Abstract
14-3-3σ, one of the 14-3-3 family members, was initially identified as a human mammary epithelium-specific marker 1. The expression of 14-3-3σ is directly regulated by p53. It has been demonstrated that 14-3-3σ stabilizes p53 and enhances its transcriptional activity through the interaction with p53, suggesting that 14-3-3σ has a positive feedback effect on p53. Our previous study showed that 14-3-3σ is a direct transcriptional target of p73 and enhances the p73-mediated transcriptional as well as pro-apoptotic activity in vitro. In the present study, we explored the tumor-suppressive effect of 14-3-3σ by establishing a breast cancer xenograft nude mouse model with an inducible expression of 14-3-3σ or with an inducible expression of p53/p73 plus 14-3-3σ with ADR treatment. Tumor formation was then assayed. Moreover, 66 primary breast cancer specimens and paired tumor-free breast specimens obtained from the female patients were examined. Results showed that the expression of p73 and 14-3-3σ in breast cancer specimens was significantly lower than the tumor-free breast specimens and that 14-3-3σ expression was positively correlated with the expression of p73. Furthermore, overexpression of 14-3-3σ counteracts tumorigenicity by positively regulating p73 in p53-mutated or -deficient cancers in vivo. Therefore, our results may lead to the use of 14-3-3σ in the therapeutic application for the p53-mutated and p73-expressed breast cancer patients.
Keywords: breast cancer, 14-3-3σ, p53, p73
Introduction
The 14-3-3 protein family, which comprises at least seven isoforms, is a class of highly conserved proteins involved in regulating signal transduction pathways, cellular proliferation, differentiation and survival (1–5). Among the 14-3-3 proteins, 14-3-3σ is the isoform most directly linked to cancer and was initially identified as a human mammary epithelium-specific marker (6). The expression levels of 14-3-3σ are significantly lower in breast cancer tissues than those in normal breast tissues, which may be due to the hypermethylation of CpG islands in the 14-3-3σ gene promoter (7–13).
14-3-3σ appears to be regulated by different mechanisms following DNA damage, which contributes directly to cancer development. The major regulator of 14-3-3σ is the tumor suppressor p53 (14). Upon DNA damage, p53 becomes dephosphorylated and is able to bind the promoter region 1.8 kb upstream of the 14-3-3σ transcription start site. The subsequent activation and increased expression of 14-3-3σ leads to the arrest of G2/M, allowing sufficient time for DNA repair (15–17). Yang et al demonstrated that 14-3-3σ stabilizes p53 and enhances its transcriptional activity through the interaction with p53, suggesting that 14-3-3σ has a positive feedback effect on p53 (18). However, p53 is mutated in over 50% of all human cancers (19–21). In our previous study, 14-3-3σ was found to be a direct transcriptional target of p73 and enhanced the p73-mediated transcriptional activation as well as pro-apoptotic activity (22). Thus, p73 may functionally replace p53 to induce 14-3-3σ in p53-deficient breast cancer cells (23). It is likely that the p73/14-3-3σ pathway plays an important role in the regulation of DNA damage-induced apoptosis in certain breast cancer cells bearing p53.
In the present study, we aimed to determine the possible link between the expression of p73 and 14-3-3σ in the clinical breast specimens and the regulating mechanism of p73/14-3-3σ in vivo.
Materials and methods
Clinical specimens
A total of 132 samples, 66 primary breast cancer specimens and paired tumor-free breast specimens, analyzed in our study were obtained from the same patients who had invasive breast cancer and underwent surgical treatment at the Breast Center, the Fourth Clinical Hospital of Hebei Medical University, China, in 2007. The patients, aged between 30 and 76 years, did not undergo preoperative adjuvant chemotherapy and radiotherapy. Following surgery, all 66 specimens were sent to the pathology department of the hospital to be fixed and paraffin-embedded for routine immunohistochemistry (IHC) analysis. Clinical data were collected retrospectively from patient files and pathology reports, including clinical stages, tumor size, pathological types, histological grading, status of axillary nodes, ER status and c-erbB2 status. Patients provided written informed consent prior to enrollment.
Immunohistochemistry
Sections (5 μm) were deparaffinized from the formalin-fixed and paraffin-embedded tissue blocks. The sections were then heated in a microwave oven for 5 min in 10 mmol/l Na-citric buffer (pH 6.0) for antigen retrieval and washed with phosphate-buffered saline (PBS, pH 7.2). The sections were immersed in 0.3% hydrogen peroxide in methanol for 20 min to suppress endogenous peroxidase activity. After washing with PBS, the sections were incubated in 1:10 diluted normal goat serum at room temperature in a humidified chamber for 30 min to prevent non-specific immuno globulin binding. The sections were then treated with the monoclonal anti-p73 (Ab-4, NeoMarkers, Fremont, CA, USA) or with polyclonal anti-14-3-3σ (C-18, Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibody at 4°C overnight. Normal IgG, which replaced the primary antibody, served as the control. A streptoavidin-biotinylated horseradish peroxidase-based detection system was used to reveal specific binding. Sections were counterstained with hematoxylin for light microscopic review and evaluation.
p73 and 14-3-3σ were always positively detected in both the cytoplasm and the nucleus of primary breast cancer specimens and tumor-free breast specimens. Immunoreactivity was measured for p73 and 14-3-3σ and scored in the following way: -, no positive cells (negative); +, <20% positive cells (‘mild reaction’); ++, 21–50% positive cells (‘moderate reaction’); and +++, >50% positive cells (‘strong reaction’). The immunoreactivity scores for p73 were denoted as either ‘negative’ or ‘positive’, with positive including mild, moderate and strong reactions. The immunoreactivity scores for 14-3-3σ were defined as either ‘negative’ or ‘positive’, with positive including moderate and strong reactions. The percentage of positive cells and staining intensity were scored by 2 independent observers.
Cell culture and cells transfection in plasmid
Human breast cancer-derived MDA-MB-231 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 50 units of penicillin and 50 g/ml streptomycin. Cells were grown at 37°C in a water-saturated atmosphere of 5% CO2 in air. Cells were transfected with empty, p53 expression, p73 expression, 14-3-3σ expression, p53 expression plasmid plus 14-3-3σ expression plasmid or with p73 expression plasmid plus 14-3-3σ expression plasmid using a Lipofectamine 2000 transfection reagent according to the manufacturer’s recommendations (Invitrogen). The expression plasmids were provided by Professor Nakagawara of the Chiba Cancer Center.
RNA extraction and RT-PCR
Total RNA of cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The RNA concentration was routinely measured by a spectrophotometer and its quality was evaluated by visualization following agarose gel electrophoresis and ethidium bromide staining. Total RNA (1μg) was used to generate the first strand cDNA using random primers and SuperScript II reverse transcriptase (Invitrogen). Reverse transcription was carried out at 42°C for 1 h. The resultant cDNAs were amplified by PCR-based strategy using rTaq DNA polymerase (Takara, Ohtsu, Japan). cDNAs of p53, p73, 14-3-3σ and GAPDH were amplified by using primers as shown in Table I. The expression of GAPDH was measured as an internal control. PCR products were separated on 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
Table I.
Primer pairs used for amplification reactions.
| Genes | Primers | Annealing (°C) | Cycles |
|---|---|---|---|
| p53 | F: 5′-CTGCCCTCAACAAGATGTTTTG-3′ R: 5′-CTATCTGAGCAGCGCTCATGG-3′ |
58 | 28 |
| p73 | F: 5′-TCTGGAACCAGACAGCACCT-3′ R: 5′-GTGCTGGACTGCTGGAAAGT-3′ |
58 | 28 |
| 14-3-3σ | F: 5′-GAGCGAAACCTGCTCTCAGT-3′ R: 5′-CTCCTTGATGAGGTGGCTGT-3′ |
58 | 28 |
| GAPDH | F: 5′-ACCTGACCTGCCGTCTAGAA-3′ R: 5′-TCCACCACCCTGTTGCTGTA-3′ |
58 | 22 |
Experimental animals
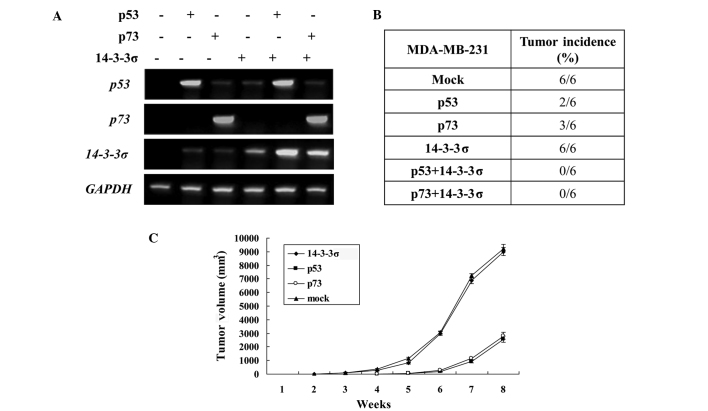
All experiments with animals were conducted according to the guidelines of our Institutional Animal Ethics Committee. Thirty-six 3-week old female nude mice were purchased from the Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences (CAMS) and maintained in the Animal Center, the Fourth Hospital of Hebei Medical University, China. The mice were randomly divided into 6 groups (n=6 per group). MDA-MB-231 cells were transfected with the indicated combination of different expression plasmids. The stable clones were subsequently established, and the effects were examined with RT-PCR (Fig. 2A). MDA-MB-231 cells (5×106) in 0.2 ml of PBS were harvested and injected subcutaneously into the flank region of the female nude mice. Mice were observed daily and palpated for tumor formation per one week. When the tumor masses reached 50–80 mm3, the mice were administered intraperitoneal injections of adriamycin (ADR) (15 mg/kg per day). Two-dimensional tumor measurements were made daily after tumor formation. The tumor volume was calculated according to the formula: volume = π (short diameter2) × (long diameter)/6. After 8 weeks, the mice were sacrificed by cervical dislocation. Tumors were excised and used for IHC as described. The growth curve of the tumors was drawn.
Figure 2.
Overexpression of 14-3-3σ suppresses tumorigenicity by positively regulating p53 or p73 in nude mice. (A) MDA-MB-231 cells were transfected with the indicated combination of different expression plasmids and the stable clones were subsequently established. The transfection efficacy was evaluated by RT-PCR. (B) Tumorigenicity. MDA-MB-231 cells (5×106) in 0.2 ml of PBS were harvested and injected subcutaneously into the flank region of female nude mice. Tumor volumes were measured and recorded per one week. After 8 weeks, the mice were sacrificed by cervical dislocation. (C) The growth curve of the tumors is shown.
Statistical analysis
Data were analyzed with SPSS 13.0 software. Pearson’s χ2 test or continuity corrected Pearson’s χ2 test was applied for the data analysis, expressed as qualitative values. The κ consistency test was used to analyze the correlation between the expression of p73 and 14-3-3σ in tumor-free breast and breast cancer specimens. One way analysis of variance (ANOVA) was adopted for determine the data of tumor growth. P<0.05 was considered to be statistically significant.
Results
General
Our results, explained in detail below, showed that the expression of both p73 and 14-3-3σ was significantly lower than that in tumor-free breast specimens. Moreover, 14-3-3σ expression was positively correlated with the expression of p73. Furthermore, overexpression of 14-3-3σ counteracts tumorigenicity by positively regulating p73 in p53-mutated or -deficient cancers in vivo. These results may lead to the use of 14-3-3σ in the therapeutic application for the p53-mutated and p73-expressed breast cancer patients.
Clinical data
A total of 132 samples were obtained as paired samples of tumor and tumor-free breast tissues from 66 individuals. The patients were females, aged between 30 and 76 years. The following clinical data were collected retrospectively from patient files and pathology reports: clinical stages, tumor size, pathological types, histological grading, status of axillary nodes, ER status and c-erbB2 status. The tumor samples included 47 infiltrating ductal carcinoma, 17 infiltrating lobular carcinoma, 1 medullary carcinoma and 1 mucinous adenocarcinoma. There were 16 cases in Stage I, 27 cases in Stage II and 23 cases in Stage III. Patient characteristics are shown in Table III.
Table III.
Relationship between p73 or 14-3-3σ expression and clinical parameters of breast cancer patients.
| p73/IHC | 14-3-3σ/IHC | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Clinical parameters | − | +, ++ | P-valuea | −, + | ++, +++ | P-valuea |
| Tumor size (cm) | 0.848 | 0.936 | ||||
| ≤2 | 5 | 3 | 6 | 2 | ||
| >2 and ≤5 | 26 | 10 | 25 | 11 | ||
| >5 | 15 | 7 | 15 | 7 | ||
| Clinical stage | 0.994 | 0.744 | ||||
| I | 11 | 5 | 10 | 6 | ||
| II | 19 | 8 | 19 | 8 | ||
| III | 16 | 7 | 17 | 6 | ||
| Pathological type of carcinoma | 0.671 | 0.817 | ||||
| Infitrating ductal carcinoma | 31 | 16 | 32 | 15 | ||
| Infiltrating lobular carcinoma | 13 | 4 | 12 | 5 | ||
| Medullary carcinoma | 1 | 0 | 1 | 0 | ||
| Mucinous adenocarcinoma | 1 | 0 | 1 | 0 | ||
| Histological grading | 0.392 | 0.151 | ||||
| I | 5 | 3 | 5 | 3 | ||
| II | 29 | 9 | 30 | 8 | ||
| III | 12 | 8 | 11 | 9 | ||
| Axillary lymph nodes | 0.671 | 0.929 | ||||
| − | 25 | 12 | 26 | 11 | ||
| + | 21 | 8 | 20 | 9 | ||
| ER | 0.793 | 0.348 | ||||
| − | 26 | 12 | 25 | 13 | ||
| + | 20 | 8 | 21 | 17 | ||
| HER-2 | 0.485 | 0.04 | ||||
| −, ++ | 21 | 11 | 17 | 15 | ||
| +++ | 25 | 9 | 29 | 5 | ||
Continuity corrected Pearson’s χ2 test. IHC, immunohistochemistry.
Expression of p73 and 14-3-3σ in tumor-free breast and breast cancer specimens
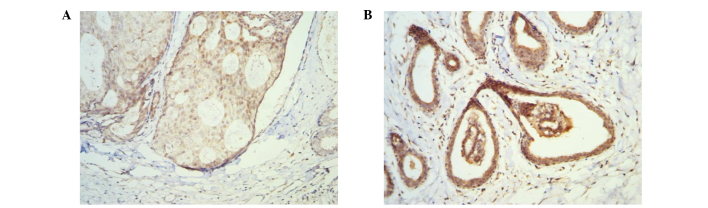
To examine the expression patterns of p73 and 14-3-3σ in breast tissues, the immunohistochemical staining with p73 and 14-3-3σ antibodies was carried out in 66 paired tumor-free breast and primary breast cancer specimens. As shown in Fig. 1, p73 and 14-3-3σ were always positively detected in the cytoplasm and the nucleus. Overall, 50 out of 66 tumor-free breast specimens (75.8%) were found to be positive (+, ++) with p73 antibody, whereas only 20 out of 66 breast cancer specimens (30.3%) were found to be positive (+, ++) with p73 antibody (Table II). The expression of p73 in tumor-free breast specimens was significantly higher than that in breast cancer specimens (P=0.000). Forty-two out of 66 tumor-free breast specimens (66.7%) were found to be positive (++, +++) with the 14-3-3σ antibody, whereas only 20 out of 66 breast cancer specimens (30.3%) were found to be positive (++, +++) with the 14-3-3σ antibody (Table II). The expression of 14-3-3σ in tumor-free breast specimens was significantly higher than that in breast cancer specimens (P=0.000).
Figure 1.
Immunohistochemical analysis of the expression of p73 (magnification, ×20) (A) and (B) 14-3-3σ (magnification, ×20) in human breast cancer specimens. Both p73 and 14-3-3σ were observed in the cytoplasm and nucleus.
Table II.
Expression of p73 and 14-3-3σ in 66 breast cancer specimens and tumor-free breast specimens detected by immuno-histochemistry.
| p73/IHC | 14-3-3σ/IHC | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Tissues | − | +, ++ | P-valuea | −, + | ++, +++ | P-valuea |
| Tumor-free breast | 16 | 50 | 24 | 42 | ||
| Breast cancer | 46 | 20 | 0.000 | 46 | 20 | 0.000 |
Pearson’s χ2 test. IHC, immunohistochemistry.
Relationship between p73 or 14-3-3σ expression and clinical parameters of breast cancer patients
The relationship between p73 or 14-3-3σ expression with clinical parameters of breast cancer patients was statistically evaluated with Pearson’s χ2 test or the continuity corrected Pearson χ2 test. As shown in Table III, no correlation was observed between p73 expression and clinical parameters of breast cancer patients such as tumor size, clinical stage, pathological types, histological grading, status of axillary lymph nodes, ER status and c-erbB2 status (P>0.05). 14-3-3σ expression was not associated with tumor size, clinical stage, pathological types, histological grading, status of axillary lymph nodes and ER status (P>0.05), but was negatively associated with the c-erbB2 status of breast cancer patients (P=0.04).
Correlations between the expression of p73 and 14-3-3σ in tumor-free breast and breast cancer specimens
In the present study, we aimed to investigate the link between p73 and 14-3-3σ in breast tissues. As shown in Table IV, 37 out of 50 (74%) p73-positive (+, ++) tumor-free breast specimens showed a positive 14-3-3σ expression (++, +++). A total of 11 out of 16 (68.8%) p73-negative (−) tumor-free breast specimens showed a negative 14-3-3σ expression (−, +). As shown in Table V, 14 out of 20 (70%) p73-positive (+, ++) breast cancer specimens showed a positive 14-3-3σ expression (++, +++). A total of 40 out of 46 (87.0%) p73-negative (−) breast cancer specimens showed a negative 14-3-3σ expression (−, +). These results suggest that the 14-3-3σ expression was positively correlated with the expression of p73 in both tumor-free breast and breast cancer specimens.
Table IV.
Correlation between the expression of p73 and 14-3-3σ in tumor-free breast specimens detected by immunohistochemistry.
| 14-3-3σ/IHC | |||
|---|---|---|---|
|
|
|||
| p73 | −, + | ++, +++ | P-valuea |
| − | 11 | 5 | |
| +, ++ | 13 | 37 | 0.000 |
κ consistency test. IHC, immunohistochemistry.
Table V.
Correlation between the expression of p73 and 14-3-3σ in breast cancer specimens detected by immunohistochemistry.
| 14-3-3σ/IHC | |||
|---|---|---|---|
|
|
|||
| p73 | −, + | ++, +++ | P-valuea |
| − | 40 | 6 | |
| +, ++ | 6 | 14 | 0.000 |
κ consistency test. IHC, immunohistochemistry.
Overexpression of 14-3-3σ counteracts tumorigenicity by positively regulating p73 and p53 in vivo
The most important tumor suppressor p53 has been successfully employed as a molecular target for cancer gene therapy. Yang et al demonstrated that an enforced expression of 14-3-3σ suppressed breast tumor growth by positively regulating p53 (5). However, p53 is highly mutated in over 50% of human cancers. The p53 family member p73 has been found to promote cell cycle arrest and/or apoptosis in response to DNA damage similar to p53, whereas it is rarely mutated in human cancers. Therefore, p73 may be an effective molecular target for cancer gene therapy in p53-mutated or -deficient cancers. Findings of our previous study indicated that 14-3-3σ increased the stability and activity of p73. Therefore, we hypothesized that the overexpression of 14-3-3σ counteracted tumorigenicity by positively regulating p73 in p53-mutated or -deficient cancers. To explore the tumor-suppressive effect of 14-3-3σ, we established a breast cancer xenograft nude mouse model with an inducible expression of 14-3-3σ or with an inducible expression of p53/p73 plus 14-3-3σ with ADR treatment. Tumor formation was then assayed (Fig. 2B and C). Tumorigenicity was found in all six mice of the mock control group and 14-3-3σ expressing group in the 2nd week. Markedly less tumorigenicity was found in the p53- (2/6) and p73- (3/6) expressing group than in the control group, and tumorigenicity was started from the 4th week, suggesting that an enforced expression of p53 or p73 was capable of eradicating tumorigenicity in breast cancer cells. However, tumorigenicity was observed not altered in the 14-3-3σ-expressing group (6/6) as compared with the control group, suggesting that an enforced expression of 14-3-3σ alone was not sufficient to eradicate tumorigenicity in breast cancer cells. No tumorigenicity was observed in the cells with an expression of 14-3-3σ plus expression of p53/p73, suggesting that the overexpression of 14-3-3σ counteracts tumorigenicity by positively regulating p73 and p53 in vivo. Taken together, our present results strongly suggested that the p73/14-3-3σ pathway plays an important role in p53-mutated or deficient breast cancer tumorigenicity.
Discussion
14-3-3σ, which belongs to the 14-3-3 family, was initially identified as a human mammary epithelium-specific marker 1 (24). By using a proteomic approach, Vercoutter-Edouart et al found that 14-3-3σ is strongly down-regulated in breast cancer in comparison to normal breast tissue (25). Mounting evidence has shown that the down-regulation of 14-3-3σ in breast cancer is due to the hypermethylation of CpG islands in the 14-3-3σ gene (7–13).
14-3-3σ has been found to be strongly induced in response to DNA damage, and its expression is directly regulated by p53. Furthermore, 14-3-3σ stabilizes p53 and directly increases its transcriptional activity through the interaction with p53, suggesting a positive feedback loop between 14-3-3σ and p53 (18). In our previous study, 14-3-3σ was found to be a direct transcriptional target of p73 that enhances the p73-mediated transcriptional as well as pro-apoptotic activity (22). The purpose of the present study was to find a possible link between p73 and 14-3-3σ expression in clinical breast specimens and to characterize the regulating mechanism of p73/14-3-3σ in vivo. We performed the immunohistochemical staining with p73 and 14-3-3σ antibodies in 66 paired tumor-free breast and primary breast cancer specimens, to analyze the expression patterns of p73 and 14-3-3σ and their correlation in breast cancer. The results show that the expression of p73 and 14-3-3σ in breast cancer specimens was significantly lower than the expression in tumor-free breast specimens. Moreover, 14-3-3σ expression is positively correlated with the expression of p73 in tumor-free breast and breast cancer specimens. Furthermore, the increased 14-3-3σ expression is associated with a low expression of c-erbB2, suggesting that a high expression of 14-3-3σ plays a protective role in the development of breast cancer.
Results of our previous study showed a positive feedback regulation between p73 and 14-3-3σ in vitro. Cosequently, we hypothesized that the overexpression of 14-3-3σ may counteract tumorigenicity by positively regulating p73 in p53-mutated or -deficient cancers. To prove our hypothesis, the breast cancer xenograft nude mouse model was established with an inducible expression of 14-3-3σ or an inducible expression of p53/p73 plus 14-3-3σ with ADR. The tumor formation assay shows that overexpression of 14-3-3σ alone is not sufficient to eradicate tumorigenicity in breast cancer cells. However, a tumor-suppressive effect was evident only in the presence of p53 or p73. Therefore, our study demonstrates that an enforced expression of 14-3-3σ may suppress breast tumor growth by positively regulating p73 in p53-mutated breast cancers.
Acknowledgements
This study was supported by the National Nature Science Foundation of China (No. 81001178). We thank Dr Nakagawara of the Chiba Cancer Center for the p53, p73 and 14-3-3σ expression plasmids. We would also like to extend our thanks to Dr Qianglin Duan, a skilled English proofreader from Tongji University for revision of this study.
References
- 1.Mhawech P. 14-3-3 proteins - an update. Cell Res. 2005;15:228–236. doi: 10.1038/sj.cr.7290291. [DOI] [PubMed] [Google Scholar]
- 2.Rosenquist M. 14-3-3 proteins in apoptosis. Braz J Med Biol Res. 2003;36:403–408. doi: 10.1590/s0100-879x2003000400001. [DOI] [PubMed] [Google Scholar]
- 3.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 4.Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 5.Van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- 6.Hermeking H. The 14-3-3 cancer connection. Nature Rev. 2003;3:931–943. doi: 10.1038/nrc1230. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson AT, Evron E, Umbricht CB, et al. High frequency of hypermethylation at the 14-3-3σ locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA. 2000;97:6049–6054. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodygin D, Hermeking H. The role of epigenetic inactivation of 14-3-3sigma in human cancer. Cell Res. 2005;15:237–246. doi: 10.1038/sj.cr.7290292. [DOI] [PubMed] [Google Scholar]
- 9.Iwata N, Yamamoto H, Sasaki S, et al. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene. 2000;19:5298–5302. doi: 10.1038/sj.onc.1203898. [DOI] [PubMed] [Google Scholar]
- 10.Gasco M, Bell AK, Heath V, et al. Epigenetic inactivation of 14-3-3 sigma in oral carcinoma: association with p16 (INK4a) silencing and human papillomavirus negativity. Cancer Res. 2002;62:2072–2076. [PubMed] [Google Scholar]
- 11.Gasco M, Sullivan A, Repellin C, et al. Coincident inactivation of 14-3-3sigma and p16INK4a is an early event in vulval squamous neoplasia. Oncogene. 2002;21:1876–1881. doi: 10.1038/sj.onc.1205256. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14-3-3 sigma gene is associated with 5V CpG island hypermethylation in human cancers. Cancer Res. 2000;60:4353–4357. [PubMed] [Google Scholar]
- 13.Ikeda K, Inoue S. Estrogen receptors and their downstream targets in cancer. Arch Histol Cytol. 2004;67:435–442. doi: 10.1679/aohc.67.435. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Zhao R, Lee MH. 14-3-3sigma, a p53 regulator, suppresses tumor growth of nasopharyngeal carcinoma. Mol Cancer Ther. 2006;5:253–260. doi: 10.1158/1535-7163.MCT-05-0395. [DOI] [PubMed] [Google Scholar]
- 15.Hermeking H, Lengauer C, Polyak K, et al. 14-3-3 σ is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 16.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 17.Laronga C, Yang HY, Neel C, Lee MH. Association of the cyclin-dependent kinases and 14-3-3sigma negatively regulates cell cycle progression. J Biol Chem. 2000;275:23106–23112. doi: 10.1074/jbc.M905616199. [DOI] [PubMed] [Google Scholar]
- 18.Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14-3-3 σ positively regulates p53 and suppresses tumor growth. Mol Cell Biol. 2003;23:7096–7107. doi: 10.1128/MCB.23.20.7096-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikawa Z, Nakagawara A, Ikawa Y. p53 family genes: structural comparison, expression and mutation. Cell Death Differ. 1999;6:1154–1161. doi: 10.1038/sj.cdd.4400631. [DOI] [PubMed] [Google Scholar]
- 20.Cattoretti G, Rilke F, Andreola S, D’ Amato L, Domenico D. p53 in breast cancer. Int J Cancer. 1988;41:178–183. doi: 10.1002/ijc.2910410204. [DOI] [PubMed] [Google Scholar]
- 21.Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci USA. 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sang M, Li Y, Ozaki T, et al. p73-dependent induction of 14-3-3 σ increases the chemo-sensitivity of drug-resistant human breast cancers. Biochem Biophys Res Commun. 2006;347:327–333. doi: 10.1016/j.bbrc.2006.06.079. [DOI] [PubMed] [Google Scholar]
- 23.Vayssade M, Haddada H, Faridoni-Laurens L, Tourpin S, Valent A, Bénard J, Ahomadegbe JC. p73 functionally replaces p53 in Adriamycin-treated, p53-deficient breast cancer cells. Int J Cancer. 2005;116:860–869. doi: 10.1002/ijc.21033. [DOI] [PubMed] [Google Scholar]
- 24.Prasad GL, Valverius EM, McDuffie E, Cooper HL. Complementary DNA cloning of a novel epithelial cell marker protein, Hmel, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ. 1992;3:507–513. [PubMed] [Google Scholar]
- 25.Vercoutter-Edouart AS, Lemoine J, Le Bourhis X, et al. Proteomic analysis reveals that 14-3-3σ is down-regulated in human breast cancer cells. Cancer Res. 2001;61:76–80. [PubMed] [Google Scholar]