Abstract
Stem cells are thought to contribute to tissue regeneration as well as carcinogenesis. Ulcerative colitis-associated colorectal cancer (UC-CRC) has shown distinct characteristics compared with those of sporadic CRC. The aim of this study was to evaluate the expression of stem cell markers CD133, OCT4 and NANOG in UC-CRC and the inflamed colonic epithelium of UC patients. Total RNAs of UC-CRC (n=6), inflamed colonic epithelium (n=24), sporadic CRC (n=37) and adjacent normal colonic epithelium (n=37) were isolated from formalin-fixed, paraffin-embedded specimens using microdissection techniques in order to purify colonic epithelial cells. Relative mRNA levels of CD133 (PROM), OCT4 (POU5F1) and NANOG were measured using real-time reverse transcription polymerase chain reaction. Three stem cell markers were also investigated immunohistochemically. PROM, POU5F1 and NANOG levels were found to be significantly lower in UC-CRC than in inflamed colonic epithelium of UC patients. By contrast, sporadic CRC showed a significantly higher expression of PROM, POU5F1 and NANOG compared with adjacent normal colonic epithelium. POU5F1 and NANOG levels were significantly lower in UC-CRC than in sporadic CRC. PROM and NANOG levels in inflamed colonic epithelium were significantly higher among younger UC patients (P<0.05). Longer disease duration was significantly associated with lower PROM expression (P=0.0117). No significant difference was found in PROM levels between UC-CRC and inflamed colonic epithelium in patients with longer disease duration. UC-CRC showed different expression profiles of stem cell markers compared with sporadic CRC. Decreases in PROM expression of inflamed colonic epithelium may identify UC patients at high risk for the development of UC-CRC.
Keywords: CD133, OCT4, NANOG, ulcerative colitis-associated colorectal cancer, ulcerative colitis
Introduction
Ulcerative colitis (UC) is a chronic inflammatory bowel disease with an increased risk of developing into colorectal cancer (CRC). The meta-analysis by Eaden et al reported that the cumulative risk probability of ulcerative colitis-associated CRC (UC-CRC) was 1.6% after 10 years, 8.3% after 20 years, and 18.4% after 30 years of disease duration (1). Risk factors for UC-CRC have been shown to be greater with the extent and duration of disease, increased severity of bowel inflammation, coexisting primary sclerosing cholangitis, young age at onset of UC and family history of sporadic CRC (2).
Typically, UC patients with a higher risk of UC-CRC undergo periodic colonoscopic surveillance with multiple biopsies for the early diagnosis and treatment of dysplasia or cancer lesions. However, such a surveillance program has a number of limitations, including its efficacy, high cost, invasiveness, incomplete patient enrollment, sampling variations and poor agreement in histopathological interpretation (3). A number of molecular markers predictive for UC-CRC have been reported (4–6), but remain unavailable in the practical management of UC patients.
Chronic inflammation in UC is considered to cause colonic epithelial injury, repair and regeneration, or the development of UC-CRC (7). Stem cell (SC) research and the cancer stem cell (CSC) hypothesis have shown that colonic SCs or CSCs are involved in tissue regeneration and colonic carcinogenesis (8–10). CSCs may greatly contribute to inflammation-associated carcinogenesis in UC.
The transcription factors OCT4 and NANOG play significant roles in maintaining pluripotency and self-renewal of embryonic stem cells as well as in adult stem cells (11). OCT4 is a nuclear protein belonging to a family of transcription factors containing the POU DNA-binding domain (12). NANOG is a homeobox transcription factor that contains DNA-binding domains involved in the regulation of key eukaryotic developmental processes (13). CD133 is a cell surface glycoprotein with five transmembrane domains. CD133 is currently regarded as one of the most significant markers of colon CSCs (14).
Subsequently, we investigated the expression of stem cell markers in UC-CRC and inflamed colonic epithelium as compared to sporadic CRC. To evaluate the expression levels of each molecule in colonic epithelial cells, microdissection techniques were used for the reduction of the contamination of stromal cells as much as possible.
In the present study, expression levels of PROM, POU5F1 and NANOG mRNAs were investigated in UC-CRC, inflamed colonic epithelium, sporadic CRC and adjacent colonic epithelium. Furthermore, the associations between disease characteristics and stem cell marker expression were also examined to evaluate their predictive value for the development of UC-CRC.
Materials and methods
Patients with UC and UC-CRC
A total of 24 UC patients and 6 patients with UC-CRC were included in the current analyses. All patients were treated at the Department of Gastrointestinal and Pediatric Surgery in the Mie University Graduate School of Medicine, Japan.
The UC diagnosis was based on the medical history, endoscopic findings, histological examination, laboratory tests and clinical disease presentation. The extent of disease was defined as proctitis, left-sided and extensive colitis. The severity of disease was classified as ‘mild’, ‘moderate’ and ‘severe’ based on clinical, endoscopic and histopathological findings as discussed above.
Patients with sporadic colorectal cancer
A total of 37 patients with sporadic CRC were also included in this study to compare the expression of stem cell markers between UC-CRC and sporadic CRC.
Sample selection criteria
For inclusion in the study, selection criteria required the availability of samples accompanying complete clinical data and the quality of isolated RNA for real-time PCR based on the expression of the β-actin (ACTB) gene as an internal control.
Microdissection of formalin-fixed, paraffin-embedded (FFPE) specimens
To purify colonic epithelial cells or cancer cells, a microdissection technique was used. Surgically resected specimens were fixed in 10% formaldehyde solution v/v and embedded in paraffin. Sections (10 μm) of FFPE specimens were stained with nuclear fast red. Manual microdissection using a scalpel was performed to collect only inflamed and non-inflamed colonic epithelial or cancer cells, excluding stromal cells as much as possible. Sample collection by microdissection typically ensured >70% of target cells for each sample.
RNA extraction from FFPE specimens
Microdissected samples were digested with proteinase K in lysis buffer containing Tris-HCl, EDTA and sodium dodecyl sulfate as previously reported with minor modifications (15). RNA was purified by phenol and chloroform extraction.
cDNA synthesis
cDNA was synthesized with random hexamer primers and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Real-time quantitative RT-PCR
Real-time quantitative RT-PCR analysis was performed with the SYBR-Green PCR master mix using an Applied Biosystems 7500 real-time PCR system according to the manufacturer’s instructions (Applied Biosystems, Inc., Foster City, CA, USA). Primers and probes for PROM (CD133), POU5F1 (OCT4), NANOG and ACTB (β-actin) were designed with primer3 software (Biology Workbench Version 3.2, San Diego Supercomputer Center, University of California, San Diego, CA, USA). Primer sequences were as follows: PROM-specific: sense, GCT TTGCAATCTCCCTGTTG and antisense, TTGATCCGG GTTCTTACCTG; POU5F1-specific: sense, CTGGAGAAGG AGAAGCTGGA and antisense, CAAATTGCTCGAGTTCT TTCTG; NANOG-specific: sense, GAGATGCCTCACACG GAGAC and antisense, CTTTGGGACTGGTGGAAGAA and ACTB-specific: sense, ACAGAGCCTCGCCTTTGC and antisense, GCGGCGATATCATCATCC. PCR was performed in a final volume of 25 μl with a SYBR-Green PCR master mix using 1 μl cDNA and 400 nM of each primer for the respective genes. Cycling conditions were 50°C for 2 min and 95°C for 10 min followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min.
Relative mRNA levels of target genes
Relative mRNA levels were determined by the standard curve method. The standard curves and line equations were generated using 5-fold serially diluted solutions of cDNA from the colon cancer cell line LoVo. Standard curves were linear in the analyzed range with an acceptable correlation coefficient (R2). The amount of target gene expression was calculated from the standard curve. Quantitative normalization of cDNA in each sample was performed using the expression of the ACTB gene as an internal control. mRNA levels of target genes were then provided as ratios to ACTB mRNA levels. Real-time PCR assays were performed in duplicate for each sample and the mean values were used for calculations of the mRNA levels.
Immunohistochemical staining for CD133, OCT4 and NANOG in UC and CRC
Sections (2 μm) of formalin-fixed, paraffin-embedded specimens were produced. After deparaffinization and dehydration for antigen unmasking, sections were brought to a boil in 10 mM sodium citrate buffer. Sections were then blocked and incubated with primary antibody overnight at 4°C. The antibody was detected by Envision reagents (Envision kit/HRP, Dako Cytomation, Denmark). Sections were counterstained with hematoxylin. Primary anti-CD133 rabbit monoclonal antibody (C24B9, Cell Signaling Technology Inc., Danvers, MA, USA), anti-OCT4 goat polyclonal antibody (C20, sc8629, Santa Cruz Biotechnology, Santa Cruz, CA, USA), and anti-NANOG rabbit polyclonal antibody (ab21603, Abcam plc, Cambridge, UK) were used at a dilution of 1:100, and detected with the streptavidin biotin method (LASB2 kit/HRP, Dako Cytomation, Denmark). Negative controls were run simultaneously with pre-immune immunoglobulin.
Statistical analysis
All statistical analyses were performed using JMP version 5 (SAS Institute Inc. Cary, NC, USA). Relative mRNA levels of each gene were expressed as median values (inter-quartile range).
Associations between gene expression levels (continuous variables) and groups (categorical variables) were evaluated using Mann-Whitney U tests for two groups or Kruskal-Wallis tests for multiple groups.
Box and whisker plots were used to summarize the distribution of mRNA levels of PROM, POU5F1 and NANOG for each group. The horizontal line in the box represents the 50th quartile (median), and the upper and lower lines of the box represent 75th and 25th quartiles, respectively. The whiskers indicate the range of the measurements. P<0.05 was considered to be statistically significant.
Results
Clinical characteristics of UC-CRC and UC patients
The mean age of six UC-CRC patients (3 men and 3 women) was 39 years (range 28–51). Among the 6 patients, 3 (50%) had extensive colitis and 3 (50%) had left-sided colitis. The median duration of disease was 11 years (range 1–19).
The mean age of the 24 UC patients (14 men and 10 women) was 32 years (range 17 to 56 years). Among the 24 patients with UC, 19 had extensive colitis and 5 had left-sided colitis. The median duration of disease was 6 years (range 0.1–28) (Table I).
Table I.
Clinical characteristics of UC-CRC and UC patients.
| Variable | UC-CRC (n=6) | UC (n=24) |
|---|---|---|
| Gender | ||
| Male | 3 (50%) | 14 (58%) |
| Female | 3 (50%) | 10 (42%) |
| Age, mean (range) | 39 (28–51) | 32 (17–56) |
| Age at diagnosis mean (range) | 31 (18–50) | 26 (13–50) |
| Duration of disease median (range) | 11 (1–19) | 6 (0.1–28) |
| Extent of disease | ||
| Proctitis | 0 (0%) | 0 (0%) |
| Left-sided colitis | 2 (33%) | 5 (21%) |
| Extensive colitis | 4 (67%) | 19 (79%) |
| Severity of imflammation | ||
| Mild | 3 (50%) | 1 (0.4%) |
| Moderate | 3 (50%) | 17 (74.6%) |
| Severe | 0 (0%) | 6 (25%) |
UC-CRC, ulcerative colitis-associated colorectal cancer; and UC, ulcerative colitis.
Clinical characteristics of UC-CRC and sporadic CRC patients
In the UC-CRC patients, the primary tumor was located in the rectum (n=3) or the colon (n=3). Pathological T status (pT) of UC-CRC was pT1 (n=2) and pT3 (n=4). The UC-CRC were well differentiated (n=2, 34%), moderately differentiated (n=1, 17%) or poorly/undifferentiated (n=3, 49%). Two patients with UC-CRC were staged as Dukes’ A and 4 patients as Dukes’ B.
In sporadic CRC patients, the mean age of 37 patients (26 men and 11 women) was 63 years (range 37–82). These tumors were located in the rectum (n=26) or the colon (n=11). Distribution of pTs were pT1 (8%), pT2 (16%), pT3 (68%) and pT4 (8%). A total of 21 patients (57%) had pathological lymph node metastases. The tumors were well-differentiated (n=19, 51%), moderately differentiated (n=15, 40%) or poorly/undifferentiated (n=3, 9%). Seven patients with sporadic CRC were staged as Dukes’ A, 7 as Dukes’ B, 12 as Dukes’ C and 11 as Dukes’ D (Table II).
Table II.
Clinicopathological characteristics of UC-CRC and sporadic CRC patients.
| Variable | UC-CRC (n=6) | Sporadic CRC (n=37) |
|---|---|---|
| Gender | ||
| Male | 3 (50%) | 26 (70%) |
| Female | 3 (50%) | 11 (30%) |
| Age, mean (range) | 39 (28–51) | 63 (37–82) |
| Tumor location | ||
| Rectum | 3 (50%) | 26 (70%) |
| Colon | 3 (50%) | 11 (30%) |
| Tumor invasion | ||
| pT1 | 2 (33%) | 3 (8%) |
| pT2 | 0 (0%) | 6 (16%) |
| pT3 | 4 (67%) | 25 (68%) |
| pT4 | 0 (0%) | 3 (8%) |
| Lymph node metastasis | ||
| Present | 0 (0%) | 21 (57%) |
| Absent | 6 (100%) | 16 (43%) |
| Histological differentiation | ||
| Well | 2 (34%) | 19 (51%) |
| Moderate | 1 (17%) | 15 (40%) |
| Poor | 3 (49%) | 3 (9%) |
| Dukes’ classification | ||
| A | 2 (33%) | 7 (19%) |
| B | 4 (67%) | 7 (19%) |
| C | 0 (0%) | 12 (32%) |
| D | 0 (0%) | 11 (30%) |
Levels of stem cell markers in UC-CRC and inflamed colonic epithelium
The relative mRNA levels of PROM, POU5F1 and NANOG were expressed as ratios between the gene of interest (PROM, POU5F1 and NANOG) and the internal reference gene (ACTB), providing a normalization factor for the amounts of the mRNAs.
Table III shows the median value (inter-quartile range) for each gene in UC-CRC and inflamed colonic epithelium. As shown in Fig. 1A, PROM, POU5F1 and NANOG were significantly lower in colitis-associated CRC than in inflamed colonic epithelium (P=0.0335 for PROM, P=0.0011 for POU5F1 and P=0.006 for NANOG).
Table III.
Stem cell markers in UC-CRC and inflamed colonic epithelium.
| Stem cell maker | UC-CRC (n=6) | Imflamed colonic epithelium (n=24) |
|---|---|---|
| PROM (CD133, range) | 0.138 (0.065–0.725) | 0.590 (0.205–1.006) |
| POU5F1 (OCT4, range) | 11.84 (8.298–18.12) | 44.59 (24.26–196.7) |
| NANOG | 19.15 (6.998–28.54) | 46.16 (30.08–130.2) |
Values of each target gene are expressed as the median value (inter-quartile range).
Figure 1.
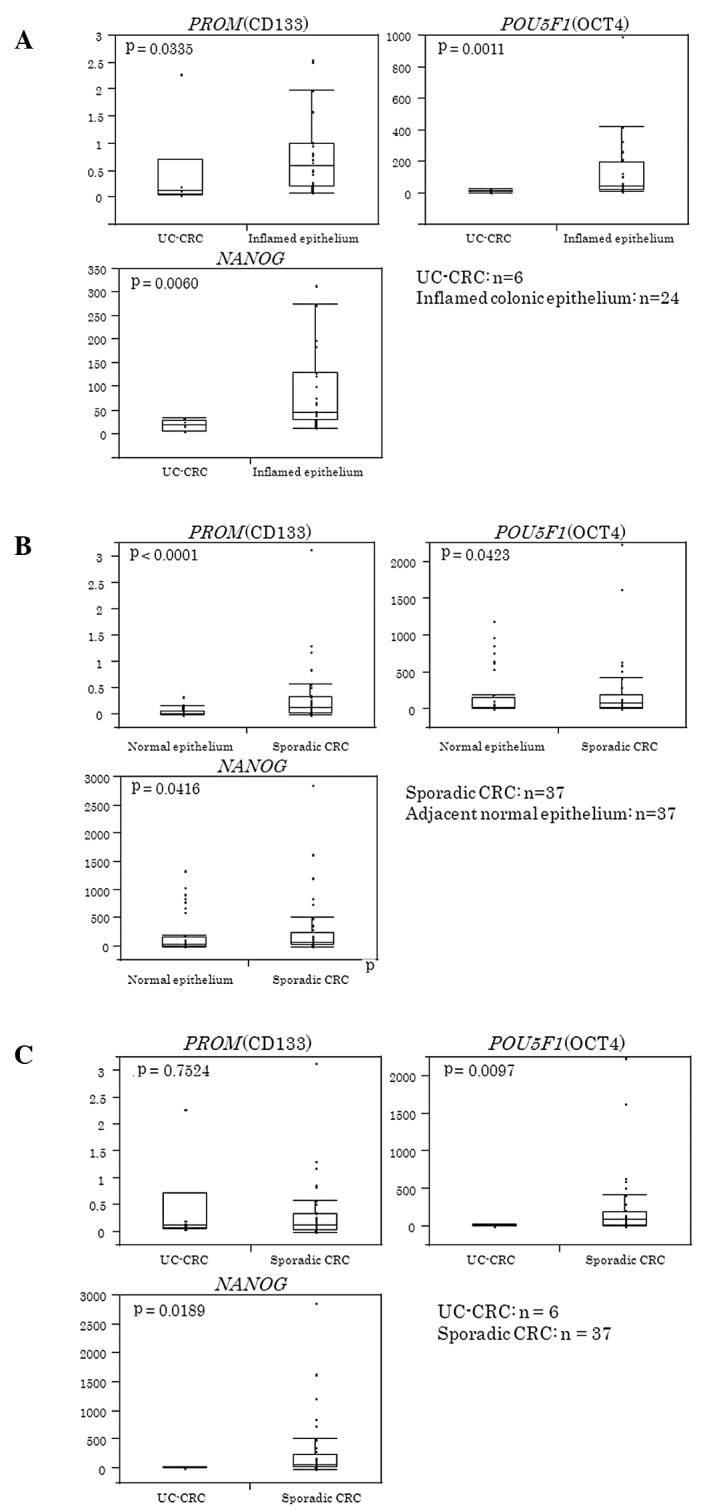
(A) Expression of stem cell markers in UC-CRC and inflamed colonic epithelium. Expression of PROM, POU5F1 and NANOG were significantly lower in UC-CRC than in inflamed colonic epithelium (P=0.0335 for PROM, P=0.0011 for POU5F1 and P=0.006 for NANOG). Box and whisker plots were used to show the distribution of mRNA levels of PROM, POU5F1 and NANOG for each group. The horizontal line in the box represents the 50th (median) and the upper and lower lines of the box represent the 75th and 25th quartiles, respectively. The whiskers indicate the range of the measurements. (B) Levels of stem cell markers in sporadic CRC and adjacent normal epithelium. PROM, POU5F1 and NANOG were significantly higher in sporadic CRC than in adjacent normal colonic epithelium (P=0.0001 for PROM, P=0.0423 for POU5F1 and P=0.0416 for NANOG). (C) Comparison of PROM, POU5F1 and NANOG in UC-CRC and sporadic CRC. POU5F1 and NANOG levels were significantly lower in colitis-associated CRC than in sporadic CRC (P=0.0097 for POU5F1, and P=0.0189 for NANOG). No significant difference was observed in PROM between colitis-associated and sporadic CRC.
Levels of stem cell markers in sporadic CRC and adjacent normal epithelium
Table IV shows the median value (inter-quartile range) of each gene in sporadic CRC and the adjacent normal epithelium. As shown in Fig. 1B, PROM, POU5F1 and NANOG were significantly higher in sporadic CRC than in adjacent normal colonic epithelium (P=0.0001 for PROM, P=0.0423 for POU5F1 and P=0.0416 for NANOG).
Table IV.
Stem cell markers in sporadic CRC and normal colonic epithelium.
| Stem cell maker | Sporadic CRC (n=6) | Normal colonic epithelium (n=37) |
|---|---|---|
| PROM (CD133, range) | 0.274 (0.137–0.830) | 0.129 (0.033–0.345) |
| POU5F1 (OCT4, range) | 29.48 (11.74–61.20) | 91.00 (24.80–197.0) |
| NANOG | 31.47 (17.44–74.69) | 70.70 (26.50–246.2) |
Values of each target gene are expressed as the median value (inter-quartile range).
Comparison of PROM, POU5F1 and NANOG in UC-CRC and sporadic CRC
Fig. 1C shows that POU5F1 and NANOG mRNA levels were significantly lower in colitis-associated CRC than in sporadic CRC (P=0.0097 for POU5F1, and P=0.0189 for NANOG). No significant difference was observed in PROM between colitis-associated and sporadic CRC.
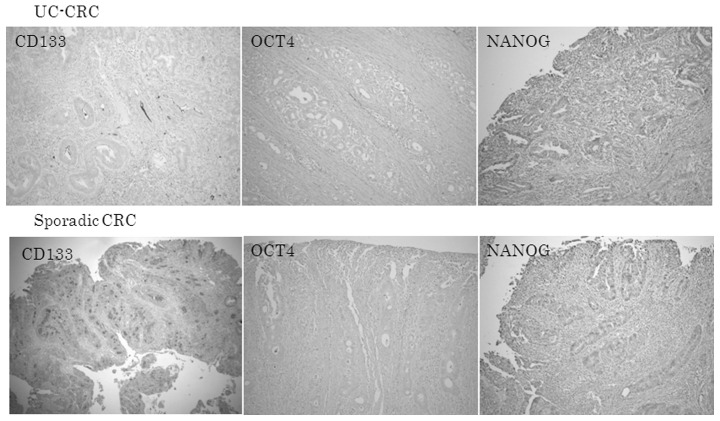
Immunoreactive CD133, OCT4 and NANOG in UC-CRC and sporadic CRC
Fig. 2 shows that the expression of CD133, OCT4 and NANOG proteins was also lower in UC-CRC than in sporadic CRC.
Figure 2.
Immunoreactive CD133, OCT4 and NANOG in UC-CRC and sporadic CRC. The expression of CD133, OCT4, and NANOG proteins was decreased in (A) UC-CRC compared with (B) sporadic CRC. Original magnification, ×40.
Immunoreactive CD133 protein was observed in a minority of CRC cells. CD133 was located at the apical/endoluminal surface (membranous), in the cytoplasm (cytoplasmic) or in both sites, for CRC cells (Fig. 4).
Figure 4.
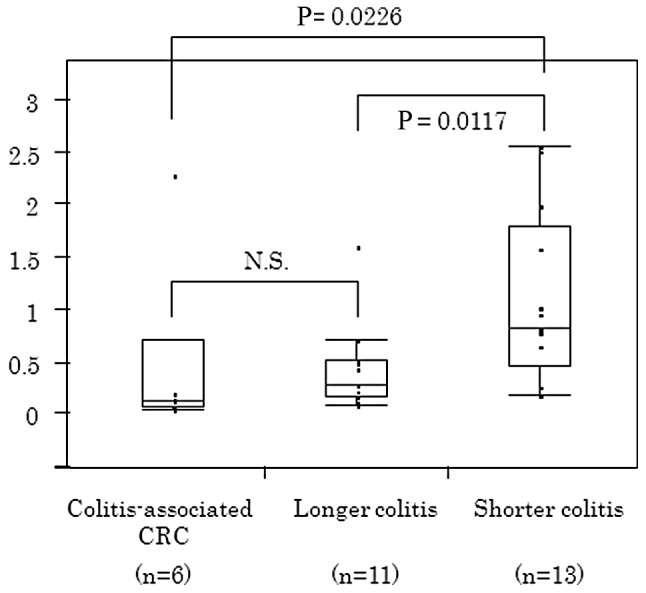
CD133 levels in inflamed epithelium according to disease duration CD133 in inflamed colonic epithelium was significantly associated with the duration of colitis. Patients with longer duration colitis showed a significantly lower CD133 expression than those with shorter duration colitis (P=0.0117).
In UC-CRC and sporadic CRC, OCT4 was found in the cytoplasm of cancer cells. Although germ cell tumors have been reported to show strong nuclear OCT4 immunostaining, colorectal cancer cells had diffuse cytoplasmic OCT4 staining.
Diffuse cytoplasmic NANOG protein was detected in UC-CRC cells. Sporadic CRC cells showed, not only cytoplasmic, but also nuclear NANOG immunoreactivity.
Association between CD133, OCT4, and NANOG in inflamed colonic epithelium and disease or patient characteristics
The expression of CD133 and NANOG proteins in inflamed colonic epithelium was significantly higher among younger UC patients (P<0.05) (Fig. 3). CD133 in inflamed colonic epithelium was significantly associated with the duration of colitis. Based on the median value of the duration of colitis, 24 patients were divided into two groups (shorter duration colitis, n=13; and longer duration colitis, n=11). As shown in Fig. 4, patients with shorter periods of colitis showed a higher CD133 expression than patients with colitis of longer duration (P=0.0117) or patients with UC-CRC (P=0.0226). No significant difference in CD133 levels was found between UC-CRC patients and longer duration colitis patients.
Figure 3.
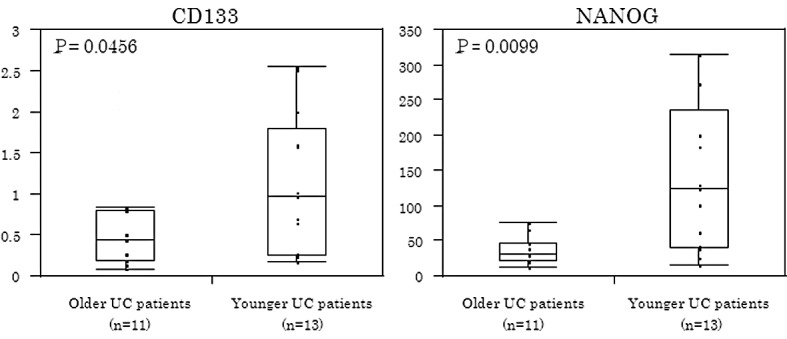
CD133 and NANOG in inflamed colonic epithelium according to patients’ age. CD133 and NANOG in inflamed colonic epithelium were expressed at significantly higher levels among younger UC patients (P<0.05).
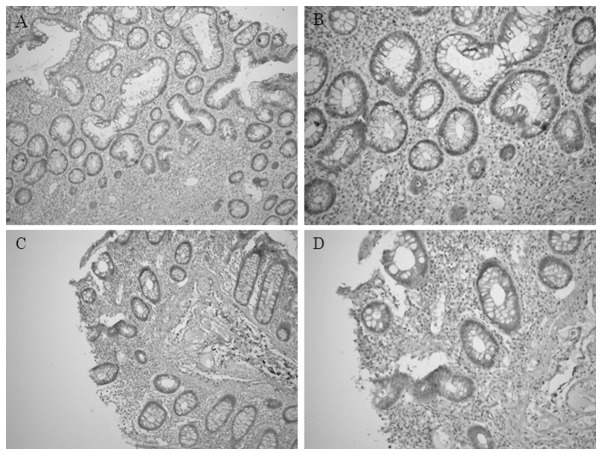
Immunohistochemical staining of CD133 in inflamed colonic epithelium
In UC patients, the minority of inflamed colonic epithelial cells expressed immunoreactive CD133 protein (Fig. 5). CD133 was observed diffusely in the cytoplasm of epithelial cells and was also located at the apical/endoluminal surface (membranous), in the cytoplasm or in both sites, of inflamed colonic epithelial cells.
Figure 5.
Immunohistochemical staining of CD133 in inflamed colonic epithelium. In inflamed colonic epithelial cells, CD133 was located at the apical/endoluminal surface (membranous) or in the cytoplasm or both sites. Original magnification, (A and C) ×100; (B and D) ×200.
Discussion
To the best of our knowledge, no report has investigated the expression of the three stem cell markers considered in this study in the context of UC-CRC or inflamed colonic epithelium, compared to sporadic CRC.
Stem cells capable of self-renewal and pluripotency contribute to normal tissue development, regeneration and disease (16). Recently, the CSC concept has been advanced through the use of isolation techniques dependent upon cell surface markers for putative CSCs and functional assays of stem/progenitor cell properties in non-obese diabetic severe combined immunodeficient mice (17,18). CD133 is one of the most significant cell surface markers of colon CSCs (19). A recent report showed that CD133 immunoreactivity was associated with poor survival in sporadic CRC (14). POU5F1 and NANOG play significant roles in maintaining pluripotency and self-renewal of SCs (11). Although each stem cell marker has been reported to be expressed in certain human malignancies, the clinical significance of these stem cell markers in UC-CRC has yet to be established.
We demonstrated that UC-CRC had a significantly lower expression of PROM, POU5F1 and NANOG genes compared to inflamed colonic epithelium, whereas sporadic CRC had a significantly higher expression of these genes compared to adjacent normal colonic epithelium. UC-CRC has been reported to develop along a different genetic pathway compared to sporadic CRC. Previous studies showed that mutations of the TP53 (p53) gene were observed in the early stage, and adenomatous polyposis coli gene or k-ras gene mutations were less frequent events in UC-CRC (20–22). Although UC-CRC appears to have a completely different carcinogenic pathway from sporadic CRC, the mechanisms underlying these differences are not understood. Our stem cell marker analysis may support this concept, showing genetic or carcinogenic differences between UC-CRC and sporadic CRC. Another possible interpretation of our results is that colonic SCs or CSCs scarcely contribute to UC-associated carcinogenesis relative to sporadic CRC. However, further analyses are required to address the reason for stem cell markers being lower in UC-CRC compared with sporadic CRC or inflamed colonic epithelium.
We also demonstrated that the CD133 level in inflamed colonic epithelium was significantly lower in UC patients with a longer duration of disease than those with a shorter duration. These results indicate that decreased CD133 levels in the colonic mucosa of UC patients may identify patients at higher risk for developing UC-CRC.
Periodic surveillance colonoscopy with two to four random biopsies collected at 10 cm intervals throughout the entire colon is now highly recommended for the early detection of UC-CRC (23). To increase the macroscopic detection rate of precancerous or dysplastic lesions, methylene blue or indigo carmine chromoendoscopy with careful inspection have also been utilized (24,25). However, current surveillance systems are unsatisfactory for the detection of dysplasia and early-stage UC-CRC due to the difficulties associated with endoscopic or histological assessment.
The analysis of the CD133 level in colonoscopic biopsy specimens in addition to conventional cancer risk evaluation may increase the identification rate of patients who are likely to develop UC-CRC. If reliable and reproducible evaluation criteria for immunohistochemical CD133 expression are established for inflamed colonic mucosa, it may be an easy and useful supplementary method to predict the development of UC-CRC.
However, our data should be interpreted with caution. A significant limitation of this study was the relatively small number of UC patients (n=30), including only six patients with UC-CRC. Large prospective trials are required to confirm our results.
In conclusion, UC-CRC showed different expression profiles of stem cell markers compared with sporadic CRC. A decreased CD133 expression in inflamed colonic epithelium may identify UC patients at a high risk for the development of UC-CRC.
References
- 1.Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526–535. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zisman TL, Rubin DT. Colorectal cancer and dysplasia in inflammatory bowel disease. World J Gastroenterol. 2008;14:2662–2669. doi: 10.3748/wjg.14.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RB, Harpaz N, Itzkowitz S, Hossain S, Matula S, Kornbluth A, Bodian C, Ullman T. Histologic inflammation is a risk factor for progression to colorectal neoplasia in ulcerative colitis: a cohort study. Gastroenterology. 2007;133:1099–1105. doi: 10.1053/j.gastro.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe T, Kobunai T, Toda E, et al. Gene expression signature and the prediction of ulcerative colitis-associated colorectal cancer by DNA microarray. Clin Cancer Res. 2007;13:415–420. doi: 10.1158/1078-0432.CCR-06-0753. [DOI] [PubMed] [Google Scholar]
- 5.Fujii S, Tominaga K, Kitajima K, et al. Methylation of the oestrogen receptor gene in non-neoplastic epithelium as a marker of colorectal neoplasia risk in longstanding and extensive ulcerative colitis. Gut. 2005;54:1287–1292. doi: 10.1136/gut.2004.062059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishikawa M, Oshitani N, Matsumoto T, Nishigami T, Arakawa T, Inoue M. Accumulation of mitochondrial DNA mutation with colorectal carcinogenesis in ulcerative colitis. Br J Cancer. 2005;93:331–337. doi: 10.1038/sj.bjc.6602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartsch H, Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch Surg. 2006;391:499–510. doi: 10.1007/s00423-006-0073-1. [DOI] [PubMed] [Google Scholar]
- 8.Boman BM, Huang E. Human colon cancer stem cells: a new paradigm in gastrointestinal oncology. J Clin Oncol. 2008;26:2828–2838. doi: 10.1200/JCO.2008.17.6941. [DOI] [PubMed] [Google Scholar]
- 9.Vermeulen L, Sprick MR, Kemper K, Stassi G, Medema JP. Cancer stem cells – old concepts, new insights. Cell Death Differ. 2008;15:947–958. doi: 10.1038/cdd.2008.20. [DOI] [PubMed] [Google Scholar]
- 10.Shipitsin M, Polyak K. The cancer stem cell hypothesis: in search of definitions, markers, and relevance. Lab Invest. 2008;88:459–463. doi: 10.1038/labinvest.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang J, Wan M, Zhang Y, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 12.Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Levasseur DN, Orkin SH. Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2008;105:6326–6331. doi: 10.1073/pnas.0802288105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horst D, Kriegl L, Engel J, Kirchner T, Jung A. CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br J Cancer. 2008;99:1285–1289. doi: 10.1038/sj.bjc.6604664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bijwaard KE, Aguilera NS, Monczak Y, Trudel M, Taubenberger JK, Lichy JH. Quantitative real-time reverse transcription-PCR assay for cyclin D1 expression: utility in the diagnosis of mantle cell lymphoma. Clin Chem. 2001;47:195–201. [PubMed] [Google Scholar]
- 16.Lanzoni G, Roda G, Belluzzi A, Roda E, Bagnara GP. Inflammatory bowel disease: Moving toward a stem cell-based therapy. World J Gastroenterol. 2008;14:4616–4626. doi: 10.3748/wjg.14.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 19.Ieta K, Tanaka F, Haraguchi N, et al. Biological and genetic characteristics of tumor-initiating cells in colon cancer. Ann Surg Oncol. 2008;15:638–648. doi: 10.1245/s10434-007-9605-3. [DOI] [PubMed] [Google Scholar]
- 20.Brentnall TA, Crispin DA, Rabinovitch PS, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107:369–378. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 21.Tarmin L, Yin J, Harpaz N, et al. Adenomatous polyposis coli gene mutations in ulcerative colitis-associated dysplasias and cancers versus sporadic colon neoplasms. Cancer Res. 1995;55:2035–2038. [PubMed] [Google Scholar]
- 22.Chaubert P, Benhattar J, Saraga E, Costa J. K-ras mutations and p53 alterations in neoplastic and nonneoplastic lesions associated with longstanding ulcerative colitis. Am J Pathol. 1994;144:767–775. [PMC free article] [PubMed] [Google Scholar]
- 23.Farrell RJ, Peppercorn MA. Ulcerative colitis. Lancet. 2002;359:331–340. doi: 10.1016/S0140-6736(02)07499-8. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto T, Nakamura S, Jo Y, Yao T, Iida M. Chromoscopy might improve diagnostic accuracy in cancer surveillance for ulcerative colitis. Am J Gastroenterol. 2003;98:1827–1833. doi: 10.1111/j.1572-0241.2003.07580.x. [DOI] [PubMed] [Google Scholar]
- 25.Rutter MD, Saunders BP, Schofield G, Forbes A, Price AB, Talbot IC. Pancolonic indigo carmine dye spraying for the detection of dysplasia in ulcerative colitis. Gut. 2004;53:56–260. doi: 10.1136/gut.2003.016386. [DOI] [PMC free article] [PubMed] [Google Scholar]