Abstract
Mutations of the epidermal growth factor receptor (EGFR) in patients with non-small cell lung cancer (NSCLC) were identified by re-sequencing all exons of this gene to evaluate the frequencies of EGFR gene mutation and identify rare or novel EGFR mutations. A total of 55 NSCLC samples from 55 patients were included in the study. Genomic DNA was extracted and exons 1–28 of the EGFR gene were sequenced to identify mutations. The cDNA of the EGFR gene with P848L and T790M double mutants was constructed by introducing point mutations into the wild-type EGFR vector using a site-directed mutagenesis kit. Among the 55 patients with NSCLC, 8 patients carried mutations of the EGFR gene. Notably, of the mutation-harboring patients with a pathological type of adenocarcinoma, 6 were non-smokers. The in vitro study demonstrated that the P848L mutant had a similar response to that of the wild-type EGFR after gefitinib treatment, and the P848L and T790M double mutant exhibited high resistance to gefitinib. These EGFR mutations preferentially occurred in lung adenocarcinoma patients, most of whom were non-smokers. In the in vitro study, P848L mutant EGFR had a similar response as the wild-type EGFR to gefitinib treatment, suggesting that lung cancer patients with a rare mutation of EGFR, such as the P848L mutation, do not respond to gefitinib treatment.
Keywords: non-small cell lung cancer, epidermal growth factor receptor, mutation, functional study
Introduction
Lung cancer is the leading cause of cancer deaths worldwide (1). Despite the improvements in diagnosis and treatment, the prognosis of patients with advanced-stage non-small cell lung cancer (NSCLC) remains a challenge, with a 5-year survival of less than 15%. Thus, novel treatment strategies are urgently required.
Studies investigating the mechanism of oncogenesis showed that NSCLC is a disease with multigenetic abnormalities. Among these abnormalities, the epidermal growth factor receptor (EGFR) mutation has received considerable attention for its marked response to small molecule tyrosine kinase inhibitor (TKI) treatment. EGFR, together with HER2/neu (erbB2), HER3 (erbB3) and HER4 (erbB4), belongs to the erbB family of receptor tyrosine kinase proteins. It contains an extracellular ligand-binding domain, a transmembrane lipophilic domain and an intracellular tyrosine kinase domain, and is critical for cell proliferation, differentiation, motility and metastasis.
Somatic mutations in the EGFR kinase domain have mainly been identified in a subset of NSCLC patients. The subset included females, never smokers, patients with adenocarcinoma histology and patients of an East Asian origin (2,3). The presence of these mutations in NSCLC correlates with the sensitivity to TKI treatment, including gefitinib and erlotinib. A number of distinct mutations have been identified, most of which cluster around the tyrosine kinase domain. The in-frame deletion in exon 19 (del-19) accounts for 45% of the mutations, while the point mutation L858R in exon 21 accounts for 40–45% of the EGFR mutations in lung cancer (4–6). Patients with these mutations have a marked response to EGFR TKIs. Thus, these mutations are usually termed classical mutations (7). Due to the optimal response in this subtype of NSCLC patients, EGFR mutation screening for the selection of TKI therapy has been recommended in the clinical care of NSCLC patients. In addition to classical mutations, other rare EGFR mutations in exons 18–21 have also been reported, and patients with these non-classical mutations have variable responses to EGFR TKIs (8,9). Among these rare mutations is the EGFR-P848L mutation, which was first reported by Sequist et al (10) in a patient of Western origin. However, such studies do not refer to the function of this mutant.
Therefore, in the present study, all of the EGFR exons were sequenced to evaluate the frequencies of EGFR gene mutations and to identify the rare or novel mutations of EGFR in 55 Chinese NSCLC patients. When a patient with a EGFR-P848L mutation was identified, the biological properties of the tumor were further investigated.
Materials and methods
Tumor samples
Tumor samples were obtained from patients suffering from NSCLC at the First Affiliated Hospital of Bengbu Medical College (Bengbu, China) and Ruijin Hospital (Shanghai, China). Clinical data for the NSCLC patients were available, including gender, age at diagnosis, tumor histology type, clinical staging, smoking status and response to treatment. All 55 patients provided written informed consent, and the study was approved by the institutional review board.
Genomic DNA extraction and direct sequencing
Genomic DNA from frozen tumor specimens was extracted using the phenol-chloroform method. Polymerase chain reaction (PCR) was used to amplify the exon 1–28 fragments of EGFR. The PCR products were purified by shrimp alkaline phosphatase/exonuclease I and then directly sequenced on ABI PRISM 3700 (Applied Biosystems, Carlsbad, CA, USA). Mutations were confirmed by at least two independent PCR amplifications and repeated sequencing reactions.
Site-directed mutagenesis of the epidermal growth factor receptor
PCDNA3.1-EGFR-WT and EGFR with a mutation of L858R or delE746-A750 were kindly provided by Dr William Pao of the Memorial Sloan-Kettering Cancer Center (New York, NY, USA). The P848L mutant EGFR construct was generated by introducing a point mutation into the wild-type EGFR vector using a site-directed mutagenesis kit (QuikChange XL, Stratagene). The primers were F: GTACTGGTGAAAACACTGCAGCATGTCAAGATC, R: GATCTTGACATGCTGCAGTGTTTTCACCAGTAC. The T790M mutation was then introduced into pCDNA3.1- EGFR-P848L using the same technique. The primers were F: ACCGTGCAGCTCATCATGCAGCTCATGCCCTTC, R: GAAGGGCATGAGCTGCATGATGAGCTGCACGGT. The mutated nucleotides are underlined.
Cell culture and Western blotting
293T cells were cultured in DMEM supplemented with 10% FBS, 100 pg penicillin/streptomycin and 2 mM glutamine. According to the manufacturer's instructions, wild-type EGFR and EGFR with different mutations were transiently transfected into cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Cells were then serum-starved for 16 h prior to treatment with various concentrations of gefitinib (Tocris, UK). Following incubation (3 h) with the inhibitors, cells were stimulated with EGF at 100 ng/ml for 30 min. Cells were then collected and the protein was extracted with RIPA lysis buffer and PMSF. Protein was electrophoresed in 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) gels, and then transferred to the nitrocellulose membrane. Anti-phospho-EGFR (pY1068) (Cell Signaling Technology, Danvers, MA, USA) was used for the detection of autophosphorylation of EGFR, anti-EGFR (Cell Signaling Technology) for total EGFR expression and anti-β-actin (abcom) as a loading control. A Western blotting luminal reagent (Thermo, Newington, NH, USA) was used to detect the signals. Results were repeated at least twice.
Computer modeling of EGFR
In order to better understand how this mutation affects the binding of EGFR with gefitinib, we constructed a three-dimensional computer model with RasMol software (version 2.6; http://www.umass.edu/microbio/rasmol/). Using the crystal structure of the wild-type EGFR (PDB code 2JITB) as a template, 321 residues (E697-L1017) of EGFR-P848L were modeled by homology using SWISS-MODEL (http://swissmodel.expasy.org/) (11) at a resolution of 3.10 Å.
Results
Clinical characteristics and EGFR mutations of lung adeno-carcinoma patients
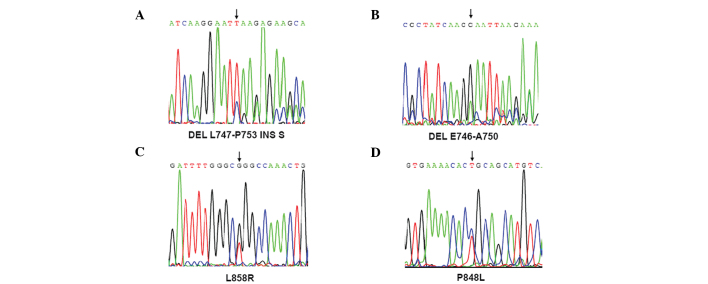
In total, 55 tumor tissues were collected from patients with NSCLC. Of these, 25 were squamous, 24 were adenocarcinomas and 6 were large cell lung cancers (Table I). The exons of the EGFR gene were sequenced in the 55 NSCLC patients using the ABI PRISM 3700 DNA sequencer. Eight (14.5%) patients harboring EGFR mutations were detected, including substitutions in exon 21 (5 patients with L858R and 1 patient with P848L) and deletions in exon 19 (1 patient with del E746-A750 and one patient with del L747-P753 INS S) (Fig. 1, Table II). Notably, all 8 patients (5 males and 3 females) with EGFR mutations had adenocarcinoma histopathology (Table II), among whom 6 were non-smokers and 2 were ever-smokers. Although only 9 female patients were enrolled in this series, 6 had adenocarcinoma and the EGFR mutation rate in female patients with lung adenocarcinoma was 50% (3/6). It was noted that no mutation in the EGFR gene was detected in patients with lung squamous carcinoma.
Table I.
Clincal data of NSCLC patients.
| Histotype | Gender (M/F) | Smoking (E/N) | Total |
|---|---|---|---|
| SCC | 22/3 | 19/6 | 25 |
| ACC | 19/5 | 13/11 | 24 |
| Other types | 5/1 | 4/2 | 6 |
Gender M/F, male/female; Smoking E/N, ever-smoking/non-smoking; SCC, squamous-cell carcinoma; ACC, adenocarcinoma.
Figure 1.
Sequencing of the different mutations of EGFR. (A and B) Two mutations in exon 19 of EGFR, leading to del L747-P753 INS S and DEL E746-A750 in EGFR, were identified. (C and D) Two missense mutations in exon 21 (L858R and P848L) were identified in the EGFR gene. Mutations are indicated by arrows.
Table II.
Summary of the patients harboring EGFR mutations.
| Patient | Gender | Age | Histology | Mutation type | Smoking | Stage |
|---|---|---|---|---|---|---|
| 1 | Female | 33 | AD | L858R | Never | T1N3M0 |
| 2 | Male | 60 | AD | L858R | Never | T2N1M0 |
| 3 | Male | 64 | AD | P848L | Never | T2N0M0 |
| 4 | Male | 49 | AD | L858R | Never | T2N2M0 |
| 5 | Male | 68 | AD | Del747-753INSS | 20 pack years | T3N2M0 |
| 6 | Female | 54 | AD | L858R | Never | T2N2M0 |
| 7 | Male | 61 | AD | Del E746-A750 | 15 pack years | T4N2M1 left humerus metatasis |
| 8 | Female | 54 | AD | L858R | Never | T4N2M1 bone metastasis |
AD, adenocarcinoma.
Response to tyrosine kinase inhibitor. The data revealed that most patients received platinum-based doublet chemotherapy. After failing this chemotherapy, 2 patients received gefitinib at a dosage of 250 mg orally once a day. The median survival following gefitinib therapy was 10.0 months. patient no. 6 is a never smoking, 54-year-old female lung adenocarcinoma patient with a L858R mutation in the EGFR gene and was in the T2N2M0 stage when she was diagnosed. She was treated with vinorelbine (36 mg on day 1 and day 8) plus cisplatin (110 mg on day 1) for 4 cycles during the initial treatment. When distant metastasis in the tempus sinistrum meningeal was detected, radiotherapy was applied. Gefitinib was then administered at a dosage of 250 mg per day, and 4 months later the patient achieved a complete response. Gefitinib treatment continued and there was a complete response status for 20 months without tempus sinistrum meningeal metatasis progression. Another patient without an EGFR mutation who received gefitinib had coronale metastasis upon diagnosis. The conventional strategy of chemotherapy for 2 cycles did not provide relief, and follow-up evidence revealed the disease had progressed. Gefitinib was administered for 3 months, and the patient had no relapse.
Sensitivity of the wild-type and the P848L mutant EGFR to TKI
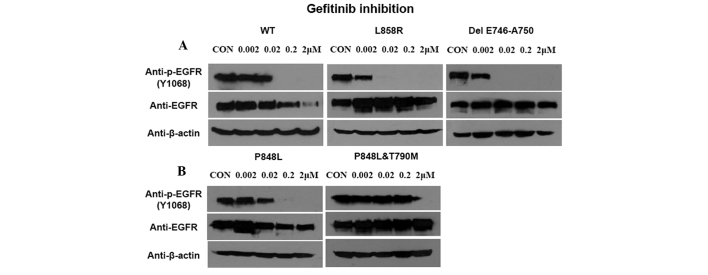
To compare the sensitivity of EGFR-P848L mutation to TKIs with the most common classical mutations, the EGFR with L858R and delE746-A750 mutation were transfected into 293T cells. In addition, the EGFR with P848L mutation or P848L and T790M double mutations were also transfected into cells to assess the change of sensitivity to TKI after T790M mutation. The EGFR-P848L mutant had a similar sensitivity to TKIs, as did the wild-type one. When the TKI concentration increased to 0.02 μM, the autophosphorylation of all three mutants was inhibited completely. Introducing T790M to P848L mutated EGFR markedly decreased the sensitivity to TKIs (Fig. 2).
Figure 2.
Sensitivity of wild-type and mutant EGFR to gefitinib. (A) 293 cells were transfected with wild-type and mutant EGFR (L858R, del 746–750), then increasing concentration of gefitinib was added. Total and phosphorylated EGFR were detected. (B) Phosphorylated EGFR of P848L with or without T790M mutation.
Computer modeling
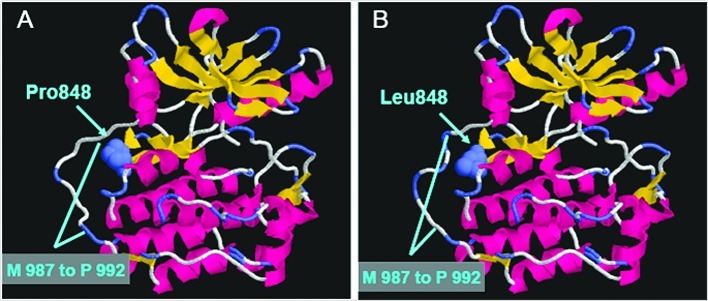
To further determine the change after the P848L mutation of EGFR, we constructed a three-dimensional computer model using the crystal structure of wild-type EGFR as the template. Fig. 3A and B reveal the crystal structure of wild-type and P848L EGFR, respectively. Although the structure of the ATP-binding pocket in the EGFR protein does not have any marked alteration, the structure of the region M987 to P992 in the EGFR gene was changed after the EGFR-P848L mutation.
Figure 3.
Molecular modeling of wild-type and P848L mutant EGFR. (A) In the wild-type EGFR, E697 to L1017 was shown in cartoon and P848 was shown in spacefill. (B) Compared to the wild-type EGFR, the structure of M987 to P992 was changed after the P848L mutation.
Discussion
A better mechanistic understanding of NSCLC would aid in the effort to find an optimal treatment for the disease. The oncogenesis of NSCLC involves numerous factors, including a large number of genetic abnormalities. Therapies targeted to these genetic abnormalities, which are different from the classical cytotoxic chemotherapy, have emerged in recent years. Culture studies have provided clear evidence that EGFR over-expression and/or mutation commonly occur in NSCLC, but only certain types of EGFR mutations have thus far been revealed to have therapeutic significance. Mutants of EGFR often show greater autophosphorylation activity, which increases the sensitivity to treatment with TKIs. The clinical observations in this study confirmed the same result as reported in the literature; patients harboring mutant EGFR show a marked response to TKI. This suggests that accurate mutation information is crucial, and only those NSCLC patients harboring an active EGFR mutation respond to the existing TKI treatment.
In the present study, all of the exons of the EGFR in 55 lung cancer patients were sequenced. In this cohort of NSCLC patients, a similar pattern of EGFR mutation was found as that reported previously. The EGFR mutations occurred in cases of adenocarcinoma, and more frequently in non-smokers, with a total gross mutation rate of 14.5%. A rare mutation in exon 21, P848L, was first identified in a patient of Chinese origin. A further functional study was carried out to observe the sensitivity of this rare mutant to gefitinib. The result showed gefitinib to have a modest inhibition of the P848L mutant EGFR, similar to that of the wild-type one. This result demonstrated that not all patients with mutations of EGFR are associated with sensitivity to gefitinib treatment. However, this does not indicate that the P848L mutant is resistant to all small molecule TKIs.
Sequist et al (10) first reported P848L and other rare point mutations in exons 18, 20 and 21 of EGFR, and in that study none of the patients with these rare point mutations responded to EGFR TKI agents. In the present study, the patient with the EGFR-P848L mutation did not adopt TKI treatment; subsequently, assessment of the clinical response to TKI was not possible. The in vitro result showed that the EGFR-P848L mutant had a similar sensitivity to gefitinib as that of the wild-type one, i.e., lower than that of the classical mutations.
With the aid of advances in computer technology to help achieve a better understanding of the alteration of this mutant, we were able to directly explore the spatial conformation using a homology model of EGFR-P848L. Marked change was not detected in the ATP-binding pocket in the EGFR-P848L mutant. However, we found that the structure of the region M987 to P992 was changed after the EGFR-P848L mutation. Combined with the in vitro analysis, it indicated that this mutation did not affect the binding of small molecule TKI with EGFR. Furthermore, we could also not exclude the possibility that mutations located around the ATP-binding pocket of the tyrosine kinase domain indirectly affect the binding affinity of mutant EGFR and gefitinib.
Although the patients with the EGFR mutation showed sensitivity to TKI at the initial stage of treatment, the majority of these patients became resistant to TKI. Two reasons could elucidate the acquired resistance in the patients treated with TKI: one reason is that a secondary mutation in the EGFR gene, T790M, occurred during the patient's TKI treatment. Another reason is that the patient treated with TKI appeared to have amplification of the MET protooncogene (12,13). However, T790M is the primary secondary mutation responsible for acquired resistance in NSCLC patients treated with TKI (14). In the present study, the EGFR gene with a double mutation, P848L and T790M, revealed a similar feature in that T790M serves a gate-residue function, even though it is a non-sensitive mutation. Therefore, it is essential to monitor in ‘real-time’ the resistant T790M mutation of the EGFR gene in the lung cancer cells of NSCLC patients. Maheswaran et al (15) have established a method to capture circulating tumor cells from the blood of NSCLC patients by using a microfluidic device containing microposts coated with antibodies against epithelial cells, leading to the probable real-time detection of the resistant mutation T790M of the EGFR gene in the lung cancer cells of NSCLC patients.
In conclusion, we studied the EGFR mutation of 55 Chinese NSCLC patients and further determined the biological properties of a rare mutation. The results revealed that the mutational hotspot was located in exons 19 and 21, which coordinates with patients of Western origin. Future functional studies are required, particularly for the rare EGFR mutations. Finally, individualized treatment of patients with different mutations is also required in our clinical work.
Acknowledgements
This study was supported in part by the National Natural Science Foundation of China (30530370, 30771017 and 30470816), the Chinese High Tech Program (863) (2006AA02Z175) and the Commission for Science and Technology of Shanghai (055407029, 06XD14016 and 06JC14054). Pacific Edit reviewed the manuscript prior to submission.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: A randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 3.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D, Eek R, Horai T, Noda K, Takata I, Smit E, Averbuch S, Macleod A, Feyereislova A, Dong RP, Baselga J. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced nonsmall-cell lung cancer (The IDEAL 1 Trial) J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 4.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Yatabe Y, Mitsudomi T. Epidermal growth factor receptor mutations in lung cancers. Pathol Int. 2007;57:233–244. doi: 10.1111/j.1440-1827.2007.02098.x. [DOI] [PubMed] [Google Scholar]
- 8.Gu D, Scaringe WA, Li K, Saldivar JS, Hill KA, Chen Z, Gonzalez KD, Sommer SS. Database of somatic mutations in EGFR with analyses revealing indel hotspots but no smoking-associated signature. Hum Mutat. 2007;28:760–770. doi: 10.1002/humu.20512. [DOI] [PubMed] [Google Scholar]
- 9.Wu JY, Wu SG, Yang CH, Gow CH, Chang YL, Yu CJ, Shih JY, Yang PC. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 10.Sequist LV, Joshi VA, Jänne PA, Muzikansky A, Fidias P, Meyerson M, Haber DA, Kucherlapati R, Johnson BE, Lynch TJ. Response to Treatment and Survival of patients with Non-Small Cell Lung Cancer Undergoing Somatic EGFR Mutation Testing. Oncologist. 2007;12:90–98. doi: 10.1634/theoncologist.12-1-90. [DOI] [PubMed] [Google Scholar]
- 11.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 12.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 13.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]