Abstract
Tumours lacking argininosuccinate synthetase-1 (ASS1) are auxotrophic for arginine and sensitive to amino-acid deprivation. Here, we investigated the role of ASS1 as a biomarker of response to the arginine-lowering agent, pegylated arginine deiminase (ADI-PEG20), in lymphoid malignancies. Although ASS1 protein was largely undetectable in normal and malignant lymphoid tissues, frequent hypermethylation of the ASS1 promoter was observed specifically in the latter. A good correlation was observed between ASS1 methylation, low ASS1 mRNA, absence of ASS1 protein expression and sensitivity to ADI-PEG20 in malignant lymphoid cell lines. We confirmed that the demethylating agent 5-Aza-dC reactivated ASS1 expression and rescued lymphoma cell lines from ADI-PEG20 cytotoxicity. ASS1-methylated cell lines exhibited autophagy and caspase-dependent apoptosis following treatment with ADI-PEG20. In addition, the autophagy inhibitor chloroquine triggered an accumulation of light chain 3-II protein and potentiated the apoptotic effect of ADI-PEG20 in malignant lymphoid cells and patient-derived tumour cells. Finally, a patient with an ASS1-methylated cutaneous T-cell lymphoma responded to compassionate-use ADI-PEG20. In summary, ASS1 promoter methylation contributes to arginine auxotrophy and represents a novel biomarker for evaluating the efficacy of arginine deprivation in patients with lymphoma.
Keywords: lymphoma, ASS1 promoter methylation, arginine, ADI-PEG20, autophagy, chloroquine
The renewed interest in cancer metabolism, and specifically arginine deprivation as a targeted approach for the treatment of cancer, has led to encouraging early-phase trials of pegylated arginine deiminase (ADI-PEG20) in patients with several solid cancers deficient in argininosuccinate synthetase-1 (ASS1).1, 2, 3, 4, 5 Essentially tumours deficient in ASS1, a rate-limiting enzyme for arginine biosynthesis, depend upon extracellular delivery of the amino acid for tumour growth and survival, known as arginine auxotrophy. It is notable that methylation-dependent silencing of the ASS1 promoter has been identified as a potential mechanism of gene repression in a subset of ASS1-deficient arginine auxotrophic solid tumours.6, 7 Moreover, while the significance for ASS1 loss in cancer is presently unclear, several groups have revealed an association with worse clinical outcome and shorter metastasis-free survival.7, 8 Although, amino-acid deprivation was established originally with asparaginase in the management of haematological cancers, few studies have focused on arginine deprivation in this context, particularly with regard to the ASS1 gene.
Work in the early 1970s revealed for the first time that arginine was required for the survival of Burkitt lymphoma cells and murine lymphosarcoma cells.9, 10 As ASS1 has not previously been studied in lymphoid malignancy, we screened for ASS1 expression in a large series of primary and relapsed lymphomas with comparative studies in normal lymphoid tissues. Although ASS1 protein was largely absent in both normal and malignant lymphoid tissues, ASS1 promoter methylation was identified specifically in the latter. Importantly, reactivation of ASS1 expression by the demethylating agent 5-Aza-dC led to resistance to ADI-PEG20, confirming that ASS1 promoter methylation modulates drug sensitivity in malignant lymphoid cells. Moreover, arginine depletion induced caspase-dependent cell death and autophagy in tumour cell lines treated with ADI-PEG20. Blocking autophagy with the antimalarial agent chloroquine enhanced the apoptotic effect of ADI-PEG20 in malignant lymphoid cell lines and primary B and T lymphoma cells. Finally, a patient with a refractory cutaneous T cell lymphoma (CTCL) is described, who responded to treatment with ADI-PEG20 on a compassionate programme. The responding tumour was tested retrospectively, with evidence for ASS1 deficiency secondary to promoter methylation. Taken together, our data reveal that ASS1 is epigenetically regulated in malignant lymphoid cells conferring arginine auxotrophy, and provide a rationale for targeting arginine deprivation to patients with ASS1-methylated lymphomas.
Results
ASS1 protein is absent in most normal and malignant lymphoid tissues
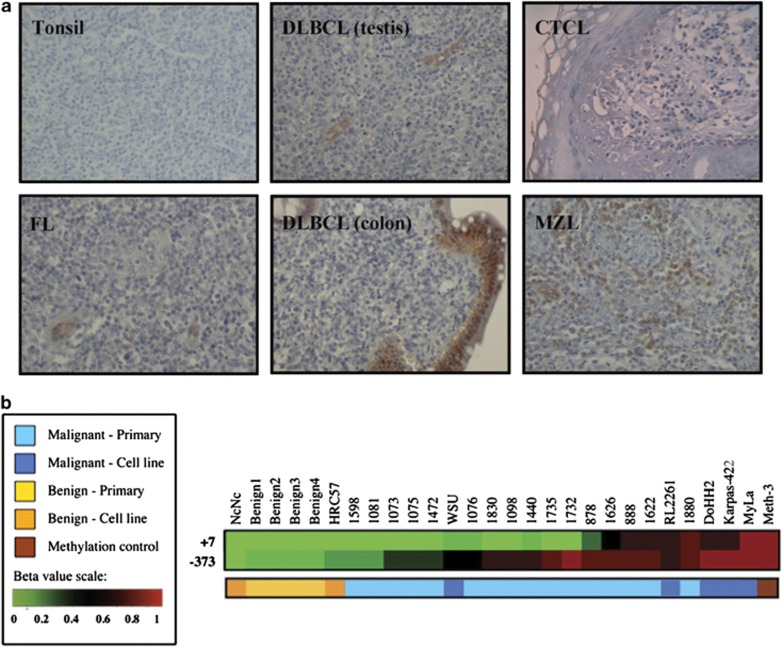
Previously, we reported loss of ASS1 expression linked to epigenetic silencing in several solid tumours.11 To extend our studies to haematological cancers, we performed an initial immunohistochemical screen of ASS1 in a wide range of primary lymphomas (Figure 1a and Table 1). Although ASS1 is considered an ubiquitously expressed protein, there was a distinct lack of ASS1 protein expression in the normal tonsil and in reactive lymph nodes (Figure 1a). We showed that most B-cell lymphomas were negative for ASS1 protein expression, while ∼35% of CTCL expressed ASS1. Notably, there was no evidence of ASS1 expression at subsequent relapse based on a large cohort of patients with serial biopsies of mantle cell and Hodgkin's lymphomas (Table 1).
Figure 1.
Lymphoid ASS1 immunostaining and methylation profiling. (a) Representative immunohistochemical stains for ASS1 as follows: ASS1-negative tonsillar lymphocytes; ASS1-negative FL, DLBCL, CTCL (Pautrier's microabcess) biopsies with internal positive controls (i.e., endothelial and epithelial cells); and an ASS1-positive MZL biopsy. Images were obtained using a Zeiss Axiophot microscope, magnification × 200, with Axiovision image acquisition software. (b) Differential methylation of the ASS1 promoter in a panel of normal and malignant lymphoid cell lines and primary tissues. Quantitative methylation values (beta values) were obtained as previously described.26 Beta values are reported for two CpG loci within the ASS1 gene (373 bp 5′ to the TSS and 7 bp 3′ to the TSS). The overall incidence of CpG island methylation was increased in malignant compared with benign samples
Table 1. ASS1 expression in normal and malignant lymphoid tissues.
| Lymphoma subtypes | % ASS1-negative cases (n) | % ASS1 promoter methylationa (n) |
|---|---|---|
| Normal tissue (spleen, reactive lymph node and tonsil) | 85 (11/13) | 0 (0/10) |
| Follicular lymphoma | 100 (38/38) | 80 (8/10) |
| Diffuse large B-cell lymphoma | 100 (66/66) | 70 (7/10) |
| Marginal zone lymphoma | 96 (26/27) | 70 (7/10) |
| Burkitt's lymphoma | 100 (8/8) | 50 (3/6) |
| Cutaneous T-cell lymphoma | 64 (18/28) | 50 (5/10) |
| Mantle cell lymphomab | 98 (90/91; 31/32)c | 70 (7/10) |
| Hodgkin's lymphomab | 97 (173/179; 50/50)d | 70 (5/7) |
Abbreviation: ASS1, argininosuccinate synthetase
Based on MS-PCR analysis of 6–10 samples
ASS1 expression in serial biopsies (baseline; relapse)
Single patient with ASS1-positive biopsy at baseline and relapse
No relapse biopsies available for baseline ASS1 expressors
Frequent ASS1 methylation in malignant but not normal lymphoid cells and tissues
As immunohistochemistry did not discriminate normal from malignant lymphoid tissues, we focused on the epigenetic status of ASS1. Quantitative methylation values were obtained for two CpG loci within the ASS1 promoter located 373 bp upstream to the transcription start site (TSS) and 7 bp downstream to the TSS, using a panel of normal and malignant lymphoid cell lines and tissues – the majority being follicular and transformed B-cell malignancies – screened with the Illumina Infinium DNA methylation platform. The normal immortalised lymphoblastoid cell lines, NcNc and HRC57, as well as normal lymphoid tissues were unmethylated at both CpG islands (Figure 1b). In contrast, our analysis revealed frequent ASS1 methylation in the intermediate promoter region (−373 bp) in lymphoma, with the pattern of CpG methylation in the proximal promoter (+7 bp) clustering samples mostly into two populations. One exhibited DNA hypermethylation in the two CpG sites tested whereas the other population was unmethylated in the proximal locus. Only two malignant lymphoma samples were fully unmethylated at both CpG loci tested. In addition, we documented a 50–80% methylation frequency of the ASS1 promoter across a broad range of lymphomas, whereas all normal lymphoid samples tested were unmethylated, by methylation-specific PCR (MS-PCR) (Table 1).
Reduced viability of ASS1-methylated lymphoid cells treated with ADI-PEG20
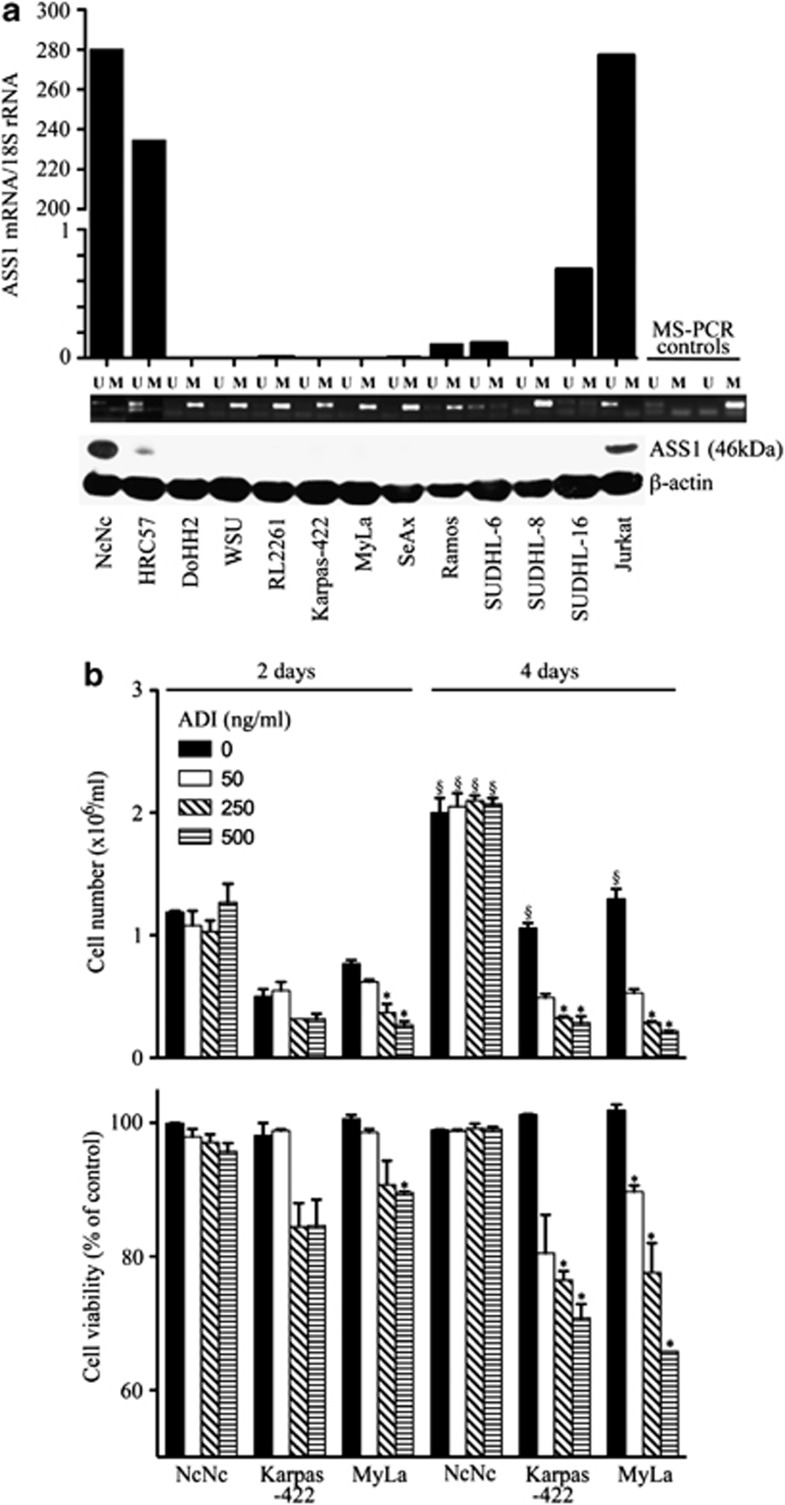
Next, we screened several lymphoid cell lines for ASS1 status by MS-PCR, quantitative real-time PCR (qPCR) and western blotting (Figure 2a). Overall, a good correlation was found between the ASS1 methylation status, and the levels of ASS1 mRNA and protein. Indeed, ASS1 mRNA and protein were detected in the unmethylated normal lymphoid cell lines, NcNc and HRC57; conversely, DNA hypermethylation was identified in the majority of malignant lymphoid cell lines with negligible levels of ASS1 mRNA and a lack of protein. Then, we addressed the impact of ASS1 methylation status on lymphoid cell sensitivity to ADI-PEG20, selecting the normal lymphoblastoid cell line, NcNc, and the lymphoma cell lines, Karpas-422 (B cell) and MyLa (T cell), respectively. Cells were treated with increasing concentrations of the arginine-degrading agent ADI-PEG20 (range 0–500 ng/ml) in the presence of citrulline, a key precursor for arginine recycling via endogenous ASS1. Cell number and viability were determined after 2 and 4 days of treatment. The ASS1-methylated tumour cell lines, Karpas-422 and MyLa, were sensitive while the normal lymphoblastoid cell line, NcNc, exhibited no difference in proliferation rate or viability with ADI-PEG20 treatment (Figure 2b).
Figure 2.
ASS1 status in normal and malignant lymphoid cell lines and sensitivity to ADI-PEG20. (a) Normal immortalised lymphoblastoid and lymphoma cell lines were screened for ASS1 mRNA and protein expression by qPCR and western blotting, respectively; ASS1 methylation was assessed using MS-PCR: HRC57, NcNc (normal lymphoblastoid cell lines); DoHH2, WSU, Karpas-422, RL2261 (FL cell lines); SUDHL-6, SUDHL-8, SUDHL-16 (DLBCL cell lines); Ramos (Burkitt's lymphoma cell line); and SeAx, MyLa (CTCL cell lines). The Jurkat leukaemia cell line was used as a positive control for ASS1 expression. A good correlation was observed between the methylation status of cell lines and the corresponding levels of ASS1 mRNA and protein. (b) Cells were treated with the arginine-depleting drug ADI-PEG20 and harvested after 2 or 4 days. Cell counting and viability were determined using the Beckman Vi-Cell cell viability analyser. Increasing concentrations of ADI-PEG20 (concentration range: 0–500 ng/ml) decreased the number of ASS1-negative lymphoma viable cells with no change in the viability of ASS1-positive cell lines by day 4. For each cell line, the viability was expressed as a percentage of untreated cells at day 2; *P<0.05 represents statistical difference to the baseline untreated cells for each cell line at each time point; §P<0.05 represents statistical difference at day 4 compared with day 2 for each cell line at each ADI-PEG20 concentration. The data are representative of three independent experiments. Error bars represent S.D.
Sensitivity to ADI-PEG20 is dependent upon methylation of the ASS1 promoter
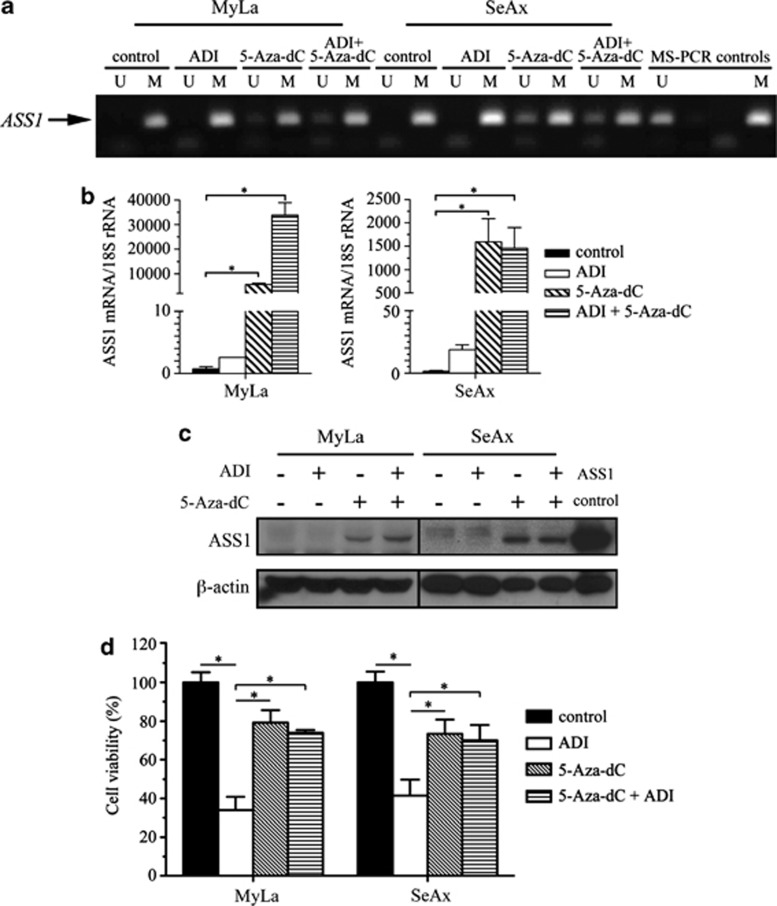
To prove that DNA methylation has a role in repressing ASS1 gene transcription, we tested whether the demethylating agent 5-Aza-dC could reactivate ASS1 in malignant lymphoid cells. We employed the CTCL cell lines, MyLa and SeAx for these experiments, as B-cells lines (Karpas, WSU and DoHH2) were highly sensitive to low doses of 5-Aza-dC (range 0.1–5 μM; data not shown). MyLa and SeAx cells were exposed to 5 μM of 5-Aza-dC for 8 days followed by either a further 4 days of treatment with 5-Aza-dC alone or combined with ADI-PEG20 (500 ng/ml). Partial DNA demethylation was detected in 5-Aza-dC-treated cells, accompanied by transcriptional activation and synthesis of ASS1 protein in the CTCL lines (Figures 3a–c). 5-Aza-dC treatment alone led to a 20% reduction in the number of viable CTCL cells compared with untreated controls (Figure 3d). However, there was no significant difference in the number of viable cells treated with 5-Aza-dC alone or in combination with ADI-PEG20, confirming that the demethylated CTCL cell lines were now insensitive to ADI-PEG20. Thus, our data indicate that ASS1 promoter methylation is an important determinant of ADI-PEG20 sensitivity and that a failure to upregulate ASS1 protein accounts for the efficacy of arginine-degrading enzymes in lymphoma cells.
Figure 3.
Induction of ASS1 expression in 5-Aza-dC-treated lymphoma cells confers resistance to ADI-PEG20. Identically treated and 5-Aza-dC (5 μM)-untreated MyLa and SeAx cells were split into new flasks and treated with or without 500 ng/ml of ADI-PEG20 for a further 4 days. (a) ASS1 promoter methylation was analysed using MS-PCR and PCR products resolved on a 2% agarose gel. For each treatment, unmethylated (U) and methylated (M) reactions are shown. (b) ASS1 mRNA expression was quantified by qPCR. *P<0.05 represents statistical significance compared with untreated cells for each cell line. (c) Detection of ASS1 expression by western blotting using a Jurkat cell line as an ASS1-positive control. (d) At the end of the 12-day experiment, cells were counted using the Beckman Vi-Cell cell viability analyser. *P<0.05 represents statistical significance compared with untreated cells for each cell line. Error bars represent S.D. The data are representative of three or more independent experiments
ADI-PEG20 induces caspase-dependent apoptosis in ASS1-methylated lymphoid cell lines
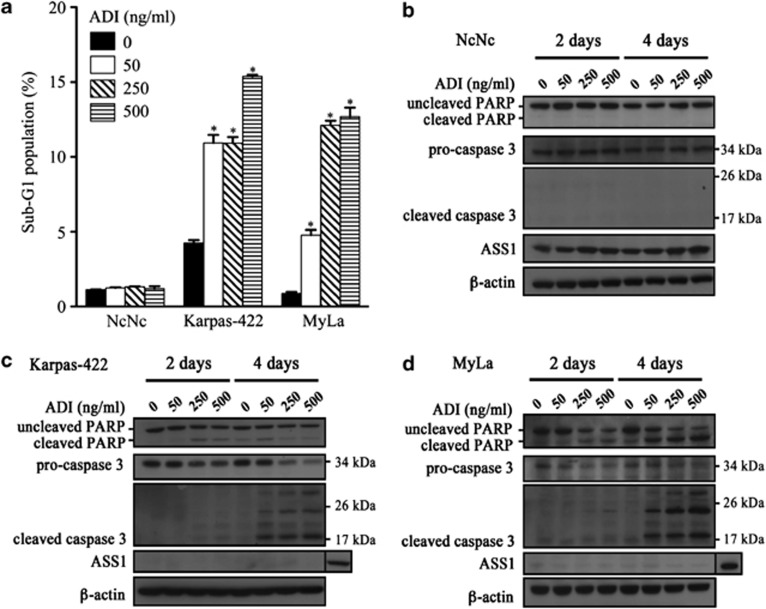
Consistent with the effects of ADI-PEG20 on cell proliferation, we observed a significant increase in the sub-G1 population of ADI-PEG20-treated ASS1-methylated cells, whereas ASS1-positive normal lymphoblastoid cells were unaffected (Figure 4a). We identified activation of caspase-3 as well as PARP cleavage in the malignant lymphoma cell lines (Figures 4c and d). Notably, ASS1-methylated lymphoma cell line susceptibility towards ADI-PEG20 was linked to the absence of ASS1 protein expression and to an inability to reinduce gene expression (Figures 4c and d). In contrast, protection from caspase-dependent cell death was associated with an increase in ASS1 protein expression in NcNc cells (Figure 4b). This is consistent with previous reports associating induction of ASS1 gene expression in low arginine conditions in tumour cell lines.12 Our data suggest that normal lymphoblastoid cells sense changes in arginine availability, and insufficiency triggers de novo production of arginine through increased ASS1 expression.
Figure 4.
ADI-PEG20 induces caspase-dependent apoptosis in ASS1-methylated lymphoma cell lines. (a) The three cell lines were treated with ADI-PEG20 (0–500 ng/ml) for 4 days. Cells were then harvested and fixed in 70% ethanol for at least 24 h. The apoptotic fraction of cells, identified as the sub-G1 population of cell cycle was measured by flow cytometry with propidium iodide staining (described in Materials and Methods); *P<0.05 represents statistical significance compared with untreated cells for each cell line; protein lysates from (b) NcNc (c) Karpas-422 and (d) MyLa cells treated with ADI-PEG20 (0–500 ng/ml) were analysed by western blotting for the expression of ASS1 and the detection of cleaved PARP and caspase-3
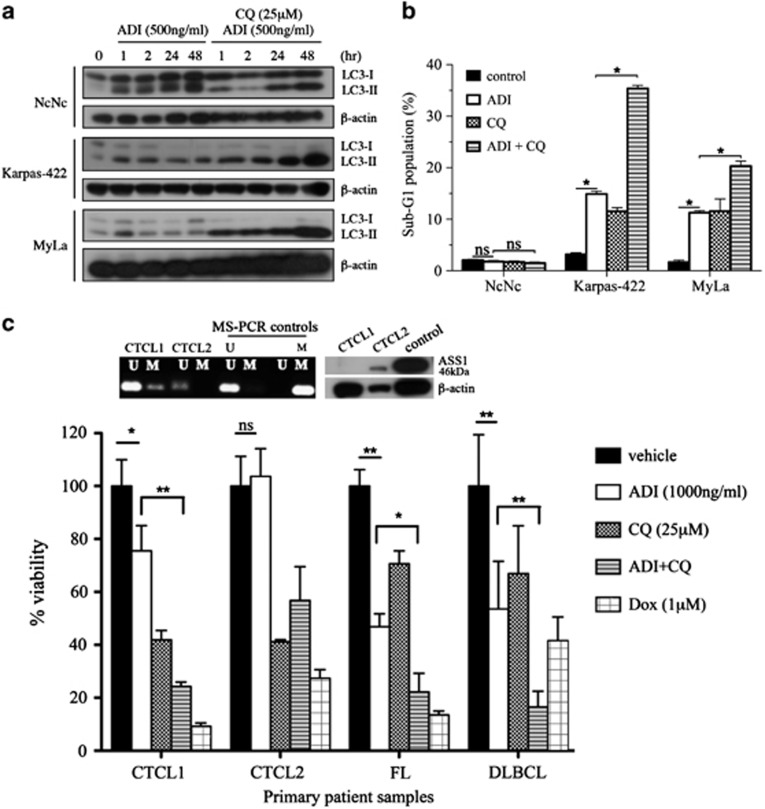
Chloroquine blocks ADI-PEG20-induced autophagy enhancing lymphoma cell apoptosis
Despite evidence of caspase-mediated cell death, the 10–20% increase in the subG1 fraction of lymphoma cells with ADI-PEG20 treatment indicated that autophagy may have a significant prosurvival role as evidenced in ASS1-deficient solid tumour cells.13 To test whether there was reciprocal induction of autophagy, we used the known autophagy modulator chloroquine and assessed levels of light chain 3 (LC3)-I and its processed (lipidated) form LC3-II in the malignant lymphoid cells. We confirmed that constitutive levels of LC3-II, a marker of ongoing autophagy, were elevated compared with constitutive LC3-I in the malignant lymphoid cells in contrast to normal lymphoblastoid cells (Figure 5a). Treatment with ADI-PEG20 increased further the levels of LC3-II in normal and malignant lymphoid lines by 1 h, whereas cotreatment with chloroquine led to an accumulation of LC3-II and apoptosis specifically in the malignant lymphoid cells (Figures 5a and b). Finally, freshly isolated CTCL and primary B (FL and DLBCL) malignant lymphoma cells responded similarly to treatment with ADI-PEG20 and chloroquine with three of four clinical samples showing sensitivity to ADI-PEG20 and marked enhancement when combined with chloroquine, using the ATP ViaLight assay (Figure 5c). We analysed the ASS1 status of the two cases of primary CTCL showing differential sensitivity to ADI-PEG20: CTCL1 was partially methylated with an absence of ASS1 protein and sensitive to ADI-PEG20, whereas CTCL2 was unmethylated, expressed ASS1 protein, and was resistant to ADI-PEG20, in good agreement with our cell line data (Figure 5c).
Figure 5.
ADI-PEG20-induced autophagy and its modulation by chloroquine in lymphoma cells. (a) Malignant lymphoid cells were treated with ADI-PEG20 (500 ng/ml) either alone or in combination with chloroquine (CQ, 25 μM) and assessed for LC3-I and LC3-II levels by western blotting. (b) Follicular (Karpas-422) and CTCL (MyLa) lymphoma cells were treated with ADI-PEG20 and CQ for 72 h, and analysed for the subG1 fraction corresponding to apoptotic cells by flow cytometry; *P<0.001. (c) Similarly, primary malignant B lymphoma and CTCL (Sezary syndrome) lymphoma cells were studied using the ViaLight assay. Doxorubicin was used as a positive control for lymphoma cell cytotoxicity. *P<0.05, **P<0.001. Error bars represent S.D.
Clinical response to therapy with ADI-PEG20 in a patient with refractory lymphoma
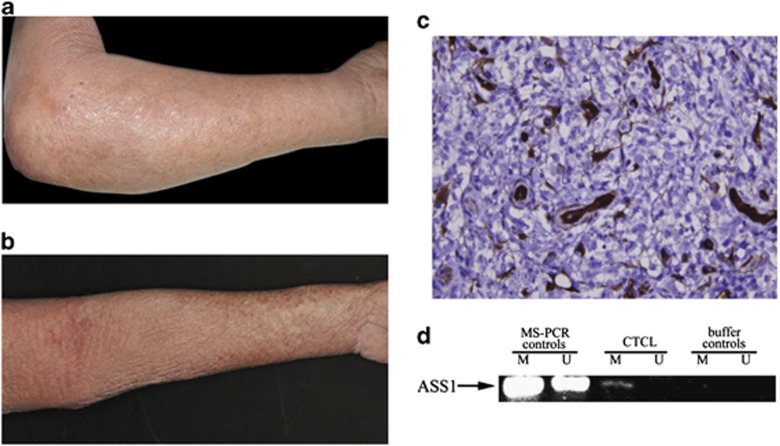
Moving forward into the clinic, a patient with a refractory CTCL (Sezary syndrome) was exposed to single-agent ADI-PEG20 on a compassionate access programme, having progressed on high-dose steroids and standard cytotoxic therapies. Following the first intramuscular dose of ADI-PEG20 (160 IU/m2/week), the patient's pruritus and skin oedema resolved (Figures 6a and b) that correlated with a minor reduction in the total white cell count (35–31 × 109/l; normal range 4–11 × 109/l). As expected, the plasma arginine concentration decreased with a corresponding increase in plasma citrulline, the degradation product of ADI-PEG20 activity (data not shown). Analysis of the patient's tumour confirmed that the T-cell lymphoma was an ASS1-negative expressor and methylated at the ASS1 promoter (Figures 6c and d). The patient was treated with three additional doses, at which point further ADI-PEG20 was discontinued due to an intercurrent infection.
Figure 6.
(a) ADI-PEG20 treatment of a patient with an ASS1-methylated CTCL. (a) Baseline appearance with marked skin oedema due to CTCL (Sezary syndrome). (b) Resolution of skin oedema after a single dose of ADI-PEG20 (160 IU/m2, i.m.). (c) Baseline tumour biopsy showing ASS1-negative CTCL cells, with skin appendageal and endothelial cells showing strong ASS1 expression ( × 400). (d) Methylated ASS1 in the tumour biopsy confirmed by MS-PCR
Discussion
Arginine deprivation is under clinical investigation as a metabolic strategy for the treatment of several solid tumours, however, its role in haematological cancers, and lymphoid malignancy in particular, has lagged behind. Furthermore, although ASS1 loss has been identified in recent years as a potential biomarker of arginine auxotrophy in cancer cells, its regulation is complex and cell type-dependent. Thus, the goal of the present study was to understand the basis for the arginine auxotrophy documented in lymphoma cells, by assessing ASS1 expression and regulation in response to the arginine-depleting drug, ADI-PEG20.9, 10 We showed that ASS1 promoter methylation differentiates lymphoid malignancy from normal lymphoid tissues and moreover that methylated ASS1 – rather than absence of ASS1 protein – may be a more sensitive indicator of lymphoma cell sensitivity to ADI-PEG20.
ASS1 promoter methylation blocked ASS1 enzyme reinduction following arginine deprivation, and although limited to CTCL lines due to their relative insensitivity to 5-Aza-dC, our demethylation studies support epigenetic silencing as a key factor accounting for the arginine auxotrophy of lymphoid cells. We have reported previously ASS1 methylation in solid tumour cells but this is the first study to reveal methylation as a determinant of sensitivity to arginine depletion in malignancy. In addition, we noted robust activation of caspase-dependent cell death in ASS1-methylated lymphoma cells, in contrast to the resistance of ASS1-unmethylated normal lymphoid cells, and the caspase-independent cell killing described previously in prostate cancer cell lines.13 The importance of cell type and response to arginine deprivation is reinforced by recent studies in melanoma cell lines, which appear to lack ASS1 promoter methylation yet are ASS1-deficient and ADI-PEG20-sensitive due to HIF-ıα-mediated suppression of promoter activity.12, 14 Moreover, our work indicates a need for additional studies of ASS1 promoter methylation using pyrosequencing and correlating the amount of methylation at specific CpG islands with cancer cell sensitivity to arginine deprivation.
In addition, we noted that in the absence of ASS1 induction, autophagy is a common mechanism of lymphoma cell resistance to arginine deprivation. First identified by Kim et al. in prostate cancer cell lines, we confirmed similar findings in lymphoma cell lines, and additionally that chloroquine may have potential as a modulator of the autophagic response in the clinic. Indeed, we observed significant efficacy of chloroquine as a single agent in lymphoma cells confirming recent studies on the inhibitory effects of the antimalarial on lymphomagenesis.15 Importantly, primary malignant B and T lymphoma cells were more sensitive to the combination of ADI-PEG20 and chloroquine than either agent alone in vitro. An alternative approach may employ PI3K inhibitors that are known to suppress autophagy and have been shown recently to potentiate the apoptotic effect of ADI-PEG20 in melanoma cells.16, 17
The apparent repression of the ASS1 enzyme that we observed in normal lymphoid tissues by immunohistochemistry suggests that exogenous arginine is also important in fuelling normal immune cell function. Indeed, many studies have highlighted the immunomodulatory effects of extracellular arginine, while few have addressed the expression and regulation of the rate-limiting enzyme for arginine biosynthesis, ASS1.18 Interestingly, increased levels of ASS1 have been noted in peripheral blood lymphocytes of patients with autoimmune systemic lupus erythematosus, with levels of the enzyme linked to disease activity.19 In contrast, arginase-induced arginine depletion has been linked to lymphocyte T hyporesponsiveness and reduced expression of the T-cell receptor CD3ζ chain in immune responses against infections, such as Leishmania, and in reduced tumour immunosurveillance.20, 21 Nevertheless, we showed that the unmethylated status of ASS1 in normal lymphoid cells was linked to the inducibility of ASS1 protein in the presence of ADI-PEG20, indicating that normal immune cells can resist arginine starvation. Importantly, citrulline is a byproduct of ADI-PEG20 catalytic activity and has been shown to overcome T-cell dysfunction secondary to arginine deprivation in cells with constitutive and inducible ASS1.22, 23 Therefore, the availability of citrulline and the unmethylated status of ASS1 in normal lymphoid cells may ameliorate any potential negative effects of arginine deprivation on the immune system. Further studies of ASS1 and arginine deprivation in normal immune cells are indicated, in addition to continued monitoring of patients for infection (see below) and modulation of tumour immunosurveillance in ongoing clinical trials of arginine-lowering agents.
Finally, we described a patient with therapy-resistant CTCL, who achieved a dramatic but brief improvement in his skin symptoms following ADI-PEG20 treatment. The patient's lymphoma displayed ASS1 promoter methylation and an absence of ASS1 expression, consistent with our lymphoma cell studies. It is noteworthy that the patient was on a tapering dose of steroids, which may have synergised with ADI-PEG20, as reported in a preclinical model of ADI-sensitive leukaemic cells.24 Furthermore, immunosuppression secondary to long-term steroid exposure and arginine deprivation may have contributed to the accompanying infection, a known complication of advanced CTCL.18 Nonetheless, the striking clinical response to ADI-PEG20 in a patient with refractory CTCL corroborates our preclinical data that ASS1 promoter methylation may be a promising biomarker for determining sensitivity to arginine deprivation in lymphomas.
In conclusion, arginine deprivation targeted to ASS1 promoter methylation may provide a novel approach to the treatment of lymphoid malignancy. Clinical trials employing arginine catabolising enzymes such as ADI-PEG20 combined with modulators of autophagy are warranted in patients with lymphoma, specifically assessing the role of ASS1 promoter methylation status as a biomarker of treatment response.
Materials and Methods
Immunohistochemistry
Normal (tonsil, reactive lymph node, and bone marrow) and malignant (follicular lymphoma, FL; diffuse large B-cell lymphoma, DLBCL; mantle cell lymphoma, MCL; marginal zone lymphoma, MZL; Burkitt's lymphoma, BL, Hodgkin's lymphoma, HL; and CTCL lymphoid paraffin sections were stained using an ASS1 antibody (BD Biosciences, Oxford, UK; dilution 1 : 100) and the Ventana Discovery XT System. Several authors scored the sections, obtained using informed patient consent and the approval of the Research Ethics Committees in London, UK and Ioannina, Greece, according to intensity and extent of ASS1 expression: FL (CO, MC, EH, AP); DLBCL, MCL, MZL, HL, BL, (EH, AP, PWS); and CTCL (SW, RC, TM, PWS), as described previously.6
Methylation profiling
Genomic DNA from 20 malignant lymphoma samples (16 primary follicular lymphomas and 4 lymphoma cell lines) and 6 benign samples (4 benign lymph nodes and 2 normal lymphoblastoid cell lines) was bisulphite converted using EZ-DNA Methylation Gold Kit (Zymo Research, Orange, CA, USA) according to protocol. Quantitative methylation profiling of bisulphite-modified DNA (500 ng) was performed using Illumina Infinium Human Methylation 27 assay (Illumina, San Diego, CA, USA).25
Methylation-specific PCR
Genomic DNA was isolated by incubating cell pellets in 500 μl SDS lysis buffer (100 mM NaCl, 10 mM Tris pH 8.0, 1 mM EDTA, 0.5% SDS) containing 100 μg/ml proteinase K at 55°C O/N and extraction with phenol/chloroform. Bisulphite conversion of genomic DNA was performed using the Zymo EZ DNA methylation kit (Cambridge Bioscience Ltd, Cambridge, UK). MS-PCR of a 143-bp fragment located within 265 bp downstream of the TSS was undertaken to determine the methylation status of ASS1 promoter. In all, 5 μl of bisulphite-modified DNA was used as template for PCR reactions with primers specific for methylated sequences (M): M forward 5′-GTAGGAGGGAAGGGG-TTTTC-3′ M reverse 5′-GCAAAAAACAAATAACCCGAA-3′ or unmethylated sequences (U): U forward 5′-GTAGGAGGGAAGGGGTTTTT-3′ U reverse 5′-ACAAAAAACAAA-TAACCCAAA-3′. CpGenome Universal Methylated DNA and Universal Unmethylated DNA (Millipore Ltd, Watford, UK) were used as positive and negative controls, respectively. PCR conditions were as follows: 8 cycles of 95°C for 2 min, 51.7°C for 30 s and 72°C for 30 s, followed by 32 cycles of 95°C for 30 s, 51.7°C for 30 s and 72°C for 30 s and a final extension at 72°C for 5 min. PCR products were run on a 2% agarose gel, stained with ethidium bromide and visualised using a transilluminator.
Cell culture and reagents
The normal lymphoblastoid B-cell line NcNc, the diffuse large B-cell lymphoma (DLBCL) cell line Karpas-422 (Clare Hall, Cancer Research UK, Potters Bar, UK), the CTCL cell lines MyLa (European Collection of Cell Cultures, Salisbury, UK) and SeAx (a kind gift from Prof Maarten Vermeer, Leiden University Medical Center, Leiden, The Netherlands) were grown in RPMI 1640 with 10% fetal bovine serum and supplemented with antibiotics. Cells were maintained in a humidified incubator containing 10% CO2 at 37°C. ADI-PEG20 (recombinant arginine deiminase, formulated with 20 000 mw polyethylene glycol molecule and a specific activity of 6.7 IU/mg) was provided by Polaris Pharmaceuticals, Inc. (San Diego, CA, USA) and used at concentrations from 0 to 500 ng/ml. Cells were harvested post-ADI-PEG20 at days 2–4 with cell line counting performed using a Beckman Vi-Cell cell viability analyser (Beckman Coulter Ltd, High Wycombe, UK). Demethylation experiments were performed using 5-aza-2′-deoxycytidine (0.5–5 μM of 5-Aza-dC, Sigma-Aldrich, Poole, UK) every other day for up to 12 days, with and without ADI-PEG20. Chloroquine (25 μM, Sigma-Aldrich) was added with ADI-PEG20 to assess the effect on LC3 protein turnover and lymphoid cell line viability. Isolated primary B lymphoma cells and CTCL (Sezary syndrome) cells, obtained using informed patient consent and Research Ethics Committee approval, were suspended in freshly prepared culture medium (IMDM; Gibco Biocult, Paisley, UK, including 10% human serum) at a concentration of 5 × 105 cells/ml, and plated into 96-well plates with the addition of soluble recombinant human CD40L (sCD40L at 100 ng/ml, Enzo Life Sciences, Exeter, UK) and recombinant interleukin-4 (IL-4 at 5 ng/ml, Sigma-Aldrich Co., UK) for 24 h before treatment with ADI-PEG20 for a further 72 h. The effect of chloroquine (25 μM) and doxorubicin (1 μM, a positive control) was assessed also, and cell viability was measured using the ViaLight assay (Lonza, Rockland, ME, USA).
Flow cytometry
Cells treated with ADI-PEG20 were harvested in cold PBS, fixed in 70% ethanol and stored at −20°C for at least 24 h. They were then washed and resuspended in propidium iodide/Triton X-100 staining solution containing RNase A. Flow cytometry was used to detect DNA content, and analyses of cell distribution in the different cell cycle phases were performed using FlowJo 7.5 (Tree Star, Inc., Ashland, OR, USA).
Real-time PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen, Crawley, UK) and retro-transcribed with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA, USA). Multiplex PCR was performed on an ABI Prism 7500 Real-Time PCR Instrument (Applied Biosystems). Primers and TaqMan probes for 18S and ASS1 were obtained from Applied Biosystems (TaqMan Gene Expression Assays).
Immunoblotting
Total protein extracts were prepared using NP40 cell lysis buffer (1% NP40, 50 mM Tris (pH 7.4), 150 mM NaCl) with Complete Protease Inhibitor Cocktail and phosSTOP tablets (Roche Diagnostics, Burgess Hill, UK). Whole-cell lysates were run on SDS-PAGE and transferred to PVDF membranes. All antibodies were purchased from Cell Signaling Technology (New England Biolabs Ltd, Hitchin, UK) except ASS1 (1 : 2500; BD Biosciences) and β-actin (1 : 20 000; Sigma-Aldrich). Immunoreactive bands were visualised by using Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer, Cambridge, UK).
Statistical analysis
GraphPad Prism version 5.0 was used to test results for statistical significance.
Acknowledgments
We would like to thank Andrew Clear, Silvia Ferreira, Chaz Mein, Keyur Trivedi and Mohammed Ikram for technical assistance. This work was supported by Cancer Research UK (C12522/A8632).
Glossary
- ASS1
argininosuccinate synthetase-1
- ADI-PEG20
pegylated arginine deiminase
- CTCL
cutaneous T-cell lymphoma
- qPCR
quantitative real-time PCR
- (MS)-PCR
methylation-specific
- LC3
light chain 3
Dr. Peter Szlosarek has received research support funding from Cancer Research UK (Clinician Scientist Fellowship) and Polaris Group (San Diego, USA). Dr. John Bomalaski is an employee of Polaris Group. The other authors have no conflicts of interest.
Footnotes
Edited by H-U Simon
References
- Izzo F, Marra P, Beneduce G, Castello G, Vallone P, De Rosa V, et al. Pegylated arginine deiminase treatment of patients with unresectable hepatocellular carcinoma: results from phase I/II studies. J Clin Oncol. 2004;22:1815–1822. doi: 10.1200/JCO.2004.11.120. [DOI] [PubMed] [Google Scholar]
- Ascierto PA, Scala S, Castello G, Daponte A, Simeone E, Ottaiano A, et al. Pegylated arginine deiminase treatment of patients with metastatic melanoma: results from phase I and II studies. J Clin Oncol. 2005;23:7660–7668. doi: 10.1200/JCO.2005.02.0933. [DOI] [PubMed] [Google Scholar]
- Glazer ES, Piccirillo M, Albino V, Di Giacomo R, Palaia R, Mastro AA, et al. Phase II study of pegylated arginine deiminase for nonresectable and metastatic hepatocellular carcinoma. J Clin Oncol. 2010;28:2220–2226. doi: 10.1200/JCO.2009.26.7765. [DOI] [PubMed] [Google Scholar]
- Yang TS, Lu SN, Chao Y, Sheen IS, Lin CC, Wang TE, et al. A randomised phase II study of pegylated arginine deiminase (ADI-PEG 20) in Asian advanced hepatocellular carcinoma patients. Br J Cancer. 2010;103:954–960. doi: 10.1038/sj.bjc.6605856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- Szlosarek PW, Klabatsa A, Pallaska A, Sheaff M, Smith P, Crook T, et al. In vivo loss of expression of argininosuccinate synthetase in malignant pleural mesothelioma is a biomarker for susceptibility to arginine depletion. Clin Cancer Res. 2006;12:7126–7131. doi: 10.1158/1078-0432.CCR-06-1101. [DOI] [PubMed] [Google Scholar]
- Nicholson LJ, Smith PR, Hiller L, Szlosarek PW, Kimberley C, Sehouli J, et al. Epigenetic silencing of argininosuccinate synthetase confers resistance to platinum-induced cell death but collateral sensitivity to arginine auxotrophy in ovarian cancer. Int J Cancer. 2009;125:1454–1463. doi: 10.1002/ijc.24546. [DOI] [PubMed] [Google Scholar]
- Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535–544. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- Osunkoya BO, Adler WH, Smith RT. Effect of arginine deficiency on synthesis of DNA and immunoglobulin receptor of Burkitt lymphoma cells. Nature. 1970;227:398–399. doi: 10.1038/227398a0. [DOI] [PubMed] [Google Scholar]
- Storr JM, Burton AF. The effects of arginine deficiency on lymphoma cells. Br J Cancer. 1974;30:50–59. doi: 10.1038/bjc.1974.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage B, Fennell DA, Nicholson L, McNeish I, Lemoine NR, Crook T, et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int J Cancer. 2010;126:2762–2772. doi: 10.1002/ijc.25202. [DOI] [PubMed] [Google Scholar]
- Feun L, You M, Wu CJ, Kuo MT, Wangpaichitr M, Spector S, et al. Arginine deprivation as a targeted therapy for cancer. Curr Pharm Des. 2008;14:1049–1057. doi: 10.2174/138161208784246199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RH, Coates JM, Bowles TL, McNerney GP, Sutcliffe J, Jung JU, et al. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res. 2009;69:700–708. doi: 10.1158/0008-5472.CAN-08-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WB, Aiba I, Lee SY, Feun L, Savaraj N, Kuo MT. Resistance to arginine deiminase treatment in melanoma cells is associated with induced argininosuccinate synthetase expression involving c-Myc/HIF-1alpha/Sp4. Mol Cancer Ther. 2009;8:3223–3233. doi: 10.1158/1535-7163.MCT-09-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- Tsai WB, Aiba I, Long Y, Lin HK, Feun L, Savaraj N, et al. Activation of Ras/PI3K/ERK pathway induces c-Myc stabilization to upregulate argininosuccinate synthetase, leading to arginine deiminase resistance in melanoma cells. Cancer Res. 2012;72:2622–2633. doi: 10.1158/0008-5472.CAN-11-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V, Zanovello P. Regulation of immune responses by ℒ-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Ohno T, Kimura Y, Sugimura K, Sagawa A, Jhodo S, Azuma I. Elevated gene expression of argininosuccinate synthetase in peripheral lymphocytes from systemic lupus erythematosus (SLE) patients. Autoimmunity. 1992;13:127–132. doi: 10.3109/08916939209001913. [DOI] [PubMed] [Google Scholar]
- Munder M, Choi BS, Rogers M, Kropf P. ℒ-arginine deprivation impairs Leishmania major-specific T-cell responses. Eur J Immunol. 2009;39:2161–2172. doi: 10.1002/eji.200839041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- Bansal V, Rodriguez P, Wu G, Eichler DC, Zabaleta J, Taheri F, et al. Citrulline can preserve proliferation and prevent the loss of CD3 zeta chain under conditions of low arginine. JPEN J Parenter Enteral Nutr. 2004;28:423–430. doi: 10.1177/0148607104028006423. [DOI] [PubMed] [Google Scholar]
- Jacoby LB. Adaptation of cultured human lymphoblasts to growth in citrulline. Exp Cell Res. 1974;84:167–174. doi: 10.1016/0014-4827(74)90393-0. [DOI] [PubMed] [Google Scholar]
- Noh EJ, Kang SW, Shin YJ, Choi SH, Kim CG, Park IS, et al. Arginine deiminase enhances dexamethasone-induced cytotoxicity in human T-lymphoblastic leukemia CCRF-CEM cells. Int J Cancer. 2004;112:502–508. doi: 10.1002/ijc.20435. [DOI] [PubMed] [Google Scholar]
- Killian JK, Bilke S, Davis S, Walker RL, Killian MS, Jaeger EB, et al. Large-scale profiling of archival lymph nodes reveals pervasive remodeling of the follicular lymphoma methylome. Cancer Res. 2009;69:758–764. doi: 10.1158/0008-5472.CAN-08-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riain C, O'Shea DM, Yang Y, Le Dieu R, Gribben JG, Summers K, et al. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 2009;2009;23:1858–1866. doi: 10.1038/leu.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]