Abstract
Colorectal cancers exhibit a high telomerase activity, usually correlated with the hypermethylation of the promoter of its hTERT catalytic subunit. Although telomerase is not expressed in normal tissue, certain proliferative somatic cells such as intestinal crypt cells have demonstrated telomerase activity. The aim of this study was to determine whether a correlation exists between telomerase activity, levels of hTERT methylation and telomere length in tumoral and normal colorectal tissues. Tumor, transitional and normal tissues were obtained from 11 patients with a colorectal cancer. After bisulfite modification of genomic DNA, hTERT promoter methylation was analyzed by methylation-sensitive single-strand conformation analysis (MS-SSCA). Telomerase activity and telomere length were measured by a fluorescent-telomeric repeat amplification protocol assay and by Southern blotting, respectively. A significant increase of hTERT methylation and telomerase activity, and a reduction of the mean telomere length were observed in the tumor tissues compared to the transitional and normal mucosa. In the transitional and normal mucosa, telomerase activity was significantly lower than that in tumor tissues, even with high levels of hTERT methylation. Nevertheless, hTERT promoter methylation was not linearly correlated to telomerase activity. These data indicate that hTERT promoter methylation is a necessary event for hTERT expression, as is telomerase activity. However, methylation is not sufficient for hTERT activation, particularly in normal colorectal cells.
Keywords: telomerase, hTERT, promoter methylation, colorectal cancer, telomere length
Introduction
Telomerase activation is a critical step in human carcinogenesis to maintain the telomere length; however, the mechanisms for its activation and regulation remain to be determined. The expression of hTERT, the catalytic subunit of the telomerase enzyme, reveals a marked correlation to telomerase activity in tumors. In the majority of tumors, active telomerase requires the expression of hTERT. Furthermore, hTERT, which is tightly regulated, is not detectable in most somatic cells where telomerase activity is not present or is at basal level (1,2). hTERT expression may be regulated by various epigenetic factors, including promoter methylation or chromatin remodeling (3,4) and by activators and repressors such as C-Myc, Sp1, USF1/2, p53, TGF-β and CTCF (5–7).
One of the most crucial mechanisms by which hTERT expression is regulated occurs throughout DNA methylation in the hTERT promoter, which is embedded in a large CpG island. Hypermethylation of hTERT was found to be necessary for its expression in cancer cells (3,8). The apparent opposition to the classic model of regulation was recently clarified by the observation that methylation plays a dual role (9,10). hTERT methylation prevents the binding and thus the repressive effect of CTCF. However, a small hypomethylated region around the transcription start site allows a weak transcription of hTERT, despite the hypermethylation of the border regions. In most normal tissues, the telomerase activity is maintained at a basal level and it has been shown that these normal somatic cells have a hypomethylated promoter (3,11,12). Certain proliferative somatic cells, such as colorectal crypts as well as gastric and endometrial cells, revealed telomerase activity and hTERT expression concomitantly with the presence of hTERT methylation (3, JB, unpublished data). An increase of telomerase activity was observed in samples of normal, transitional and tumor mucosa from patients with sporadic colorectal cancer (13). Since in the colon hTERT methylation was detected in normal and tumor tissues, we aimed to investigate whether a relationship exists between the level of telomerase activity and the degree of hTERT promoter methylation in colorectal tissues. Similarly, a potential relationship between hTERT promoter methylation and telomere length was also determined.
Materials and methods
Tissue samples
This study involved 11 patients who underwent surgery for colorectal cancer at the Lleida Arnau de Vilanova Hospital. Of the 11 patients, 7 were male and 4 were female and their average age was 75.40 years (range 65–82). Surgically resected samples of cancer tissue and transitional and normal mucosa were immediately frozen in liquid nitrogen and stored at −80°C. The transitional mucosa was defined as having a macroscopically normal appearance and lying <1 cm away from the tumor, while the normal mucosa was obtained 10 cm away from the tumor. The research protocol was approved by the Hospital Clinic Research Ethics Committee of Lleida. Microdissection of two normal colon samples was performed as previously described in order to independently analyze the colon and stromal cells (14).
Fluorescent-telomeric repeat amplification protocol assay
Telomerase activity was measured using a quantitative system of the fluorescent-telomeric repeat amplification protocol assay (TRAP-F), using a TRAPeze® telomerase detection kit (InterGen) (15). The analysis was performed as described in a previous study (13).
Telomere length measurement by Southern blotting
The telomere restriction fragment (TRF) length was determined by Southern blotting (Telo TAGGG Telomere length assay, Roche Diagnostics GmbH, Mannheim, Germany) in the tumor, transitional and normal tissues. The analysis was performed as previously described (16). The telomere length was expressed in kbp. The telomere length ratio (TLR) was determined as the quotient between the TL in the tumor tissue and the TL in normal mucosa from the same patient. Telomere shortening and elongation were defined as TLs of carcinoma <80% and >120% of the corresponding normal mucosa, respectively.
Methylation-sensitive single-strand conformation analysis (MS-SSCA)
Bisulfite modification of 1 μg of genomic DNA (or 10 ng of DNA extracted from microdissected normal tissues) was performed using the EpiTect® Bisulfite kit (Qiagen) according to the manufacturer’s instructions. One twenty-fifth of modified DNA was used to amplify a 220 bp fragment of the hTERT promoter (17). PCR products were analyzed by methylation-sensitive single-strand conformation analysis (MS-SSCA) as previously described (18).
Results
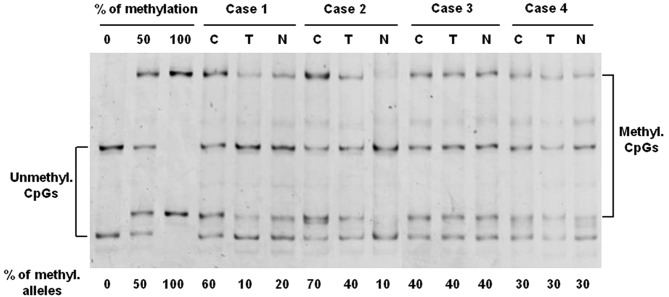
The methylation status of the hTERT promoter was analyzed, and the telomerase activity and telomere length were determined in normal, transitional and tumor samples from 11 patients who underwent surgery for a sporadic colorectal cancer. The analysis of hTERT promoter methylation was performed by MS-SSCA after bisulfite modification of genomic DNAs. Four cases are shown in Fig. 1. In all samples, a mixture of unmethylated and fully methylated alleles at varying ratios was observed. In cases 1 and 2, a significant difference in the level of hTERT methylation between tumor and normal tissues was detected. For example, in case 2, the level of methylation was approximately 70% in the tumor sample, whereas the methylation level reached only 40% in the transitional mucosa and only 10% in the normal mucosa. A similar level of methylation was observed in normal and tumor samples in cases 3 and 4. As shown in Table I, the mean level of hTERT methylation was higher in tumor samples (51%) compared with transitional (35%) and normal (20%) samples. After separation by microdissection of epithelial cells and stromal cells from two normal colon tissues, no methylation was detected in the hTERT promoter of stromal cells (data not shown). Therefore, the hTERT methylation observed in the normal colorectal tissues evidently originated from the colonic cells.
Figure 1.
Analysis of the hTERT promoter methylation by MS-SSCA. Four representative cases, with tumor, transitional and normal mucosa, are shown. Square brackets indicate the migration of the unmethylated DNA (Unmethyl. CpGs) and the hypermethylated DNA (Methyl. CpGs). The percentage of methylated alleles is reflected by the ratio of intensity between the methylated and unmethylated bands. The percentage of methylation is shown under each sample. C, cancer tissue; T, transitional tissue; N, normal tissue. 0, 50 and 100% is the scale of methylation.
Table I.
Mean age, methylation rate, telomerase activity and telomere length in tumor, transitional and normal mucosa.
| Characteristics | Mean | Range |
|---|---|---|
| Age | 75.4 years | 65–82 years |
| Methylation rate | ||
| Tumor mucosa | 51% | 10–100% |
| Transitional mucosa | 35% | 10–50% |
| Normal mucosa | 20% | 5–40% |
| Telomerase activity | ||
| Tumor mucosa | 44.20 | 0–164 |
| Transitional mucosa | 1.12 | 0–8 |
| Normal mucosa | 0.53 | 0–3 |
| Telomere length | ||
| Tumor mucosa | 7.66 kbp | 5.66–10.64 kbp |
| Transitional mucosa | 8.82 kbp | 6.80–10.95 kbp |
| Normal mucosa | 9 kbp | 6.99–11.02 kbp |
The telomerase activity was measured in arbitrary units.
Since all of the samples in this study contained at least 40–50% of colorectal cells, it is evident that in a certain number of samples only a subset of colorectal cells (normal or tumoral) harbored a methylated hTERT promoter. Therefore, in order to avoid insignificant methylation levels, a low level of hTERT methylation was arbitrarily considered when the percentage of methylation was <25%. As shown in Table II, the samples were divided into two categories: low level (<25%) and high level (≥25%) of methylation. In tumors, all but one of the samples exhibited a high level of methylation. The percentage of cases with a high level of methylation decreased to 73% in transitional samples, and 36% in normal samples.
Table II.
Low and high level of hTERT methylation in tumor and normal colorectal tissues.
| Met <25 (%) | Met ≥25 (%) | Total (%) | |
|---|---|---|---|
| Tumor mucosa | |||
| TA− | 0 (0) | 2 (18) | 2 (18) |
| TA+ | 1 (9) | 8 (73) | 9 (82) |
| Total | 1 (9) | 10 (91) | 11 (100) |
| Transitional mucosa | |||
| TA− | 2 (18) | 3 (27) | 5 (45) |
| TA+ | 1 (9) | 5 (46) | 6 (55) |
| Total | 3 (27) | 8 (73) | 11 (100) |
| Normal mucosa | |||
| TA− | 4 (36) | 3 (27) | 7 (64) |
| TA+ | 3 (27) | 1 (9) | 4 (36) |
| Total | 7 (64) | 4 (36) | 11(100) |
The level is considered as low when the percentage of hTERT methylation (Met) was <25% and as high when the percentage was >25%. Telomerase activity was measured in arbitrary units. Met, hTERT methylation; TA, telomerase activity.
The mean telomerase activity was 40–80 times higher in tumor samples than in transitional and normal mucosa, respectively, whereas the mean level of hTERT methylation was only 1.5–2.5 times higher in tumor samples (Table I). In normal and transitional tissues, a low level of telomerase activity was observed even in the presence of a high level of methylation (Table III).
Table III.
Means of telomerase activity and telomere length in tumor and normal colorectal tissues according to the degree of hTERT methylation.
| Met <25% | Met ≥25% | |
|---|---|---|
| Tumor mucosa | ||
| Telomerase activity | 15 | 47.1 |
| Telomere length | 5.66 kpb | 7.87 kpb |
| Transitional mucosa | ||
| Telomerase activity | 0.13 | 1.75 |
| Telomere length | 7.31 kpb | 8.46 kpb |
| Normal mucosa | ||
| Telomerase activity | 0.40 | 0.75 |
| Telomere length | 8.55 kpb | 9.69 kpb |
The level was considered as low when the percentage of hTERT methylation (Met) was <25%, and as high when the percentage was >25%. Telomerase activity was measured in arbitrary units.
A reduction in the mean telomere length was observed between normal (9 kbp), transitional (8.82 kbp), and tumor mucosa (7.66 kbp) (Table I). These results confirmed that tumor cells have shorter telomeres compared to normal cells (19). Notably, in the tumor, transitional and normal samples, the mean telomere length was always shorter in the samples with a low level of methylation than in those with a high level of methylation (Table III).
An increase of hTERT methylation and telomerase activity, and a reduction of mean telomere length were observed in the transitional mucosa in comparison to the distant normal mucosa. However, the significant increase of methylation in the transitional tissues was not concomitant with high telomerase activity, as observed in the tumor samples (Tables II and III). These data indicate that hTERT promoter methylation is not linearly correlated to telomerase activity in colorectal tissues.
Discussion
A significant increase of hTERT methylation and telomerase activity, and a reduction of mean telomere length were observed from normal to tumor tissue. This observation also proves to be true of transitional to tumor mucosa, but to a lesser extent. A correlation between the degree of methylation and telomerase activity was found in colorectal tumor samples. In other words, tumors with a high degree of hTERT methylation revealed high telomerase activity. An apparently different behavior occurred in transitional and normal mucosa: the telomerase activity was low in the two cases regardless of the methylation status of the hTERT promoter. Nevertheless, when the level of hTERT methylation was high, a higher level of telomerase activity was observed in transitional and normal samples. Therefore, in tumor and normal tissues, the higher the number of colorectal cells harboring an hTERT methylated promoter, the more significant the level of telomerase activity.
This study showed that hTERT promoter methylation is required for hTERT expression, and thus telomerase activity, but it is not sufficient to allow hTERT expression in normal colorectal cells. The data observed in this study are in accordance with those previously obtained in in vitro transcription experiments with hTERT reporter gene constructs. The transcriptional activity of the hTERT core promoter was found to be significantly higher in cancer cells than in normal cells (9). Considering the significance of this enzyme, it is clear that other factors should be recruited to modulate hTERT expression.
hTERT methylation detected in normal mucosa may come from the few stem cells and more largely from the daughter stem cells, known as niche cells, contained in the colorectal crypts. In previous studies, the cell expression of hTERT determined by immunohistochemistry indicated that hTERT is mainly expressed at the base of the crypts (20). The fact that hTERT in transitional mucosa was more methylated than in normal mucosa (mean of 35 versus 20%) indicates that some significant changes occur in normal colorectal cells in the vicinity of a tumor. This phenomenon may be due either to high cell renewal or to the occurrence of epigenetic modifications in the colorectal cells close to a cancer. Aberrant methylation of CpG island promoters is known to be an early event in colorectal carcinogenesis (21). Therefore, a favorable hTERT methylation context may initiate the first phase of carcinogenesis.
hTERT methylation was related, to a certain extent, to the telomerase activity in normal and tumor colorectal tissues. Low but significant levels of hTERT methylation and weak telomerase activity are already present in the normal colorectal cells. A slight increase of hTERT methylation, concomitantly with a great change in telomerase activity level, occurs in tumor cells. In conclusion, this study indicates that hTERT promoter methylation is required but not sufficient for hTERT expression and telomerase activity in colorectal cells.
Acknowledgements
This study was supported in part by grant 2006BE-00187 from the Generalitat de Catalunya (AGAUR).
References
- 1.Meyerson M, Counter CM, Eaton EN, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura TM, Morin GB, Chapman KB, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 3.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101:335–341. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
- 4.Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goueli BS, Janknecht R. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene. 2003;22:8042–8047. doi: 10.1038/sj.onc.1206847. [DOI] [PubMed] [Google Scholar]
- 6.Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- 7.Renaud S, Loukinov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005;33:6850–6860. doi: 10.1093/nar/gki989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nomoto K, Maekawa M, Sugano K, et al. Methylation status and expression of human telomerase reverse transcriptase mRNA in relation to hypermethylation of the p16 gene in colorectal cancers as analyzed by bisulfite PCR-SSCP. Jpn J Clin Oncol. 2002;32:3–8. doi: 10.1093/jjco/hyf001. [DOI] [PubMed] [Google Scholar]
- 9.Renaud S, Loukinov D, Abdullaev Z, et al. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35:1245–1256. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- 11.Dessain SK, Yu HY, Reddel RR, Beijersbergen RL, Weinberg RA. Methylation of the human telomerase gene CpG island. Cancer Res. 2000;60:537–541. [PubMed] [Google Scholar]
- 12.Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–6090. [PubMed] [Google Scholar]
- 13.Bautista CV, Felis CP, Espinet JM, Garcia JB, Salas JV. Telomerase activity is a prognostic factor for recurrence and survival in rectal cancer. Dis Colon Rectum. 2007;50:611–620. doi: 10.1007/s10350-006-0820-y. [DOI] [PubMed] [Google Scholar]
- 14.Baisse B, Bian YS, Benhattar J. Microdissection by exclusion and DNA extraction for multiple PCR analyses from archival tissue sections. Biotechniques. 2000;28860:856–8. 862. [PubMed] [Google Scholar]
- 15.Kim NW, Piatyszek MA, Prowse KR, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 16.Bautista CV, Felis CP, Rene Espinet JM, Garcia JB, Salas JV. Telomere length is a prognostic factor for overall survival in colorectal cancer. Colorectal Dis. 2010 Sep 27; (E-pub ahead of print) [Google Scholar]
- 17.Clement G, Benhattar J. A methylation sensitive dot blot assay (MS-DBA) for the quantitative analysis of DNA methylation in clinical samples. J Clin Pathol. 2005;58:155–158. doi: 10.1136/jcp.2004.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian YS, Yan P, Osterheld MC, Fontolliet C, Benhattar J. Promoter methylation analysis on microdissected paraffin embedded tissues using bisulfite treatment and PCR-SSCP. Biotechniques. 2001;30:66–72. doi: 10.2144/01301st02. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Aranda C, De JC, Diaz-Lopez A, et al. Correlations of telomere length, telomerase activity, and telomeric-repeat binding factor 1 expression in colorectal carcinoma. Cancer. 2006;106:541–551. doi: 10.1002/cncr.21625. [DOI] [PubMed] [Google Scholar]
- 20.Yan P, Benhattar J, Seelentag W, Stehle JC, Bosman FT. Immunohistochemical localization of hTERT protein in human tissues. Histochem Cell Biol. 2004;121:391–397. doi: 10.1007/s00418-004-0645-5. [DOI] [PubMed] [Google Scholar]
- 21.Kim YH, Petko Z, Dzieciatkowski S, et al. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45:781–789. doi: 10.1002/gcc.20341. [DOI] [PubMed] [Google Scholar]