ABSTRACT
Preimplantation genetic diagnosis (PGD) is a genetic screening of embryos conceived with assisted reproduction technologies (ART). A single blastomere from an early-stage embryo is removed and molecular analyses follow to identify embryos carrying genetic defects. PGD is considered highly successful for detecting genetic anomalies, but the effects of blastomere biopsy on fetal development are understudied. We aimed to determine whether single blastomere removal affects steroid homeostasis in the maternal-placental-fetal unit during mouse pregnancy. Embryos generated by in vitro fertilization (IVF) were biopsied at the four-cell stage, cultured to morula/early blastocyst, and transplanted into the oviducts of surrogate mothers. Nonbiopsied embryos from the same IVF cohorts served as controls. Cesarean section was performed at term, and maternal and fetal tissues were collected. Embryo biopsy affected the levels of steroids (estradiol, estrone, and progesterone) in fetal and placental compartments but not in maternal tissues. Steroidogenic enzyme activities (3beta-hydroxysteroid dehydrogenase, cytochrome P450 17alpha-hydroxylase, and cytochrome P450 19) were unaffected but decreased activities of steroid clearance enzymes (uridine diphosphate-glucuronosyltransferase and sulfotransferase) were observed in placentas and fetal livers. Although maternal body, ovarian, and placental weights did not differ, the weights of fetuses derived from biopsied embryos were lower than those of their nonbiopsied counterparts. The data demonstrate that blastomere biopsy deregulates steroid metabolism during pregnancy. This may have profound effects on several aspects of fetal development, of which low birth weight is only one. If a similar phenomenon occurs in humans, it may explain low birth weights associated with PGD/ART and provide a plausible target for improving PGD outcomes.
Keywords: assisted reproduction technologies, embryo, in vitro fertilization, placenta, steroid hormones
Embryo biopsy deregulates steroid metabolism during mouse pregnancy and leads to decreased fetal body weight.
INTRODUCTION
Preimplantation genetic diagnosis (PGD) is a method for genetic screening of embryos conceived with assisted reproduction technologies (ART). Cleavage-stage embryo biopsy and subsequent single-cell analysis is the most widespread general approach. PGD allows identification of all common and many rare Mendelian single gene defects as well as chromosomal abnormalities, late-onset conditions, predisposition to cancer, and human leukocyte antigen (HLA) matching [1]. It also allows embryo sexing, which is of importance when sex chromosome-specific genetic mutations, such as Duchenne muscular dystrophy that affects primarily males, are involved.
PGD is now offered as a well-established clinical service worldwide, and the method is considered highly successful in respect to its accuracy in detecting genetic anomalies. Thousands of children have been born after PGD, and thus far striking increases in the rates of congenital abnormalities or syndromes have not been attributed to the procedure, as compared to children born with ART without biopsy [2]. However, it was recently reported that PGD increased incidence of stillbirths in multiple ART pregnancies [3]. Moreover, the ongoing debate about negative effects of ART overall, combined with increasing evidence pointing to several problems that may be associated with these techniques [4–7], validates reassessment of PGD effects on reproductive outcomes.
In a mouse model of PGD, it has been recently shown that although the biopsy procedure does not affect preimplantation embryo development and global pattern of gene expression, biopsied embryos undergo premature and sometimes abnormal hatching and exhibit developmental delay [8]. The endpoint of this study was blastocyst stage, so it was not clear whether subsequent embryonic and fetal development were normal.
We have recently shown that in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) alter steroid metabolism and transport in mouse pregnancies at the level of the placenta [9, 10], with a likely contributing factor being placental inflammation and oxidative stress [11]. This evidence indicates that higher rates of obstetric and neonatal complications observed in pregnancies achieved with ART may be mediated, at least partially, through abnormal placental function.
Here, we examined whether blastomere removal from IVF-generated cleavage-stage mouse embryos alters fetal development and endocrinology in murine pregnancy. We generated pregnancies with biopsied and matching nonbiopsied embryos, and examined the levels of steroid hormones as well as activities of steroidogenic and steroid metabolizing enzymes. We demonstrated that embryo biopsy led to abnormal steroidogenesis and impaired steroid clearance in placentas and fetal tissues.
MATERIALS AND METHODS
Reagents
Mineral oil was purchased from Squibb and Sons (Princeton, NJ); pregnant mares' serum gonadotropin (eCG), human chorionic gonadotropin (hCG), and estradiol (E2) kits for cytochrome P450 19 (CYP19) analysis were purchased from Calbiochem (Spring Valley, CA); estrone (E1), E2, and progesterone kits were purchased from Alpco Diagnostics (Salem, NH). All other chemicals were obtained from Sigma Chemical Co. (St. Louis, MO) unless otherwise stated.
Animals
B6D2F1 (C57BL/6 x DBA/2) and Swiss Webster mice were obtained from the National Cancer Institute (Raleigh, NC) and CD-1 mice from Charles River Laboratories (Wilmington, MA), at 6–8 wk of age. B6D2F1 mice were used as sperm and oocyte donors for IVF, and CD-1 or Swiss Webster mice were used as surrogate mothers and vasectomized males for embryo transfer. Mice were fed ad libitum with a standard diet and maintained in a temperature- and light-controlled room (22°C, 14L:10D), in accordance with the guidelines of the Laboratory Animal Services at the University of Hawaii and guidelines presented in National Research Council's Guide for the Care and Use of Laboratory Animals published by the Institute for Laboratory Animal Research of the National Academy of Science, Bethesda, MD, 2011. The protocol for animal handling and treatment procedures was reviewed and approved by the Animal Care and Use Committee at the University of Hawaii.
Media
Medium T6 [12] was used for IVF, and HEPES-buffered CZB medium (HEPES-CZB [13, 14]) was used for gamete handling and embryo micromanipulation. Medium CZB [13] was used for embryo culture. Both CZB and T6 were maintained in an atmosphere of 5% CO2 in air, and HEPES-CZB was maintained in air.
In Vitro Fertilization
Sperm capacitation and IVF were performed as reported by us before [15]. Briefly, the oocytes were collected from females induced to superovulate with injections of 5 IU eCG and 5 IU hCG given 48 h apart. Epididymal sperm were collected by release from cauda epididymis directly into T6 medium, and were capacitated for 1.5 h at 37°C in a humidified atmosphere of 5% CO2. The gametes were coincubated for 4 h. After gamete coincubation, the oocytes were washed with HEPES-CZB, followed by at least one wash with CZB medium. Only morphologically normal oocytes were selected for culture.
Embryo Culture, Biopsy, and Transfer
Fertilized eggs (zygotes with two well-developed pronuclei and extruded second polar body) were cultured in 50-μl drops of CZB medium pre-equilibrated overnight with humidified 5% CO2 in air. After ∼48 h of culture, four-cell embryos were transferred into Ca2+- and Mg2+- free CZB for 10–20 min to disrupt cell adhesion, and were then transferred to microdrops of Ca2+- and Mg2+-free HEPES-CZB on the micromanipulation dish. Embryo biopsy was performed using Eppendorf Micromanipulators (Micromanipulator TransferMan; Eppendorf, Hamburg, Germany) with a piezoelectric actuator (PMM Controller, model PMAS-CT150; PrimeTech, Tsukuba, Japan). The zona pellucida was penetrated with a micropipette (20 μm internal diameter) and one blastomere was aspirated. Control (nonbiopsied) embryos from the same IVF cohorts were preincubated in Ca2+- and Mg2+-free CZB medium but were not transferred into Ca2+- and Mg2+-free HEPES-CZB medium and were not micromanipulated. Biopsied and nonbiopsied embryos were cultured under the same conditions for the subsequent 24 h until they developed to morula/early blastocyst stage. The embryos were then transferred into the oviducts (four to eight per oviduct) of CD-1 females mated during the previous night with vasectomized CD-1 males. Biopsied and nonbiopsied embryos were transferred into separate females. Cesarean section was performed on Day 18. In an additional experiment we also generated matching biopsied and sham-biopsied embryos; sham-biopsied embryos were subjected to exactly the same manipulations and culture conditions as biopsied embryos but the blastomeres were not removed. The surrogate mothers for these embryos were Swiss Webster females.
Tissue Collection and Processing
Tissues were collected and processed as previously described [9–11]. Briefly, fetuses were killed by decapitation and whole blood was collected into 0.5-ml tubes containing a small amount of heparin (1 μl of 1000 U/ml heparin lithium salt in water). Whole blood was also collected from mothers through cardiac puncture under anesthesia immediately prior to cesarean section. Maternal livers and ovaries, placentas, and fetal livers were collected after cesarean section, washed briefly in Dulbecco PBS (D-PBS), drained, and placed singly into tubes. During collection all tissues were kept on ice for up to 30 min, and were subsequently frozen at −80°C until use.
Maternal livers and ovaries, placentas, and fetal livers were thawed, wet weight recorded, cut in half, and one half homogenized 1:4 in Tris-HCl buffer containing 5 mM MgCl2 and 2 mM PMSF (pH 7.4) using a handheld Tissue Tearor rotor-stator for 30 sec (Biospec, Bartesville, OK). Subsequently, tissue homogenates were divided in half, and one half was centrifuged at 10 000 × g for 20 min at 4°C to remove mitochondria, nuclei, and cellular cytoskeleton (called an S9 fraction). Aliquots of tissue lysate and S9 fraction for each tissue, as well as the remaining organ halves, were frozen at −80°C until use. Before use, all tissue homogenates were normalized for protein concentration to 2.0 mg/ml using the bicinchoninic acid method [16].
Quantification of Steroid Hormone Levels
The levels of three major sex steroids, progesterone, E1, and E2, were measured in duplicate for each tissue sample using commercial ELISAs (Alpco Diagnostics). All ELISAs were validated with external controls (manufacturer supplied) and results for each plate were accepted if the derived concentration of the external controls fell within 15% of the theoretical concentration. Moreover, tissue results were accepted if derived concentrations in the duplicates varied by less than 15%. The average interday variation of external calibrators was progesterone, 4.1% ± 1.8%; E1, 5.0% ± 2.7%; and E2, 5.6% ± 3.7%.
Biochemical Assays for Steroidogenic Enzymes
Assays for 3-β-hydroxysteroid dehydrogenase (3β-HSD), cytochrome P450 17α-hydroxylase (CYP17), and CYP19 were performed using maternal livers, placentas, and fetal livers.
3β-HSD assay.
The activity of 3β-HSD was determined by measuring the conversion of pregnenolone to progesterone as previously described [10]. Briefly, 10 μl of protein (0.1 mg/ml liver or placenta), 79 μl of assay buffer (0.1 M Tris-HCl buffer with 50 mM MgCl2, pH 7.4) and 1 μl of pregnenolone (500 μM stock) were added to a glass tube. An aliquot, 10 μl, of NAD+ (10 mM stock) was added to initiate the reaction, and tubes were covered and incubated in a 37°C water bath for 10 min. Reactions were terminated by plunging tubes into ice. Progesterone product in the supernatant of each reaction was quantified in duplicate using a commercial ELISA as per manufacturer's instructions.
CYP17 assay (17α-hydroxylase activity).
The published meta-dinitrobenzene assay for 17-ketosteroids [17] was adapted to specifically determine the conversion of 17α-hydroxypregnenolone to dehydroepiandrosterone as an index of CYP17 activity, as previously described by us [10]. Briefly, in glass tubes the following were added: 10 μl of protein (0.1 mg/ml liver or placenta), 79 μl of 0.1 M Tris-HCl buffer containing 50 mM MgCl2, pH 7.4, 1 μl of 17α-hydroxypregnenolone (50 mM stock), and 10 μl of NADPH (10 mM). Tubes were capped and incubated at 37°C for 5 min, then reactions were terminated by addition of 100 μl KOH (5 M). Immediately, 200 μl of 2% m-dinitrobenzene was added and mixed, and then solution was transferred to an Eppendorf tube and centrifuged for 2 min at 10, 000 rpm. Triplicate aliquots (100 μl) were transferred to a 96-well plate and optical density at λ = 520 nm determined in a Spectramax 340plus spectrometer (Molecular Devices, Sunnyvale, CA).
CYP19 assay (aromatase activity).
Activity of CYP19 was determined by measuring the conversion of testosterone to 17β-E2. Briefly, glass tubes were kept on ice while 10 μl of protein (0.1 mg/ml liver or placenta) in 0.1 M Tris-HCl buffer, pH 7.4, containing 50 mM MgCl2 and 10 ng/ml of testosterone, was added. Tubes were preincubated at 37°C in a hot water bath. Reactions were initiated through the addition of 1 mM NADPH and incubated at 37°C for 10 min. After incubation the reaction was terminated by plunging the tubes into ice for 5 min. Detection of 17β-E2 was determined in duplicate with a commercial ELISA and converted to fg min−1 (mg protein)−1.
Biochemical Assays for Steroid Metabolizing Enzymes
Assays for steroid metabolism and clearance enzymes were performed using maternal livers, placentas, and fetal livers.
Uridine diphosphate-glucuronosyltransferase.
Total uridine diphosphate-glucuronosyltransferase (UGT) activity was determined using the method of Collier [18] with the exception that alamethicin (50 μg/mg protein in dimethyl sulfoxide [DMSO]) was used as the UGT activator and 5 mM saccharolactone was included in each reaction to inhibit β-G activity. DMSO was never more than 1% of the reaction by volume. Fluorescence was monitored continuously at 355 nm excitation/460 nm emission and results transformed to pmol min−1 (mg protein)−1 using a standard curve generated with 4-methyl umbelliferone.
β-G.
β-G activity was determined using the method of Trubetskoy and Shaw [19]. Microplates on ice were loaded with protein (10 μg) and buffer; plates were preincubated at 37°C for 3 min and the reaction initiated with 100 μM 4-methyl umbelliferone glucuronide. Fluorescence was continuously monitored at 355 nm ex/460 nm em. Results were transformed to pmol min−1 (mg protein)−1 using a standard curve generated with 4-MU.
General sulfotransferase.
The para-nitrophenol substrate (400 μM) was used to determine general sulfotransferase (SULT) activity [20, 21]. Protein (10 μg) in 0.05 M potassium phosphate buffer pH 6.5 and para-nitrophenol were added to microcentrifuge tubes on ice and then preincubated for 5 min at 37°C in a hot water bath. Reactions were initiated with the addition of 60 μM 3′-phosphoadenosine 5′-phosphosulfate and the tubes were incubated at 37°C in the water bath for 1 h. Reactions were quenched with equal volume of 1 M NaOH. Absorbance was read in triplicate microplate wells (100 μl) at λ = 400 nm with the spectrophotometer and results transformed using a standard curve generated with para-nitrophenol.
Arylsulfatase C (steroid sulfatase).
Activity of the arylsulfatase (AS) isoform C was determined using a modification of the method of Roy [22, 23] as previously described [9]. Briefly, microplates were kept on ice and 50 μg of protein in 0.1 M Tris-HCl buffer pH 7.4 was added to each well. The samples were preincubated for 5 min at 37°C, then reactions were initiated with the addition of 100 μM para-nitrophenyl sulfate. Absorbance was monitored continuously at λ = 400 nm and results generated using a standard curve of para-nitrophenol. AS C activity was significantly reduced through decrease in the pH of the assay buffer to pH 6.5 [24, 25]
Statistical Analyses
Statistical analyses were performed using Prism 5.0 (GraphPad Prism, San Diego, CA) with α = 0.05. Steroid levels were compared between all organs using one-way ANOVA and between biopsied and nonbiopsied groups for the same organ with paired sample t-tests. Bland-Altman tests for variance were carried out on raw means and, when it was revealed that standard deviation increased as mean increased, the natural log of each data point was taken and one-way ANOVA of the natural logs performed with Dunnet post hoc multiple comparison tests [26]. The purpose of this log transformation was to decorrelate variances from the means so that data were normalized, thereby making means appropriate [20, 21].
RESULTS
Generation of Fetuses after Blastomere Biopsy
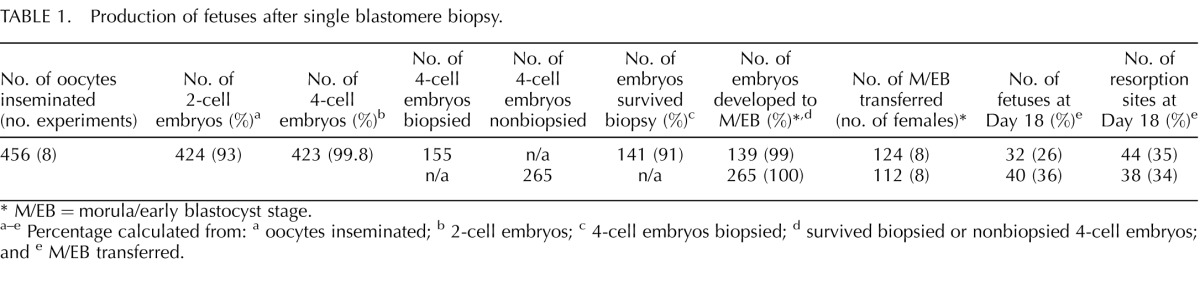
Eight IVF experiments were performed, which yielded a total of 423 four-cell embryos, 155 of which were subjected to blastomere biopsy (Table 1). More than 90% of biopsied embryos survived the procedure and all but two of those (99%) developed to morula/early blastocyst stage. Similarly to Duncan et al. [8], we observed developmental delay of biopsied embryos; after 72 h of culture, fewer embryos developed to early blastocyst stage in the biopsied as compared to the nonbiopsied group (36%, 51 of 141, vs. 56%, 149 of 265, P < 0.001). Biopsied and nonbiopsied embryos were transferred to 16 surrogate mothers (two per experiment) and all females became pregnant. There were no statistically significant differences between the average number of fetuses (mean ± SD, 4 ± 2.56 vs. 5 ± 2.07) and embryos that implanted per female (mean ± SD, 9.5 ± 3.51 vs. 9.75 ± 3.01) in biopsied and nonbiopsied pregnancies, respectively. Hence the effects observed in this study cannot be attributed to overcrowding. Postimplantation development measured by proportion of live fetuses and resorption sites was similar in both groups. Totals of 32 and 40 fetuses were obtained from biopsied and nonbiopsied pregnancies, respectively (Table 1).
TABLE 1. .
Production of fetuses after single blastomere biopsy.
M/EB = morula/early blastocyst stage.
a–e Percentage calculated from: a oocytes inseminated; b 2-cell embryos; c 4-cell embryos biopsied; d survived biopsied or nonbiopsied 4-cell embryos; and e M/EB transferred.
Maternal and Fetal Body and Organ Weights
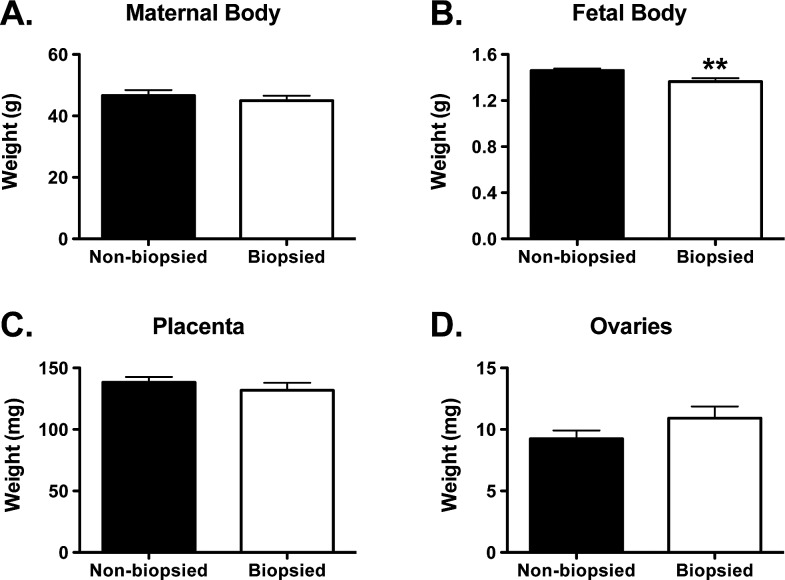
Body or organ size can affect production of steroids and of steroidogenic and steroid clearance enzymes [27, 28]. We therefore compared biopsied and nonbiopsied groups in respect to maternal and fetal whole body weights as well as weights of maternal ovaries and placentas (Fig. 1). No differences were found between the two examined groups in maternal body, placental, and ovarian weights, but biopsied fetuses had significantly lower weights than their nonbiopsied counterparts (Fig. 1B, P = 0.004, t-test).
FIG. 1. .
Maternal and fetal body and organ weights. A) Maternal whole body (n = 7 for nonbiopsied and n = 8 for biopsied). B) Fetal whole body (n = 40 for nonbiopsied and n = 32 for biopsied). C) Placentas (n = 39 for nonbiopsied and n = 32 for biopsied). D) Ovaries (n = 8 for both biopsied and nonbiopsied). Each graph represents a mean ± SEM. Statistical significance: **P < 0.01 (t-test).
ELISA Quantification of Steroid Hormone Levels
Levels of three hormones, E2, E1, and progesterone, were determined in maternal livers, maternal ovaries, placentas, and fetal livers. Additionally, maternal blood and fetal blood were assessed.
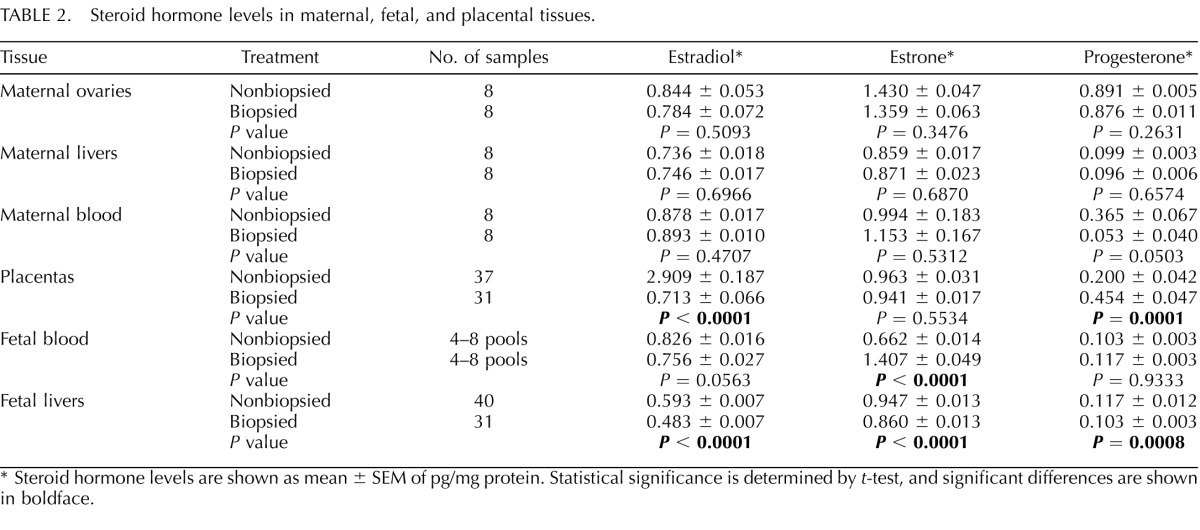
Significant differences were observed in steroid hormone levels between biopsied and nonbiopsied groups (Table 2). Although maternal production and systemic levels of E2 did not differ between the two examined groups, placentas and fetal livers from biopsied pregnancies had significantly lower levels of E2 as compared to controls (P < 0.0001, Table 2). The levels of E1 produced in maternal ovaries as well as E1 levels in the maternal-placental compartments did not differ. Fetuses originating from biopsied embryos had significantly more E1 in their blood but less in the liver (P < 0.0001, Table 2). Progesterone levels were significantly higher in placentas from biopsied pregnancies (P = 0.0001, Table 2) and lower in fetal livers (P = 0.0008) with no changes in maternal ovaries, maternal blood, or fetal blood (Table 2).
TABLE 2. .
Steroid hormone levels in maternal, fetal, and placental tissues.
Steroid hormone levels are shown as mean ± SEM of pg/mg protein. Statistical significance is determined by t-test, and significant differences are shown in boldface.
Overall, this analysis demonstrated that blastomere biopsy resulted in decreased levels of all steroid hormones in fetal livers. Moreover, levels of E2 and progesterone in placenta and of E1 in fetal blood were also altered.
Biochemical Assays for Steroidogenic Enzymes
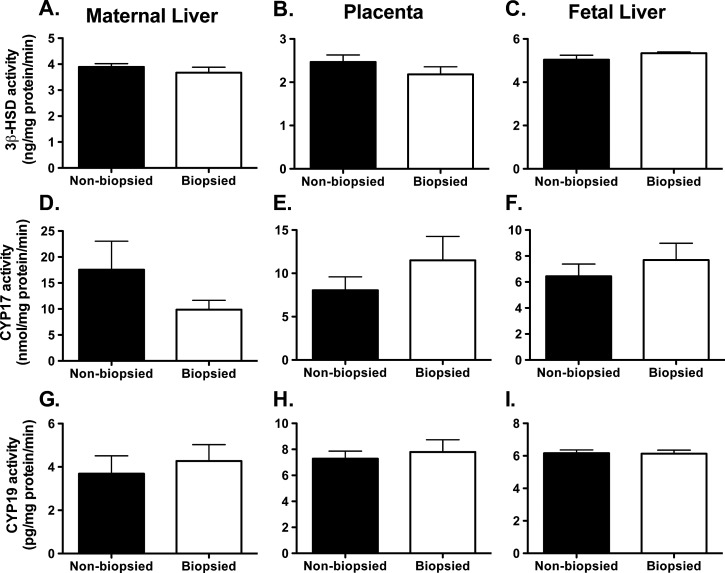
The activity of steroidogenic enzymes was measured individually in maternal livers (n = 8 per group), placentas (n = 14–15 per group; 1–2 per female) and fetal livers (n = 14–15 per group; 1–2 per female). The analysis of three steroidogenic enzymes, 3β-HSD, CYP17, and CYP19, revealed no differences between biopsied and nonbiopsied groups in any tissue or any enzyme tested (Fig. 2).
FIG. 2. .
The activities of steroidogenic enzymes: 3β-HSD (A–C), CYP17 (D–F), and CYP19 (G–I). A, D, G) Maternal liver. B, E, H) Placenta. C, F, I) Fetal liver. Each graph represents a mean ± SEM, with n = 8 (maternal livers), n = 14–15 (placenta and fetal livers). No statistical differences were observed between biopsied and nonbiopsied groups.
Biochemical Assays for Steroid Clearance/Regeneration Enzymes
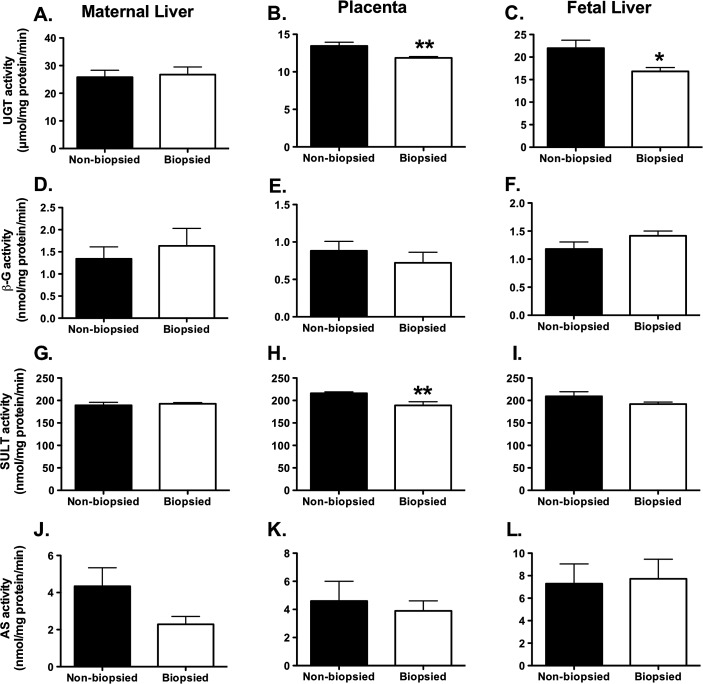
The activity of steroid clearance/regeneration enzymes was measured in maternal livers (n = 8 per group), placentas (n = 15 per group) and fetal livers (n = 11–15 per group). Steroid clearance generally proceeds through two pathways: glucuronidation and sulfonylation. The two enzyme families regulating glucuronidation, UGT and beta-glucuronidase (β-G), and the two families balancing sulfonylation, SULT and AS, were examined. For the glucuronidation pathway, UGT and β-G activities were, on per milligram of tissue basis, highest in maternal and fetal livers and decreased by ∼50% in placentas. UGT activity was lower in placentas and fetal livers (Fig. 3, B and C, P = 0.006 and P = 0.01, respectively) in the biopsied group as compared with the nonbiopsied group. No differences in enzymatic activity in maternal livers were observed. The activities of β-G, which partners UGT, cleaving UGT metabolites and returning them to active form, did not differ between the biopsied and nonbiopsied groups. The ratio of glucuronidation to cleavage was similar for all tissues. In the sulfonylation pathway, SULT and AS activities were at comparable levels in the biopsied and nonbiopsied groups across all tissues tested except for SULT activity in placentas in the biopsied group, which was significantly lower (Fig. 3H, P = 0.005). The activity of regenerating AS was similar in all tissues. Clearance by SULT occurred at approximately 40-fold higher rates than recycling activity by AS. The SULT:AS ratio for placentas was similar in the nonbiopsied (47:1) and the biopsied group (49:1), but a significant difference between the nonbiopsied and biopsied groups was present in the SULT:AS ratio in maternal livers with a ∼50% decrease in available steroids and the ratio shifting from 44:1 to 84:1, demonstrating greater steroid removal by SULT in maternal liver.
FIG. 3. .
The activities of steroid clearance enzymes: UGT (A–C), β-G (D–F), SULT (G–I), and AS (J–L). A, D, G, J) Maternal liver. B, E, H, K) Placenta. C, F, I, L) Fetal liver. Each graph represents a mean ± SEM, with n = 8 (maternal livers), n = 15 (placenta) and n = 11–15 (fetal livers). Statistical significance: *P < 0.05, **P < 0.001 (t-test).
Testing for the Effects of Sham Biopsy on Steroid Metabolism
Several different types of controls can be envisioned in studies on the effects of blastomere biopsy. In previous works variable controls were used, such as 1) intact controls, represented by embryos cultured without interruptions in parallel to biopsied embryos [29–33]; 2) CMF-exposed controls, represented by embryos that, similarly to biopsied embryos, were transiently exposed to Ca2+- and Mag2+-free medium, which facilitates biopsy [34–36], but were not manipulated; and 3) sham-biopsied controls, represented by embryos manipulated in exactly the same way as biopsied embryos but without the blastomere removal [37, 38]. Occasionally, several treatment groups and several controls were included [8, 39]. No differences were found between intact and CMF-exposed controls [39] or between intact and sham-biopsied controls [37, 38]. In our study we initially used as controls embryos that were subjected to the same culture conditions (CMF-exposed) but were not manipulated. To exclude the possibility that observed differences were due to manipulations, we performed additional experiments in which sham-biopsied embryos were used as controls for biopsied embryos. The results of these additional experiments are shown as Supplemental Data (all Supplemental Data are available online at www.biolreprod.org). Out of 248 four-cell embryos obtained, 103 were subjected to blastomere biopsy and 88 to sham biopsy (Supplemental Table S1). No differences between the two groups were noted in the progression of preimplantation development. Biopsied and sham-biopsied embryos were transferred to 11 surrogate mothers (3–4 per experiment). There were no statistically significant differences between the average number of fetuses (mean ± SD, 3.4 ± 1.5 vs. 4.0 ± 2.7) and embryos that implanted (mean ± SD, 8.7 ± 2.3. vs. 10.3 ± 5.9) per pregnant female, and postimplantation development measured by proportion of live fetuses and embryos that implanted was also similar in both groups (Supplemental Table S1). No differences were found between biopsied and sham-biopsied groups in maternal body, placental, and ovarian weights (Supplemental Fig. S1). The fetal body weight, which was lower in biopsied embryos compared to nonbiopsied counterparts (Fig. 1), was now similar between sham-biopsied and biopsied groups. The analysis of steroidogenic enzymes (3β-HSD, CYP17, and CYP19) revealed no differences between biopsied and sham-biopsied groups in any tissue or any enzyme tested (Supplemental Fig. S2), in agreement with earlier biopsied versus nonbiopsied comparison. The analysis of steroid clearance/regeneration enzymes activities demonstrated a decrease in UGT activity in placentas and fetal livers (Supplemental Fig. S3, B and C, P = 0.018 and P = 0.047, respectively), and a decrease in SULT activity in placenta (Supplemental Fig. S3H, P = 0.0019), and matched previous data from biopsied versus nonbiopsied comparison (Fig. 3). We also noted decreased β-G activity in fetal livers (Supplemental Fig. S3F, P = 0.0072), which we had not observed before, and which strengthened our argument that steroid clearance and regeneration is deregulated.
Overall, the comparison of biopsied and sham-biopsied groups yielded similar results to those originating from comparison of biopsied and nonbiopsied counterparts, and provided additional support for our conclusion that embryo biopsy deregulates placental steroid metabolism.
DISCUSSION
Here we report changes in placental steroid metabolism resulting from single blastomere removal from cleavage-stage mouse embryos. As far as we are aware, this is the first study assessing placental function and steroid homeostasis in pregnancies conceived after embryo biopsy in any species.
Embryo biopsy is used in conjunction with PGD, which is a common and vital part of ART. Previous studies linking ART with placental dysfunction are limited. In humans, ART procedures have been shown to influence expression of placental genes and proteins associated with placentation [40, 41]. It has also been suggested that placentas from ART have more frequent pathological findings such as villous edema and microcalcification [42] or ultrastructural anomalies in placental blood barrier, which may lead to maternal-fetal traffic downregulation [43]. In the mouse, our own work pointed to altered steroid metabolism and transport in placentas from pregnancies conceived with IVF and ICSI [9, 10]. Because blastomere biopsy is an additional, and rather invasive, embryo manipulation, we speculated that it might extend the effect caused by IVF on embryonic and fetal well-being.
Preimplantation biopsy of embryos can be performed at any stage from the two-cell to the blastocyst. Although trophectoderm biopsy is slowly gaining popularity, cleavage-stage embryo still remains the primary approach [1, 44]. In humans, biopsy is routinely performed in the morning on Day 3 after fertilization, at which time embryos are at the six- to eight-cell stage [45, 46]. In this study we performed biopsy at the four-cell stage to avoid interfering with embryo compaction, which in mice begins at the eight-cell stage [32, 47–49], whereas in humans it does not occur before the 16–32-cell stage [50]. Performing biopsy during or after compaction is likely to have negative effects on developing embryos because it would interfere with cell-cell adhesion [51, 52], gap and tight junctions [53–55], and cytoplasmic polarization [56], which all first appear at the eight-cell stage in the mouse. The four-cell stage in mice is similar to the eight-cell stage in humans also because of the timing in embryonic genome activation, which is initiated at the two- to four-cell stage in mice [57] and the four- to eight-cell stage in humans [58].
When we assessed the efficiency of preimplantation and postimplantation embryo development, we did not observe any impairment resulting from a single blastomere removal. Similar proportions of morula/early blastocysts after embryo culture, and fetuses after embryo transfer, were obtained in biopsied and nonbiopsied groups. However, in agreement with previous reports [8, 32], we noted that biopsied embryos exhibited developmental delay during preimplantation development. This was thought to be due to insufficient volume of biopsied embryos. Indeed, it has been shown that blastomere removal increased frequency of contraction and expansion before hatching as if the embryos required more time, and effort, to break through the zona [32]. This latter study also reported on premature compaction of biopsied embryos at six- rather than eight-cell stage, which, as the authors suggested, might have reflected disturbances in polarity between blastomeres. Here, when fetuses were delivered at term via cesarean section, they were all morphologically normal. However, those originating from biopsied embryos had lower whole-body weights, which could either be a remnant of their previous developmental delay or a result of impaired development in utero, perhaps as a consequence of insufficient steroid action. Interestingly, we did not observe the differences in progression of preimplantation development and in fetal body weights between biopsied and sham-biopsied groups, supporting the notion that zona pellucida manipulation contributes to developmental delay and fetal weight decrease of biopsied embryos.
It has been previously reported that implantation rate was decreased after embryo transfer with biopsied embryos [36]. Here, we did not observe a similar reduction. The discrepancy between our study and this past study was likely due to the differences in embryo transfer procedure. We transferred biopsied and nonbiopsied embryos into the oviducts of recipients on Day 0.5 of pseudopregnancy, whereas Wilton et al. [35, 36] transferred embryos to the uteri on the third day of pseudopregnancy. Our approach shortened the exposure to in vitro culture and provided embryos with a benefit of extended time in a physiological environment (oviducts) prior to implantation. Moreover, in our study uterine tissue was not subjected to any manipulations.
Fetuses derived from biopsied embryos had decreased levels of steroids (E2, E1, and progesterone) in their livers, and altered levels of some steroids in placenta and blood. Because the maternal production and maternal systemic steroid levels were not altered, the observed differences were not due to changes in steroid production in the maternal ovaries or elsewhere. The activities of steroidogenic enzymes that produce progesterone, E2, and E1—3β-HSD, CYP19 and CYP17—were not affected by biopsy, further supporting that observed differences were not due to impaired steroidogenesis.
The levels of progesterone were higher in term placentas from biopsied pregnancies. In humans, high levels of progesterone negatively affect trophoblast invasion and movement during vascular remodeling at early stages of pregnancy [59]. Therefore, if increases in placental progesterone occur early in the pregnancy, this would be expected to affect placentation and might be a mechanism for the delayed development of fetuses originating from biopsied embryos. Progesterone is also a key player in parturition. In most mammals, including mice, uterine quiescence throughout pregnancy is maintained by elevated progesterone, and parturition is associated with a marked decline of the hormone [60]. Consequently, higher levels of progesterone late in pregnancy, as observed in this study, could lead to problems initiating labor. Unfortunately, we do not have evidence supporting this prediction because cesarean section was performed to determine postimplantation fetal loss and to exclude the possibility that oxidative stress and parturition signaling were responsible for changes observed. Although mice and humans differ in that there is no clear progesterone withdrawal in term women, a “functional” withdrawal is thought to occur at the onset of labor via expression changes of progesterone receptors and coactivators [61]. Thus, progesterone regulation is also important for ending pregnancy and initiating parturition in humans [60].
Blood is a transient compartment and delivers steroids to the site of action. Maternal and fetal livers as well as the placenta are organs of metabolism and clearance. So the differences in hormones observed between maternal liver, maternal blood, placentas, fetal blood, and fetal livers likely represent a transient state between blood delivery and organ removal of steroids. Another possible explanation for decreased E2 in placentas and increased E1 in fetal blood occurring together is that, as steroids transit the placenta, the interconversion of E2/E1 (performed by the enzyme 17-β-hydroxysteroid dehydrogenase) is shunted towards conversion of E2 to E1. Because the contribution of fetal ovaries/adrenals/testes to fetal levels of steroids was not studied, we cannot exclude that fetal organ steroidogenesis is impaired.
Glucuronidation and sulfonation are the two major pathways for steroid clearance and elimination. Biopsy resulted in decreased activities of both UGT and SULT in placentas and fetal livers, but not in maternal livers. However, because glucuronidation and sulfonation have extremely high capacity, they are seldom, if ever, saturated. Therefore, decreased activities of glucuronidation or sulfonation may be statistically significant as observed here, but not translate to changes in steroid elimination because of high capacity. Moreover, other enzymes, including the cytochromes P450 and catechol-O-methyl transferases, also biotransform these steroids (phase I metabolism), and if these enzymes are also induced in the fetus unit this could also account for lower steroid levels [62].
The impairment in steroid clearance in placenta may be due to abnormal function of this organ overall, or may represent shift in time. If the delays observed during preimplantation development of fetuses originating from biopsied embryos had continued during pregnancy, it could have led to placental and fetal steroid levels being different (lower) than they should be at the same development stage. This is supported by Delle Piane et al. [63], who also reported developmental delay but no structural differences in placentas after IVF and ICSI. If fetuses and placentas are retarded (late) in their development, but otherwise normal, this could mean that the whole suite of enzymes and pathways responsible for steroid homeostasis is delayed as well. This delay may very well be expressed as decreased fetal steroid levels, deregulation of 17-β-hydroxysteroid dehydrogenase activity, and impaired steroid removal/clearance.
We cannot exclude that observed differences between biopsied and nonbiopsied groups represent a rodent-specific effect, because in the mouse steroids are made in the maternal ovary, then join the blood circulation, pass through the liver, and transit the placenta to the fetus. In humans, the fetus and placenta combine to produce steroids during pregnancy, and alterations caused by maternal liver would not be as relevant. Regardless of these endocrine differences and the structural differences between mouse and human placenta (the mouse being hemotrichorial and the human hemomonochorial), metabolism and transport performed by the placentas of both species are believed to be extremely similar, and lessons from mice can provide novel insights into human placental function [64]. Future work will be necessary to establish the mechanism by which embryo biopsy in mice alters steroid levels, focusing for example on the catechol-O-methyl transferase or cytochrome P450 enzymes in the livers and placentas and/or on fetal adrenal, ovarian, and testicular functions. It will also be necessary to investigate whether similar dysfunction of placental steroid metabolism occurs in humans.
To summarize, our data point to the possibility that preimplantation embryo biopsy deregulates placental steroid metabolism. If a similar effect takes place in humans, its characterization and understanding may lead to improving pregnancy outcomes where PGD is applied through proper monitoring of steroid delivery to the fetus, with supplementation if necessary—analogous to the use of corticosteroids to develop premature fetal lungs.
ACKNOWLEDGMENT
The authors thank Yasuhiro Yamauchi from M.A.W.'s lab for his initial work on setting up embryo biopsy protocol, Jonathan Riel from M.A.W.'s lab for help with sample collection, and John Grove from the Department of Public Health at the University of Hawaii for discussion on statistical analyses and experimental design.
Footnotes
This material is based on work supported by NIH RR024206 (Project 4) to A.C.C. and NIH RR024206 (Project 2) and NIH HD058059 to M.A.W.
REFERENCES
- Handyside AH. Preimplantation genetic diagnosis after 20 years. Reprod Biomed Online 2010; 21: 280 282 [DOI] [PubMed] [Google Scholar]
- Goossens V, Harton G, Moutou C, Traeger-Synodinos J, Van Rij M, Harper JC. ESHRE PGD. Consortium data collection IX: cycles from January to December 2006 with pregnancy follow-up to October 2007. Hum Reprod 2009; 24: 1786 1810 [DOI] [PubMed] [Google Scholar]
- Liebaers I, Desmyttere S, Verpoest W, De Rycke M, Staessen C, Sermon K, Devroey P, Haentjens P, Bonduelle M. Report on a consecutive series of 581 children born after blastomere biopsy for preimplantation genetic diagnosis. Hum Reprod 2010; 25: 275 282 [DOI] [PubMed] [Google Scholar]
- Grace KS, Sinclair KD. Assisted reproductive technology, epigenetics, and long-term health: a developmental time bomb still ticking. Semin Reprod Med 2009; 27: 409 416 [DOI] [PubMed] [Google Scholar]
- Hansen M, Bower C, Milne E, de Klerk N, Kurinczuk JJ. Assisted reproductive technologies and the risk of birth defects—a systematic review. Hum Reprod 2005; 20: 328 338 [DOI] [PubMed] [Google Scholar]
- Reddy UM, Wapner RJ, Rebar RW, Tasca RJ. Infertility, assisted reproductive technology, and adverse pregnancy outcomes: executive summary of a National Institute of Child Health and Human Development workshop. Obstet Gynecol 2007; 109: 967 977 [DOI] [PubMed] [Google Scholar]
- Savage T, Peek J, Hofman PL, Cutfield WS. Childhood outcomes of assisted reproductive technology. Hum Reprod 2011; 26: 2392 2400 [DOI] [PubMed] [Google Scholar]
- Duncan FE, Stein P, Williams CJ, Schultz RM. The effect of blastomere biopsy on preimplantation mouse embryo development and global gene expression. Fertil Steril 2009; 91: 1462 1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AC, Miyagi SJ, Yamauchi Y, Ward MA. Assisted reproduction technologies impair placental steroid metabolism. J Steroid Biochem Mol Biol 2009; 116: 21 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raunig JM, Yamauchi Y, Ward MA, Collier AC. Assisted reproduction technologies alter steroid delivery to the mouse fetus during pregnancy. J Steroid Biochem Mol Biol 2011; 126: 26 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raunig JM, Yamauchi Y, Ward MA, Collier AC. Placental inflammation and oxidative stress in the mouse model of assisted reproduction. Placenta 2011; 32: 852 858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn P, Barros C, Whittingham DG. Preservation of hamster oocytes to assay the fertilizing capacity of human spermatozoa. J Reprod Fertil 1982; 66: 161 168 [DOI] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86: 679 688 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biol Reprod 1995; 52: 709 720 [DOI] [PubMed] [Google Scholar]
- Ajduk A, Yamauchi Y, Ward MA. Sperm chromatin remodeling after intracytoplasmic sperm injection differs from that of in vitro fertilization. Biol Reprod 2006; 75: 442 451 [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem 1985; 150: 76 85 [DOI] [PubMed] [Google Scholar]
- Holtroff AF, Koch FC. The colorimetric estimation of 17-ketosteroids and their application to urine extracts. J Biol Chem 1940; 135: 377 392 [Google Scholar]
- Collier AC, Tingle MD, Keelan JA, Paxton JW, Mitchell MD. A highly sensitive fluorescent microplate method for the determination of UDP-glucuronosyl transferase activity in tissues and placental cell lines. Drug Metab Dispos 2000; 28: 1184 1186 [PubMed] [Google Scholar]
- Trubetskoy OV, Shaw PM. A fluorescent assay amenable to measuring production of beta-D-glucuronides produced from recombinant UDP-glycosyl transferase enzymes. Drug Metab Dispos 1999; 27: 555 557 [PubMed] [Google Scholar]
- Mulder GJ, van Doorn AB. A rapid NAD+-linked assay for microsomal uridine diphosphate glucuronyltransferase of rat liver and some observations on substrate specificity of the enzyme. Biochem J 1975; 151: 131 140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrett CA, Coughtrie MW. Phenol sulfotransferase 1A1 activity in human liver: kinetic properties, interindividual variation and re-evaluation of the suitability of 4-nitrophenol as a probe substrate. Biochem Pharmacol 2003; 66: 2089 2097 [DOI] [PubMed] [Google Scholar]
- Roy AB. Comparative studies on the liver sulphatases. Biochem J 1958; 68: 519 528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AB. On the relation between steroids, vitamin E and cancer, with introduction to a new biochemical test for an early diagnosis of neoplastic diseases. Acta Unio Int Contra Cancrum 1958; 14: 941 945 [PubMed] [Google Scholar]
- Chang PL, Moudgil G. A specific ultrastructural stain for arylsulfatase A activity in human cultured fibroblasts. J Histochem Cytochem 1984; 32: 617 624 [DOI] [PubMed] [Google Scholar]
- Roy AB. Sulphatases, lysosomes and disease. Aust J Exp Biol Med Sci 1976; 54: 111 135 [DOI] [PubMed] [Google Scholar]
- Snedecor GW, Cochran WG. Statistical Methods. Ames, IA: Iowa State University Press; 1989. [Google Scholar]
- Bates GW, Whitworth NS. Effects of obesity on sex steroid metabolism. J Chronic Dis 1982; 35: 893 896 [DOI] [PubMed] [Google Scholar]
- Goodman DS, Smith FR, Seplowitz AH, Ramakrishnan R, Dell RB. Prediction of the parameters of whole body cholesterol metabolism in humans. J Lipid Res 1980; 21: 699 713 [PubMed] [Google Scholar]
- Liu J, Van den Abbeel E, Van Steirteghem A. The in-vitro and in-vivo developmental potential of frozen and non-frozen biopsied 8-cell mouse embryos. Hum Reprod 1993; 8: 1481 1486 [DOI] [PubMed] [Google Scholar]
- Ludwig M, Muschalla H, Al-Hasani S, Diedrich K. The effect of multiple cryopreservation procedures and blastomere biopsy on the in-vitro development of mouse embryos. Hum Reprod 1998; 13: 3165 3168 [DOI] [PubMed] [Google Scholar]
- Pierce KE, Michalopoulos J, Kiessling AA, Seibel MM, Zilberstein M. Preimplantation development of mouse and human embryos biopsied at cleavage stages using a modified displacement technique. Hum Reprod 1997; 12: 351 356 [DOI] [PubMed] [Google Scholar]
- Ugajin T, Terada Y, Hasegawa H, Velayo CL, Nabeshima H, Yaegashi N. Aberrant behavior of mouse embryo development after blastomere biopsy as observed through time-lapse cinematography. Fertil Steril 2010; 93: 2723 2728 [DOI] [PubMed] [Google Scholar]
- Yu Y, Wu J, Fan Y, Lv Z, Guo X, Zhao C, Zhou R, Zhang Z, Wang F, Xiao M, Chen L, Zhu H. et al. Evaluation of blastomere biopsy using a mouse model indicates the potential high risk of neurodegenerative disorders in the offspring. Mol Cell Proteomics 2009; 8: 1490 1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Sandow BA, Morsy M, Kaufmann RA, Beebe SJ, Hodgen GD. Preclinical models for human pre-embryo biopsy and genetic diagnosis. I. Efficiency and normalcy of mouse pre-embryo development after different biopsy techniques. Fertil Steril 1992; 57: 425 430 [DOI] [PubMed] [Google Scholar]
- Wilton LJ, Shaw JM, Trounson AO. Successful single-cell biopsy and cryopreservation of preimplantation mouse embryos. Fertil Steril 1989; 51: 513 517 [DOI] [PubMed] [Google Scholar]
- Wilton LJ, Trounson AO. Biopsy of preimplantation mouse embryos: development of micromanipulated embryos and proliferation of single blastomeres in vitro. Biol Reprod 1989; 40: 145 152 [DOI] [PubMed] [Google Scholar]
- Krzyminska U, O'Neill C. The effects of cryopreservation and thawing on the development in vitro and in vivo of biopsied 8-cell mouse embryos. Hum Reprod 1991; 6: 832 835 [DOI] [PubMed] [Google Scholar]
- Krzyminska UB, Lutjen J, O'Neill C. Assessment of the viability and pregnancy potential of mouse embryos biopsied at different preimplantation stages of development. Hum Reprod 1990; 5: 203 208 [DOI] [PubMed] [Google Scholar]
- Santalo J, Grossman M, Egozcue J. Does Ca2+/Mg(2+)-free medium have an effect on the survival of the preimplantation mouse embryo after biopsy? Hum Reprod Update 1996; 2: 257 261 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cui Y, Zhou Z, Sha J, Li Y, Liu J. Altered global gene expressions of human placentae subjected to assisted reproductive technology treatments. Placenta 2010; 31: 251 258 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang YL, Feng C, Wu YT, Liu AX, Sheng JZ, Cai J, Huang HF. Comparative proteomic analysis of human placenta derived from assisted reproductive technology. Proteomics 2008; 8: 4344 4356 [DOI] [PubMed] [Google Scholar]
- Lalosevic D, Tabs D, Krnojelac D, Vejnovic T, Radunovic N. Histological characteristics of placentas from assisted reproduction programs. Med Pregl 2003; 56: 521 527 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao W, Jiang Y, Zhang R, Wang J, Li C, Zhao H, Gao L, Cui Y, Zhou Z, Sha J, Liu J. et al. Ultrastructural study on human placentae from women subjected to assisted reproductive technology treatments. Biol Reprod 2011; 85: 635 642 [DOI] [PubMed] [Google Scholar]
- Harper JC, Sengupta SB. Preimplantation genetic diagnosis: state of the art 2011. Hum Genet 2012; 131: 175 186 [DOI] [PubMed] [Google Scholar]
- Harper JC, Coonen E, De Rycke M, Harton G, Moutou C, Pehlivan T, Traeger-Synodinos J, Van Rij MC, Goossens V. ESHRE PGD. Consortium data collection X: cycles from January to December 2007 with pregnancy follow-up to October 2008. Hum Reprod 2010; 25: 2685 2707 [DOI] [PubMed] [Google Scholar]
- Harton GL, Magli MC, Lundin K, Montag M, Lemmen J, Harper JC. ESHRE PGD. Consortium/Embryology Special Interest Group—best practice guidelines for polar body and embryo biopsy for preimplantation genetic diagnosis/screening (PGD/PGS). Hum Reprod 2011; 26: 41 46 [DOI] [PubMed] [Google Scholar]
- Ducibella T, Albertini DF, Anderson E, Biggers JD. The preimplantation mammalian embryo: characterization of intercellular junctions and their appearance during development. Dev Biol 1975; 45: 231 250 [DOI] [PubMed] [Google Scholar]
- Levy JB, Johnson MH, Goodall H, Maro B. The timing of compaction: control of a major developmental transition in mouse early embryogenesis. J Embryol Exp Morphol 1986; 95: 213 237 [PubMed] [Google Scholar]
- Ziomek CA, Johnson MH. Cell surface interaction induces polarization of mouse 8-cell blastomeres at compaction. Cell 1980; 21: 935 942 [DOI] [PubMed] [Google Scholar]
- Hardy K, Warner A, Winston RM, Becker DL. Expression of intercellular junctions during preimplantation development of the human embryo. Mol Hum Reprod 1996; 2: 621 632 [DOI] [PubMed] [Google Scholar]
- Damsky CH, Richa J, Solter D, Knudsen K, Buck CA. Identification and purification of a cell surface glycoprotein mediating intercellular adhesion in embryonic and adult tissue. Cell 1983; 34: 455 466 [DOI] [PubMed] [Google Scholar]
- Johnson MH, Maro B, Takeichi M. The role of cell adhesion in the synchronization and orientation of polarization in 8-cell mouse blastomeres. J Embryol Exp Morphol 1986; 93: 239 255 [PubMed] [Google Scholar]
- Lee S, Gilula NB, Warner AE. Gap junctional communication and compaction during preimplantation stages of mouse development. Cell 1987; 51: 851 860 [DOI] [PubMed] [Google Scholar]
- Lo CW, Gilula NB. Gap junctional communication in the preimplantation mouse embryo. Cell 1979; 18: 399 409 [DOI] [PubMed] [Google Scholar]
- Ducibella T, Anderson E. Cell shape and membrane changes in the eight-cell mouse embryo: prerequisites for morphogenesis of the blastocyst. Dev Biol 1975; 47: 45 58 [DOI] [PubMed] [Google Scholar]
- Johnson MH, Maro B. A dissection of the mechanisms generating and stabilizing polarity in mouse 8- and 16-cell blastomeres: the role of cytoskeletal elements. J Embryol Exp Morphol 1985; 90: 311 334 [PubMed] [Google Scholar]
- Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays 1993; 15: 531 538 [DOI] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev 1990; 26: 90 100 [DOI] [PubMed] [Google Scholar]
- Chen JZ, Wong MH, Brennecke SP, Keogh RJ. The effects of human chorionic gonadotrophin, progesterone and oestradiol on trophoblast function. Mol Cell Endocrinol 2011; 342: 73 80 [DOI] [PubMed] [Google Scholar]
- Ratajczak CK, Fay JC, Muglia LJ. Preventing preterm birth: the past limitations and new potential of animal models. Dis Model Mech 2010; 3: 407 414 [DOI] [PubMed] [Google Scholar]
- Mendelson CR. Minireview: fetal-maternal hormonal signaling in pregnancy and labor. Mol Endocrinol 2009; 23: 947 954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu BT. Catechol-O-methyltransferase (COMT)-mediated methylation metabolism of endogenous bioactive catechols and modulation by endobiotics and xenobiotics: importance in pathophysiology and pathogenesis. Curr Drug Metab 2002; 3: 321 349 [DOI] [PubMed] [Google Scholar]
- Delle Piane L, Lin W, Liu X, Donjacour A, Minasi P, Revelli A, Maltepe E, Rinaudo PF. Effect of the method of conception and embryo transfer procedure on mid-gestation placenta and fetal development in an IVF mouse model. Hum Reprod 2010; 25: 2039 2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet 2001; 2: 538 548 [DOI] [PubMed] [Google Scholar]