Abstract
MicroRNAs (miRNAs) are involved in a number of biological processes, including tumour biology. Pre-clinical studies have shown that miRNA-126 regulates signalling downstream of vascular endothelial growth factor receptor 2 (VEGFR-2) and, consequently, angiogenesis. The aim of this study was to analyse the possible relationship between miRNA-126, VEGFR-2 and angiogenesis in tumour tissue from patients with colorectal cancer (CRC). Tumour tissue was obtained from 81 patients. The miRNA-126 and VEGFR-2 gene expression levels were analysed by PCR and the protein concentrations of VEGFR-2 were analysed by ELISA. Angiogenesis, visualised by the endothelial cell marker CD105 combined with caldesmon, was assessed by immunohistochemistry and the microvessel density (MVD) technique. In situ hybridisation was performed for miRNA-126. Tumours were classified as low or high miRNA-126-expressing using the median as the cut-off. The median gene expression levels of VEGFR-2 were significantly lower in the tumours expressing low levels of miRNA-126, 0.30 (95% CI, 0.24–0.36), compared to those expressing high levels of miRNA-126, 0.48 (95% CI, 0.28–0.60), p=0.02. A positive association was observed with VEGFR-2 protein concentrations, p=0.06. The median MVD was significantly lower in the tumours expressing low levels of miRNA-126, 5.8 (95% CI, 5.33–6.67), compared to those expressing high levels, 8.0 (95% CI, 6.33–9.00), p<0.01. miRNA-126 was detected in endothelial cells by in situ hybridisation analysis. These results suggest that high levels of miRNA-126 in CRC are associated with high VEGFR-2 mRNA and protein levels and a higher density of newly formed microvessels. However, further studies should be conducted to analyse the clinical value of miRNA-126 in CRC.
Keywords: angiogenesis, colorectal neoplasm, microRNAs, microvessel density, vascular endothelial growth factor receptor 2
Introduction
Angiogenesis, the development of capillaries from pre-existing blood vessels, is essential for the growth of malignant tumours (1). The vascular endothelial growth factor (VEGF) system plays a significant role in regulating this process (2), and the clinical benefit from targeting this system in patients with metastatic colorectal cancer (mCRC) is well documented (3).
Pre-clinical studies demonstrated a role of microRNAs (miRNAs) in the regulation of angiogenesis and the VEGF system (4,5). miRNA-126 has been reported to play an essential role in angiogenesis by modulating VEGFR-2-related signal transduction through the RAS/ERK and PI3K/AKT pathways by inhibiting regulatory units (6–9).
miRNAs are a group of small single-stranded non-coding RNAs with a length of approximately 22 nucleotides. They negatively regulate gene transcripts at a post-transcriptional level by targeting mRNAs leading to mRNA degradation or translational repression, depending on the complementarities between miRNA and its targets (10). It has been demonstrated that one miRNA may target several different mRNAs, and that one mRNA may be targeted by several miRNAs; several other principles have been suggested to explain miRNA-mediated gene regulation.
Approximately 1,000 unique human miRNAs have been identified (11). Most of the miRNAs are located in introns of mRNA transcripts, whereas others have been identified in exons, in the 3′ untranslated region (3′UTR) of a gene transcript or in non-coding transcripts, including miRNA clusters. A large fraction of the identified miRNAs are located in areas of the genome known to be involved in the development of cancer and are consequently thought to play a prominent role in malignant transformation (10). The expression levels of miRNAs are often dysregulated in malignant tissue compared to the corresponding normal tissue, which enables miRNAs to act as either tumour suppressors or oncogenes depending on the function of their target networks (10). An increasing number of reports on the clinical significance of miRNAs in various types of cancer, including CRC, have been published. Differences in expression levels between CRC and normal colorectal tissue have been described (12–14), and certain studies have also provided evidence of prognostic (15,16) and predictive (17) value related to miRNAs.
At present, little is known about the possible relationship between miRNAs and the VEGF system in patients with CRC. The aim of this study was to analyse the relationship between miRNA-126, VEGFR-2 and neo-angiogenesis in patients with CRC.
Materials and methods
Study population
This study included 81 consecutive patients with CRC during the period between February 2004 and July 2005, all undergoing surgical resection for histologically verified adenocarcinomas of the colon or rectum at the Department of Surgery, Vejle Hospital, Denmark. Patients who had received preoperative chemoradiation for rectal cancer were excluded. Pre-treatment examinations included a chest X-ray and ultrasound or CT scan of the abdomen. Postoperatively, the tumours were histologically classified and staged according to the pTNM system. Information regarding patient characteristics was based on patient records and registries. The study was approved by the Regional Scientific Ethical Committee for Southern Denmark according to Danish law, J.nr. S-VF-20040047. Informed consent was obtained from all patients enrolled in the study.
Tissue sampling
Immediately after surgery the removed bowel segment was brought to the Department of Clinical Pathology and tissue from the tumour was sampled by a pathologist. Samples for mRNA and miRNA analyses were placed in RNAlater™ (Qiagen, CA, USA) and stored at −20˚C. Samples for quantitative protein analysis were frozen and stored at −80˚C. The samples were frozen within 30 min of surgical removal. Samples intended for later immunohistochemistry (IHC) and in situ hybridisation (ISH) followed routine fixation and paraffin embedding. Samples for quantitative protein analysis and IHC were sampled in close proximity to each other. Based on a microscopic examination of H&E-stained FFPE tumour sections, it was semi-quantitatively estimated that the tissue used for quantitative protein analyses was dominated by carcinoma cells (>50%).
VEGFR-2 mRNA and protein analyses
The VEGFR-2 gene expression and protein analyses have previously been described (18). Briefly, tissue samples were homogenised and total RNA was isolated according to the RNeasy® mini handbook of June 2001 (Qiagen, MD, USA). RNA was quantified using spectrophotometry (Eppendorf, Hamburg, Germany) followed by cDNA synthesis using a M-MLV RT kit (Invitrogen Co., Carlsbad, CA, USA). The geometrical mean of β-2-microglobulin and β-actin was used for normalisation. Gene expression analyses were performed using fluorescence-based real-time reverse transcription-polymerase chain reaction (RT-PCR). The analyses were performed on the ABI PRISM 7900 HT fast real-time PCR system, TaqMan (Applied Biosystems, Foster City, CA, USA).
After protein extraction, VEGFR-2 was analysed using Quantikine ELISA kits (DVR200; R&D Systems, Minneapolis, MN, USA). The controls were also purchased from R&D Systems. The assay employs the quantitative sandwich enzyme immunoassay technique. Results are presented in pg/mg of total protein. Tissue samples, standards and controls were assayed in duplicate and the mean was recorded. The total coefficients of variation on three levels (low, medium and high concentrations of VEGFR-2) were 11.3, 10.1 and 8.3%, respectively.
CD105 and caldesmon immunostaining and MVD counting
The staining and counting procedures have previously been described (19). Briefly, MVD was measured using tissue sections stained by antibodies against CD105 and caldesmon in order to visualise immature microvessels. A kit from Dako (Glostrup, Denmark; code K5361, EnVision™ G|2 doublestain system, rabbit/mouse, DAB+/permanent red) was used for the detection of primary antibodies. The anti-CD105 antibody was obtained from Novocastra (Bristol, UK; endoglin NCL-CD105, clone 4G11) and used at a dilution of 1:25. The anti-caldesmon antibody was obtained from Dako (code M3557, clone h-CD) and used at a dilution of 1:50.
Microvessels were counted at the invasive tumour front by two observers unaware of the clinical parameters. The mean MVD from three hotspots was used for statistical analysis. Any stained endothelial cell or endothelial cell cluster clearly separated from adjacent microvessels by tumour cells and/or stroma elements was considered a single countable microvessel. Vessel lumen was not necessary for a structure to be counted as a microvessel. Consensus counts resolved any discrepancy between the observers. Regarding the CD105 (brown) and caldesmon (red) staining, only microvessels without associated red staining were counted in order to preferentially estimate the MVD of the immature vessels.
miRNA-126 gene expression analyses
miRNA quantification was carried out using predesigned TaqMan miRNA assays (Applied Biosystems). cDNA synthesis, pre-amplification and qPCR were performed according to the manufacturer's instructions. In brief, RNA was reversely transcribed using the TaqMan miRNA reverse transcription kit in combination with the stem-loop Megaplex™ primer pool A v2.0, allowing simultaneous reverse transcription of 377 unique miRNAs. Subsequently, the Megaplex RT product was pre-amplified using TaqMan PreAmp master mix and Megaplex PreAmp primer pool A v2.0. RT-PCR amplification of miRNA-126 (part no. 4395339) was performed in TaqMan Universal PCR master mix. Cycling conditions were as follows: 95˚C for 10 min followed by 40 cycles of 95˚C for 15 sec and 60˚C for 1 min. PCR reactions were performed on the ABI PRISM 7900 HT Fast RT-PCR system (Applied Biosystems). Raw Cq values were calculated using the SDS software v2.1 using automatic baseline and threshold settings. Expression levels of Hsa-miR-340 (part no. 4427975) were used for normalisation, since this miRNA was found by the NormFinder algorithm (20), a rigorous statistical approach for evaluating expression variation of normalisation gene candidates, to be the most stably expressed transcript throughout the tissue samples.
miRNA-126 in situ hybridisation
ISH was performed as described elsewhere (21). In brief, ISH was carried out on 6-μm tissue sections containing CRC and normal colorectal tissue using double DIG-labelled LNA probes for human miRNA-126 (Exiqon A/S, Vedbaek, Denmark). The DIG-labelled probe was detected with alkaline phosphatase-conjugated sheep anti-DIG Fab fragments followed by NBT-BCIP chromogenic staining and nuclear fast red staining.
Statistical analysis
The Fisher's exact test was used for two-group comparisons. Median values were compared using the Wilcoxon rank sum test. Disease-free survival (DFS) was defined as the time from surgery until the first documented tumour recurrence or death. Progression-free survival (PFS) was defined as the time from surgery until the first documented tumour recurrence, progression or death. Overall survival (OS) was defined as the time from surgery until death. Survival curves were determined according to the Kaplan-Meier method, and the log-rank test was used to test for differences between the groups. Survival data from patients diagnosed with a new malignancy following their surgical resection for CRC (5 patients) were censored from the date of their new cancer diagnosis. These data were excluded to prevent any potential bias related to the presence of a new cancer or the chemotherapeutic treatments used. Statistical calculations were carried out using NCSS statistical software (NCSS Statistical Software, version 2007; Kaysville, UT, USA). P<0.05 was considered to be statistically significant and all tests were two-sided.
Results
Patient characteristics
The patient characteristics, along with miRNA-126 expression levels in CRC obtained by PCR analysis, are shown in Table I. Female patients presented with a significantly lower median miRNA-126 expression in their tumour tissue compared to their male counterparts. Furthermore, non-significant associations were observed with regard to age at diagnosis and tumour localisation. Younger patients and patients with left-sided colon cancers presented with a higher miRNA-126 expression. No differences were observed between the patient characteristics, as listed in Table I, and gene or protein levels of VEGFR-2 or MVD. The 5-year survival rate for the entire cohort was 60%.
Table I.
Patient characteristics.
| No. (%) (n=81) |
miRNA-126 | ||
|---|---|---|---|
|
| |||
| Median (95% CI) | p-value | ||
| Gender | 0.04 | ||
| Male | 37 (46) | 1.00 (0.87–1.28) | |
| Female | 44 (54) | 0.83 (0.59–1.08) | |
| Age (years) | 0.09 | ||
| Mean (SD) | 71.7 (11.1) | ||
| Range | 47–91 | ||
| >mean | 44 (54) | 0.85 (0.89–1.48) | |
| <mean | 37 (46) | 1.02 (0.89–1.48) | |
| T category | 0.72 | ||
| 1–3 | 63 (78) | 0.97 (0.82–1.12) | |
| 4 | 18 (22) | 0.79 (0.59–1.48) | |
| N category | 0.81 | ||
| 0 | 46 (57) | 0.99 (0.72–1.13) | |
| 1–2 | 35 (43) | 0.96 (0.77–1.28) | |
| M category | 0.57 | ||
| 0 | 65 (80) | 0.96 (0.81–1.12) | |
| 1 | 16 (20) | 0.90 (0.70–1.69) | |
| Stage | 0.73 | ||
| I–II | 43 (53) | 1.01 (0.68–1.13) | |
| III–IV | 38 (47) | 0.95 (0.77–1.28) | |
| Localisation | |||
| Rectum | 20 (25) | 0.88 (0.70–1.48) | 0.88 |
| Colon | 61 (75) | 0.98 (0.77–1.23) | 0.07 |
| Left colon | 28 (46) | 1.05 (0.94–1.47) | |
| Right colon | 33 (54) | 0.77 (0.56–1.13) | |
| MSI status | 0.33 | ||
| MSI | 15 (19) | 0.72 (0.38–1.24) | |
| MSS | 66 (81) | 0.96 (0.82–1.08) | |
| Tumour grade | 0.74 | ||
| 1 and 2 | 61 (75) | 0.95 (0.79–1.08) | |
| 3 | 20 (25) | 0.96 (0.59–1.58) | |
| Vascular invasion | 0.42 | ||
| Yes | 10 (12) | 1.08 (0.70–1.73) | |
| No | 71 (88) | 0.96 (0.79–1.02) | |
| Neuronal invasion | 0.18 | ||
| Yes | 10 (12) | 1.32 (0.70–1.87) | |
| No | 71 (88) | 0.94 (0.77–1.02) | |
| Peritoneal perforation | 0.33 | ||
| Yes | 17 (21) | 0.77 (0.59–1.36) | |
| No | 64 (79) | 0.97 (0.83–1.12) | |
CI, confidence interval; SD, standard deviation; MSI, microsatellite instable; MSS, microsatellite stable.
miRNA-126 expression
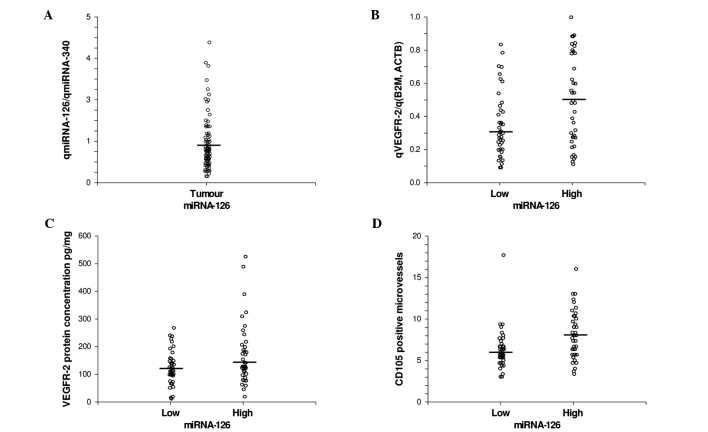
Paired samples of CRC and normal colorectal tissue were obtained from 28 patients. The median gene expression of miRNA-126 in CRC, 0.85 (95% CI, 0.71–0.97), did not differ significantly from the median gene expression in normal colorectal tissue, 1.01 (95% CI, 0.80–1.11), p=0.56. The distribution of miRNA-126 expression levels in the CRC samples for the entire patient cohort is shown in Fig. 1A. Tumours were classified as low or high miRNA-126-expressing tumours using the median as a cut-off.
Figure 1.
(A) Distribution of miRNA-126 gene expression in CRC tissue. (B) Distribution of VEGFR-2 gene expression in CRC tissue according to miRNA-126 levels, p=0.02. (C) Distribution of VEGFR-2 protein concentration in CRC tissue according to miRNA-126 levels, p=0.06. (D) Distribution of CD105-positive microvessel density in CRC tissue according to miRNA-126 levels, p<0.01. Black bars, medians, n=81.
The distribution of VEGFR-2 gene expression in CRC based on miRNA-126 levels is shown in Fig. 1B. The median gene expression level of VEGFR-2 was significantly lower in the tumours expressing low levels of miRNA-126, 0.30 (95% CI, 0.24–0.36), compared to those expressing high levels, 0.48 (95% CI, 0.28–0.60), p=0.02.
The distribution of VEGFR-2 protein concentrations in CRC based on miRNA-126 levels is shown in Fig. 1C. The median protein concentration of VEGFR-2 was slightly lower, although not significant, in the tumours expressing low levels of miRNA-126, 115 pg/mg (95% CI, 98–140), compared to those expressing high levels, 134 pg/mg (95% CI, 121–180), p=0.06.
The median MVD was significantly lower in the tumours expressing low levels of miRNA-126, 5.8 (95% CI, 5.33–6.67), compared to those expressing high levels, 8.0 (95% CI, 6.33–9.00), p<0.01 (Fig. 1D).
miRNA-126 in situ hybridisation
ISH analyses using high-affinity LNA probes revealed intense ISH signals in endothelial cells. miRNA-126-positive vessels were observed in the tumour stroma, as well as in the lamina propria of normal mucosa and in the deep unaffected bowel wall (Fig. 2).
Figure 2.
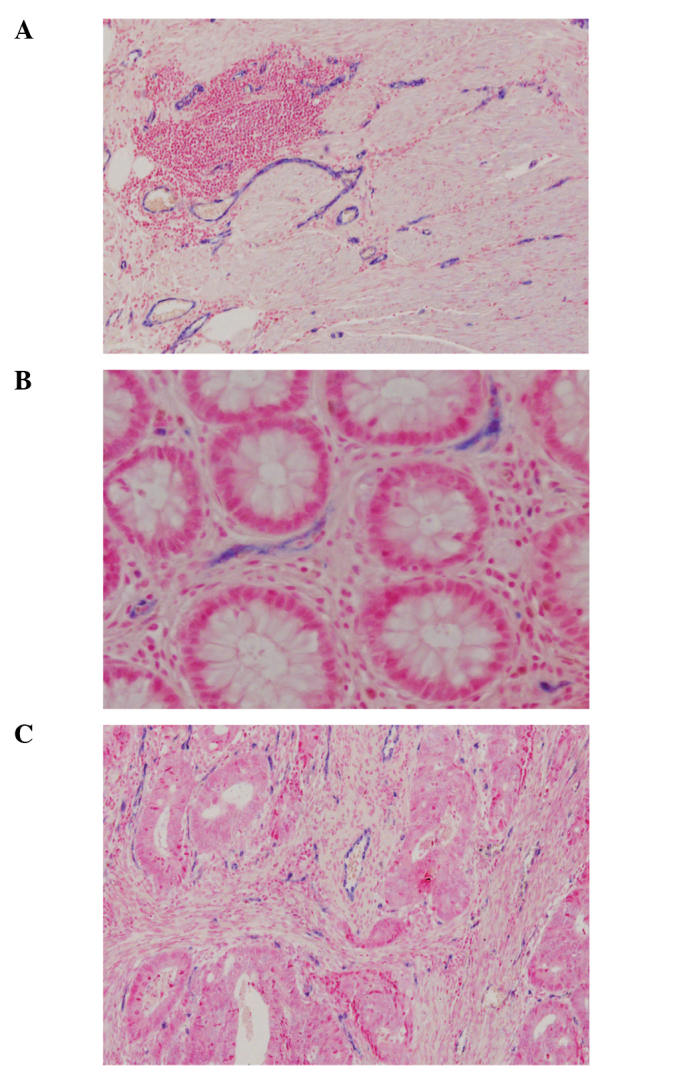
miRNA-126 ISH. (A and B) Expression of miRNA-126 in normal colon tissue. (C) Expression in colon cancer tissue. The miRNA-126 ISH signal is observed in endothelial cells and is not confined to tumour-associated vessels.
miRNA-126 and prognosis
The possible prognostic value of miRNA-126 expression in CRC was assessed by comparing patients with a low expression of miRNA-126 to those with a high expression, using the median as the cut-off. Neither PFS [hazard ratio 0.85 (95% CI, 0.44–1.64)] nor OS [hazard ratio 0.99 (95% CI, 0.49–1.98)] differed significantly between the two groups (p=0.64 and p=0.98, respectively).
Our patient cohort included a total of 28 patients with microsatellite stable (MSS) stage II CRC and, within this subgroup, short DFS was significantly linked to high miRNA-126 expression levels (Fig. 3).
Figure 3.
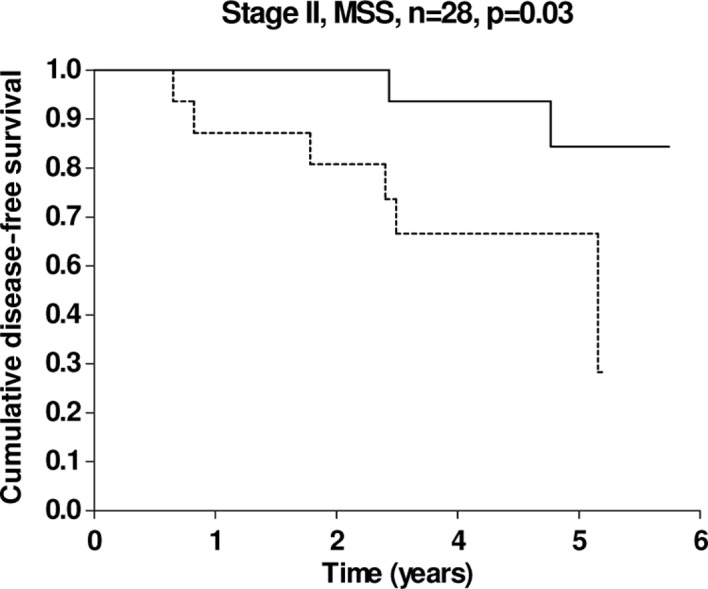
Kaplan-Meier DFS curves according to miRNA-126 expression in patients with stage II and MSS tumours, n=28 (14 patients in each group). The solid line pertains to patients with a low miRNA-126 expression and the dashed line to patients with a high miRNA-126 expression, p=0.03.
Discussion
In the present pilot study, a direct correlation was found between miRNA-126, VEGFR-2 and angiogenesis in samples from patients with CRC.
Paired samples of CRC and normal colorectal tissue were obtained from 28 patients, and the median miRNA-126 expression level in the two tissue types did not differ significantly. Guo et al (7) demonstrated that the miRNA-126 level was significantly lower in colon cancer compared to normal colon tissue in a panel of 6 paired samples. In both cases, the results are based on a rather limited number of paired samples and should therefore be interpreted with caution. Furthermore, the use of different normalisation methods (Guo et al used 5S rRNA) may also be significant in this context.
The miRNA-126 expression in CRC was significantly lower in females compared to males, and a possible, albeit non-significant, difference was also observed regarding age and tumour localisation. The miRNA-126 expression tended to be lower in the older group of patients and in patients with right-sided colon cancers.
Based on these findings, a correlation with microsatellite instable (MSI) status was expected. However, no such correlation was found in the present study. Díaz et al (15) analysed several miRNAs in 110 patients who underwent surgery for CRC. miRNA-126 expression was found to be significantly lower in the youngest group of patients and no significant association with gender was documented. These discrepancies are difficult to explain since the two studies involved a similar number of patients, included Caucasians only, used almost the same methodology, although Díaz et al used 5S for normalisation, and also included patients with stage I–IV disease. The study by Díaz et al used fresh-frozen tumour tissue compared to the present study in which miRNA was extracted from tissue preserved in RNAlater. A possible effect on miRNA-126 expression levels by directly comparing preservation in RNAlater and freezing of the tissue specimens remains to be clarified.
A high miRNA-126 expression was correlated to a high gene expression of VEGFR-2 and likely also to high protein concentrations of VEGFR-2 in CRC, although the latter correlation did not reach statistical significance (p=0.06). Furthermore, a high miRNA-126 expression was significantly correlated to a higher density of newly formed micro vessels. These results, obtained in a clinical setting, suggest a relationship between the VEGF system, neo-angiogenesis and miRNA-126, and correlate well with results reported in the pre-clinical literature. Two recent studies (6,9) have demonstrated the significant role of miRNA-126 in media-ting developmental angiogenesis and identified two specific targets for miRNA-126, i.e., the sprouty-related, EVH1 domain-containing protein 1 (SPRED-1) and the phospho-inositol-3 kinase regulatory subunit 2 (PIK3R2), both acting as negative regulators of the RAS/ERK and PI3K/AKT pathways, respectively. miRNA-126 is capable of potentiating the angiogenic signalling downstream of VEGFR-2 by modulating the signals through these pathways. Other studies have demonstrated a similar regulatory role of miRNA-126 with regard to these pathways (7,22). In 2010, Nicoli et al (8) published results suggesting that blood flow stimulates angiogenesis in the zebrafish by a genetic pathway in which a transcription factor (klf2a) induces the expression of miRNA-126 and thereby enhances VEGF-A signalling. These pre-clinical studies, confirm that high levels of miRNA-126 increase signalling downstream of VEGFR-2, ultimately resulting in increased angiogenesis. To a certain extent, this increase is supported by the present results obtained in a clinical setting.
Our results showed a relationship between miRNA-126, VEGFR-2 and neo-angiogenesis estimated by CD105 MVD, suggesting that miRNA-126 expression was correlated to tissue vascularisation. The miRNA-126 ISH analyses revealed an expression in endothelial cells in malignant and normal colorectal tissue similar to results from various in vitro studies (5,6,8,9). These observations indicate high and specific miRNA-126 expression levels in endothelial cells. The clinical value of the miRNA-126 ISH analyses, which may be an alternative to the MVD technique based on IHC, deserves further analysis using larger patient cohorts.
The possible prognostic value of miRNA-126 expression in CRC was analysed. However, no significant correlation with survival was found in the unselected patient cohort, which corresponds to the results obtained by Díaz et al (15). A subgroup analysis consisting of patients with stage II disease only and MSS tumours is relevant due to the need to identify new prognostic markers in stage II CRC, and the well-known prognostic difference between MSI and MSS tumours (23) justifies this selection. Although this subgroup only consisted of 28 patients in the present study, it is significant that our observations are consistent with the findings reported by Schepeler et al (16). These authors demonstrated that miRNA-126, among other miRNAs, was up-regulated in the primary tumour tissue from patients with stage II MSS colon cancers who experienced relapse significantly later compared to those that did not. A meaningful multivariate survival analysis was not conducted due to the small sample size. Therefore, although miRNA-126 expression in CRC does not appear to be of prognostic value in the entire patient cohort, it may harbour a prognostic impact for a selected group of CRC patients. The meta-analysis by Des et al demonstrated a correlation between high MVD and poor prognosis in CRC (24). The present relationship between high MVD and high miRNA-126 expression in the present study provides a plausible explanation for these findings, although this hypothesis requires validation in a larger patient cohort.
In conclusion, significant associations were found between miRNA-126 levels in CRC, VEGFR-2 gene expression and neo-angiogenesis, although a great overlap was observed between the compared groups. Conclusions should therefore be drawn with caution, and additional testing in larger and more homogeneous study populations is necessary to validate the results. The present results correspond well with the pre-clinical literature, indicating a pivotal role of miRNA-126 in the modulation of angiogenesis (6–9,25). Thus, miRNA-126 may harbour predictive information with regard to anti-angiogenetic therapy. A better understanding of how this miRNA is regulated is of great significance. Further studies that analyse the clinical value of miRNA-126 in CRC, are therefore required.
Acknowledgements
The authors are grateful for the technical assistance provided by Lone Frischknecht, Lone Hartmann Hansen, Sara Egsgaard, Birgit Roed Sørensen, Pamela Celis and Karin Larsen. This study was supported by The Cancer Foundation, which had no influence on any part of the study.
References
- 1.Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 3.Wagner AD, Arnold D, Grothey AA, Haerting J, Unverzagt S. Anti-angiogenic therapies for metastatic colorectal cancer. Cochrane Database Syst Rev. 2009;8:CD005392. doi: 10.1002/14651858.CD005392.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hua Z, Lv Q, Ye W, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. Plos One. 2006;1:E116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 6.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 12.Faber C, Kirchner T, Hlubek F. The impact of microRNAs on colorectal cancer. Virchows Arch. 2009;454:359–367. doi: 10.1007/s00428-009-0751-9. [DOI] [PubMed] [Google Scholar]
- 13.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 14.Yamamichi N, Shimomura R, Inada K, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15:4009–4016. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 15.Díaz R, Silva J, Garcia JM, Lorenzo Y, Garcia V, Pena C, Rodriguez R, Munoz C, Garcia F, Bonilla F, Dominguez G. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 16.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 17.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen TF, Spindler KL, Lorentzen KA, Olsen DA, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A. Quantitative analysis of vascular endothelial growth factor receptors 1 and 2 in colorectal cancer. Mol Med Reports. 2009;2:787–792. doi: 10.3892/mmr_00000173. [DOI] [PubMed] [Google Scholar]
- 19.Hansen TF, Sorensen FB, Spindler KL, Olsen DA, Andersen RF, Lindebjerg J, Brandslund I, Jakobsen A. Microvessel density and the association with single nucleotide polymorphisms of the vascular endothelial growth factor receptor 2 in patients with colorectal cancer. Virchows Arch. 2010;456:251–260. doi: 10.1007/s00428-009-0878-8. [DOI] [PubMed] [Google Scholar]
- 20.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen S, Baker A, Moller S, Nielsen BS. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 2010;52:375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 23.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 24.Des GG, Uzzan B, Nicolas P, Cucherat M, Morere JF, Benamouzig R, Breau JL, Perret GY. Microvessel density and VEGF expression are prognostic factors in colorectal cancer. Meta-analysis of the literature. Br J Cancer. 2006;94:1823–1832. doi: 10.1038/sj.bjc.6603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuehbacher A, Urbich C, Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci. 2008;29:12–15. doi: 10.1016/j.tips.2007.10.014. [DOI] [PubMed] [Google Scholar]