
- 28.Kalkhoven E, Roelen BA, de Winter JP, Mummery CL, van den Eijnden-van Raaij AJ, van der Saag PT, van der Burg B. Resistance to transforming growth factor beta and activin due to reduced receptor expression in human breast tumor cell lines. Cell Growth Differ. 1995;6:1151–1161. [PubMed] [Google Scholar]