Abstract
According to the concept of lipotoxicity, ectopic accumulation of lipids in non-adipose tissue induces pathological changes. The most prominent effects are seen in fatty liver disease, lipid cardiomyopathy, non-insulin-dependent diabetes mellitus, insulin resistance and skeletal muscle myopathy. We used the MCK(m)-hLPL mouse distinguished by skeletal and cardiac muscle-specific human lipoprotein lipase (hLPL) overexpression to investigate effects of lipid overload in skeletal muscle. We were intrigued to find that ectopic lipid accumulation induced proteasomal activity, apoptosis and skeletal muscle damage. In line with these findings we observed reduced Musculus gastrocnemius and Musculus quadriceps mass in transgenic animals, accompanied by severely impaired physical endurance. We suggest that muscle loss was aggravated by impaired muscle regeneration as evidenced by reduced cross-sectional area of regenerating myofibers after cardiotoxin-induced injury in MCK(m)-hLPL mice. Similarly, an almost complete loss of myogenic potential was observed in C2C12 murine myoblasts upon overexpression of LPL. Our findings directly link lipid overload to muscle damage, impaired regeneration and loss of performance. These findings support the concept of lipotoxicity and are a further step to explain pathological effects seen in muscle of obese patients, patients with the metabolic syndrome and patients with cancer-associated cachexia.
Keywords: lipotoxicity, lipoprotein lipase, muscle atrophy, muscle regeneration
Higher metazoa deposit their spare energy mainly in lipid droplets of adipocytes as neutral lipids.1, 2 If adipocytes are overloaded, not abundant enough, or regulatory mechanisms are impaired, fat is stored ectopically, which frequently induces pathological changes such as fatty liver, lipid cardiomyopathy, non-insulin-dependent diabetes mellitus, insulin resistance and skeletal muscle myopathy.3 The pathophysiological concept underlying these conditions is termed lipotoxicity, that is, ectopic accumulation of lipids in non-adipose tissue negatively affecting cells, tissues and organs that store it.1, 4, 5, 6 Neutral lipids are probably not directly lipotoxic but can be taken as a good measure for overall lipid load, whereas other lipid species such as free fatty acids (FFAs) or ceramides are more likely to cause lipotoxicity.6
Lipoprotein lipase (LPL) EC 3.1.1.34 facilitates entry of lipids into muscle by catalyzing the hydrolysis of triacyl glyerol (TAG) present in very low-density lipoproteins or chylomicrons in the blood stream at the surface of the capillary endothelium. Released FFAs are subsequently taken up by the tissue parenchyma cells.7 An intriguing murine model system to study direct effects of lipid overload in muscle has been established earlier, the muscle creatin kinase (MCK) promoter; medium expressor(m); human(h) LPL-expressing mouse.8 A fully functional hLPL minigene is expressed exclusively in skeletal and cardiac muscle using the MCK promoter. hLPL expression results in decreased plasma TAG levels, elevated FFA uptake by muscle tissue, weight loss and premature death. Transgenic mice show severe myopathy, fiber atrophy, enhanced glycogen storage, as well as mitochondrial and peroxisomal proliferation.8 This model has been used by several groups showing increased alpha-tocopherol,9 intracellular FFAs10 and TAG levels11 in muscle. Peroxisomal enzymes are induced,10 whereas no inhibition of insulin-stimulated whole-body and muscle-specific glucose uptake could be detected.11
Skeletal muscle is second only to bone marrow with regards to its regenerative capacity.12 Muscle regeneration can be triggered by muscle injury, which may be caused by mechanical trauma, extensive physical activity such as resistance training, or by genetic defects such as muscle dystrophies. Muscle regeneration is characterized by an initial degenerative period followed by a regenerative phase triggered primarily by a mononuclear cell population termed satellite cells.13, 14
The C2C12 murine myoblast model can be used to recapitulate some of the basic mechanisms of muscle regeneration, and studies with C2C12 myoblasts have provided valuable molecular insights into this process. C2C12 cells undergo rapid proliferation when maintained in medium with a high serum concentration (i.e. 10% fetal bovine serum (FBS)) and differentiate into myotube-like cells within 2–6 days when shifted to low-serum conditions (i.e. 2% horse serum, HS).15, 16
On the basis of previous morphological observations in skeletal muscle of LPL-overexpressing mice,8 we hypothesized that lipid overload induces lipoapoptosis17 and proteolysis and that impaired recovery of skeletal muscle damage due to lipotoxic effects might occur.1, 4, 6
Results
MCK-driven hLPL expression leads to muscle loss and impaired physical stamina
Previously, it was shown that MCK-driven hLPL expression leads to approximately eight-fold enhanced LPL activity in skeletal muscle, resulting in severe histologically traceable pathological changes in adult MCK(m)–hLPL mice.8, 9, 10, 11 We suspected that these animals might be physically impaired. Indeed, in treadmill experiments, 4–6-month-old hLPL transgenic animals showed >6-fold reduced physical endurance compared with isogenic wild-type (wt) mice (Figure 1a).
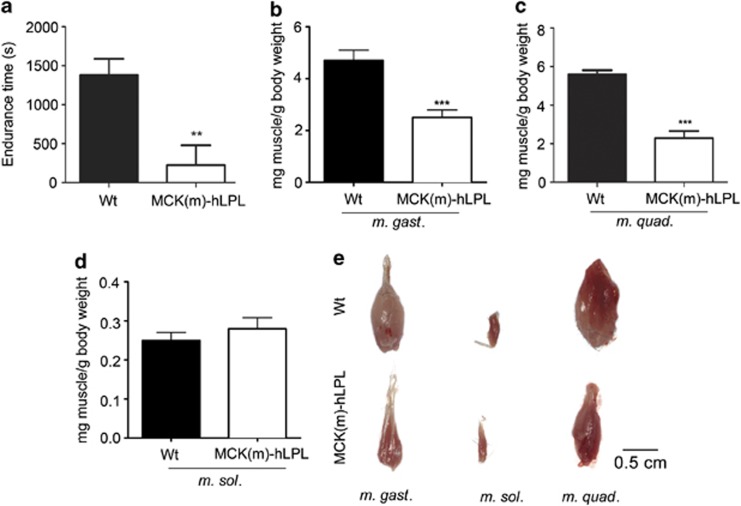
Figure 1.
(a) Physical endurance of MCK(m)-hLPL and isogenic wt mice (n=3) was tested using a treadmill (V=0.23 m/s). A mild electric stimulus was delivered each time an animal reached the rear end of the treadmill lane. The time that elapsed until 10 stimuli occurred is presented as mean value ±SD. Endurance of MCK(m)-hLPL mice was significantly (**P<0.005) lower than that of wt mice (226±252 versus 1379±209 s). (b and c) Mice were killed and muscles extracted and weighed. Muscle weights are presented in mg muscle per g body weight ±SD. M. gast. and M. quad. weights of MCK(m)-hLPL compared with wt mice were both significantly (***P<0.0001) decreased (2.51±0.28 versus 4.70±0.39 mg/g (n=5) and 2.29±0.9 mg/g (n=3) versus 5.59±0.77 mg/g (n=6), respectively). (d) No significant differences in M. sol. weights between MCK(m)-hLPL and wt mice was found (0.028±0.002 versus 0.025±0.003 mg/g (n=5)). (e) Representative images of muscle preparations. Upper panel, wt and lower panel, MCK(m)-hLPL
To study the effect of hLPL overexpression on skeletal muscle we killed mice and carefully anatomically prepared three different skeletal muscles, M. gastrocnemius (M. gast.), Musculus quadriceps (M. quad.) and Musculus soleus (M. sol.). First, we analyzed hLPL mRNA levels as measure for transgene expression. Quantitative reverse transcription real-time PCR (qPCR) confirmed strong expression of hLPL (mRNA) in M. gast. (3.81±0.39-fold lower signal than 18s rRNA), M. quad. (3.7±0.46-fold lower signal than 18s rRNA) and M. sol. (10.33±3.5-fold lower signal than 18s rRNA) of MCK(m)-hLPL mice. Noticeable, expression in M. sol. however, was approximately three-fold less compared with M. gast. and M. quad. of MCK(m)-hLPL mice. No hLPL (mRNA) was detectable in wt mice (n=5). Examples of amplification blots and melting curves are shown in Supplementary Figure 1. Whole muscle weights of M. gast. and M. quad. from MCK(m)-hLPL mice were significantly (***P<0.0001) decreased compared with wt mice (2.51±0.28 versus 4.70±0.39 mg/g and 2.29±0.9 versus 5.59±0.77 mg/g, respectively, Figures 1b and c). There were however, no significant differences in M. sol. weights (Figure 1d). Images from isolated muscles are shown to demonstrate anatomically correct preparation (Figure 1e). The hypotrophic appearance of M. gast. and M. quad. from MCK(m)-hLPL mice, illustrates the differences between genotypes.
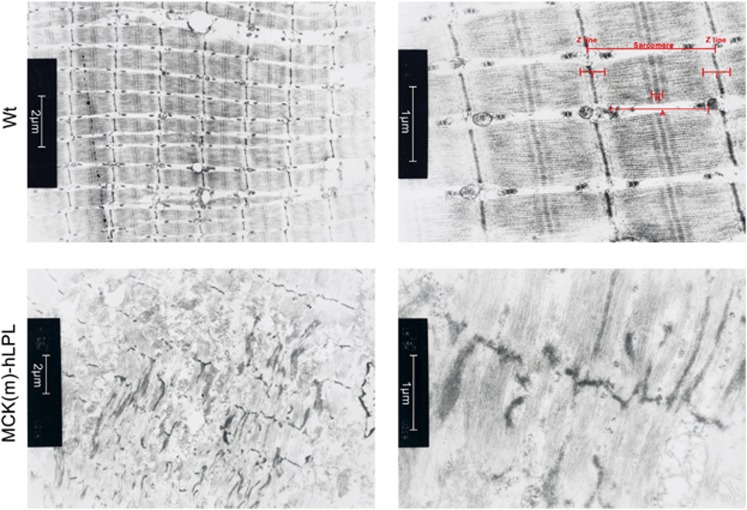
Previous electron microscopy (EM) studies of muscles from MCK-hLPL mice showed increased numbers of mitochondria in subsarcolemmal, perinuclear and intermyofibrillar positions.8 We speculated that performance deficiencies and muscle mass loss of MCK(m)-hLPL mice (Figure 1) might correspond to sarcomeric aberrations. Indeed, samples from MCK(m)-hLPL mice showed focal disruption of sarcomere units with ultrastructural irregularities, whereas such areas were absent in M. gast. from wt animals (Figure 2) and M. sol. samples from MCK(m)-hLPL mice (Supplementary Figure 2) as well as M. sol. samples from wt animals (data not shown).
Figure 2.
M. gast. samples from MCK(m)-hLPL and wt mice (n=3) were analyzed by EM. Representative images are shown. MCK(m)-hLPL M. gast. is characterized by severely disrupted sarcomeres and hardly defined myofibrillar structures (lower panels). A full sarcomere unit, as readily apparent in M. gast. of wt mice (upper panels), is used to illustrate normal muscle structure depicting Z-lines, H-band, A-bands and I-bands in the upper right panel. Inserts: size bars
Muscles of MCK(m)-hLPL mice contain increased lipids and show increased protein degradation and lipoapoptosis
To assess muscle lipid content we used anatomically prepared (Figure 1e) entire muscles from 12 h fasted MCK(m)-hLPL and wt mice. In M. gast. of LPL transgenic mice FFA levels were elevated >3-fold (***P<0.0001) and TAG levels >4-fold (**P<0.005) compared with wt animals (Figures 3a and b, respectively). These findings are in accordance with previous studies.8, 11 FFA and TAG levels were elevated to a lesser degree in M. sol. of MCK(m)-hLPL compared with wt mice (Figures 3c and d, respectively).
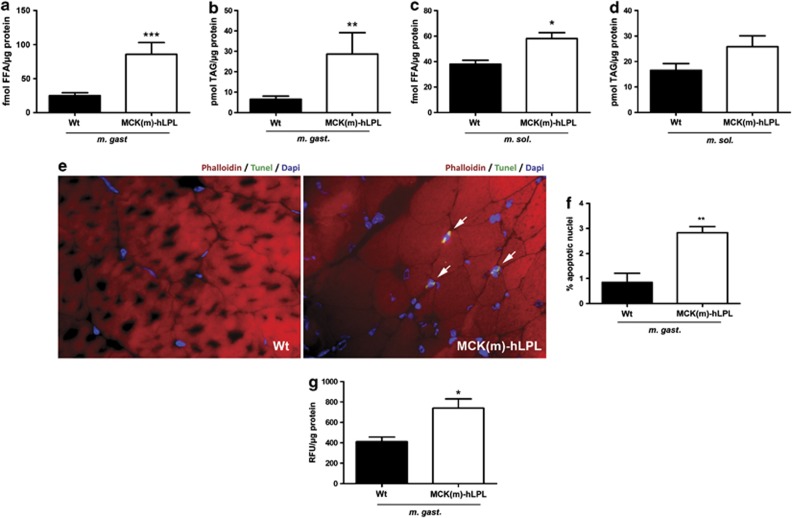
Figure 3.
(a–d) Lipid analysis of muscles from 12 h fasted MCK(m)-hLPL and wt mice (n=6). Muscles were prepared as described in Figure 1. FFA and TAG were determined biochemically from extracts of transversal cross sections. (a) FFA levels were significantly (***P<0.0001) increased in M. gast. of MCK(m)-hLPL compared with wt mice (85.84±17.39 versus 25.0±4.3 fmol/μg). (b) TAG levels were significantly (**P<0.005) increased in M. gast. of MCK(m)-hLPL compared with wt mice (28.71±10.49 versus 6.48±1.62 pmol/μg). (c) FFA levels were significantly (*P<0.05) increased in M. sol. of MCK(m)-hLPL compared with wt mice (58.79±4.34 versus 39.06±2.19 fmol/μg). (d) TAG levels were not significantly increased in M. sol. of MCK(m)-hLPL compared with wt mice (25.82±4.47 versus 16.88±2.65 fmol/μg). (e) TdT-mediated dUTP nick end label (TUNEL) staining of frozen transverse sections of M. gast. TUNEL-positive apoptotic nuclei stain bright green. Representative sections are shown and positive nuclei are highlighted with arrows. Phalloidin and DAPI were used as counterstain for actin filaments and nuclei, respectively. (f) A total of 10 fields of view ( × 100 magnification) encompassing >300 nuclei were counted from each section (n=4). MCK(m)-hLPL M. gast. showed a significantly (**P<0.005) higher percentage of apoptotic nuclei compared with wt M. gast. (2.83±0.24% versus 0.84±0.37%). (g) ATP-depended proteasomal enzymatic activity was measured biochemically in extracts of transversal cross sections of M. gast. and is presented as relative fluorescence units (RFU)/μg protein ±SD. Significantly (*P<0.05) higher 26S-protesome activity was detected in M. gast. of MCK(m)-hLPL compared with wt mice (739.5±91.5 versus 411.4±45.1 RFU/μg protein; n=3)
To investigate lipoapoptosis and protein degradation we performed tunel staining and analyzed 26S proteasomal activity. Tunel staining showed a significantly (**P<0.005) higher percentage of apoptotic nuclei in M. gast. of MCK(m)-hLPL mice compared with wt (2.83±0.24% versus 0.84±0.37% Figures 3e and f). 26S proteasome-dependent proteolysis was increased significantly (*P<0.05) on average by 79.8% in LPL transgenic animals compared with wt (Figure 3g). To investigate whether some of the effects we recorded might be just a consequence of fiber-type differences between LPL transgenic and wt mice, we performed fiber-type analysis. As expected, the percentage of fast twitch (type II) fibers was much higher in M. gast. and M. quad. compared with M. sol.. However, no differences regarding the percentage of type I and type II fibers, was found between MCK(m)-hLPL and wt mice within each muscle group (M. gast., M. sol. and M. quad.; Supplementary Figure 3A and B).
Overexpression of hLPL in muscle cells reduces their regenerative/myogenic capacity
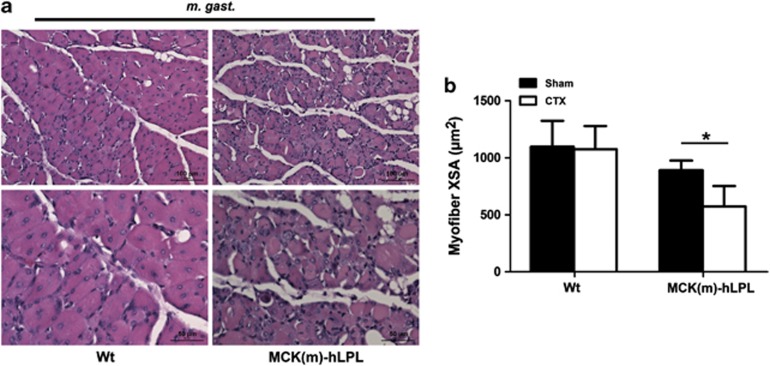
Skeletal muscle satellite cells are activated upon muscle injury or disease. Under normal circumstances their progenies, muscle myoblasts, subsequently undergo proliferation and eventually differentiate to form new muscle fibers, allowing for proper muscle regeneration.14, 18 We hypothesized that in addition to protein degradation and apoptosis in skeletal muscle, MCK(m)-hLPL mice might lack muscle regeneration due to lipotoxic effects. Therefore, we investigated the regeneration potential of skeletal muscle from MCK(m)-hLPL and wt mice by carefully injecting M. gast. with cardiotoxin (CTX) or PBS (Sham). Mice were killed 10 days later. M. gast. and two control muscles from the hind limb (M. extensor digitorum longus (M. edl.) and M. sol.) were isolated to study their regenerative response. Indeed, regeneration potential was significantly decreased in MCK(m)-hLPL mice as evidenced by histological examination of M. gast. specimens (Figure 4a). Average cross section area (XSA) of muscle fibers was significantly (*P<0.05) reduced in the MCK(m)-hLPL CTX versus the Sham group after 10 days of regeneration (64.5±20.1% Figure 4b), whereas wt muscle fibers had already reached almost their normal XSA (98.1±18.3% Figure 4b). No tissue damage was detected in M. edl. and M. sol. of CTX-injected mice (Supplementary Figure 4) or in sham-injected muscles (data not shown).
Figure 4.
Mice were injected with CTX or vehicle PBS (Sham) directly into M. gast. and killed 10 days later (n=4 for MCK(m)-hLPL and n=6 for wt and equal numbers for Sham). M. gast. were excised, transverse tissue sections prepared and H&E-stained. Representative images of regenerating areas are shown in (a). Inserts represent size bars. (b) Images of myofibers were captured and XSA of 100–260 myofibers within a fixed area (289 319 μm2) were measured. Measurements of myofibers in CTX-injected animals was performed in regenerating areas as defined by centrally localized nuclei. Average XSA of muscle fibers in MCK(m)-hLPL mice was significantly (*P<0.05) reduced 10 days after CTX versus sham-injected mice (64.5±20.1%). No statistical significant difference was observed in wt mice between CTX- and sham-injected mice (98.1±18.3%)
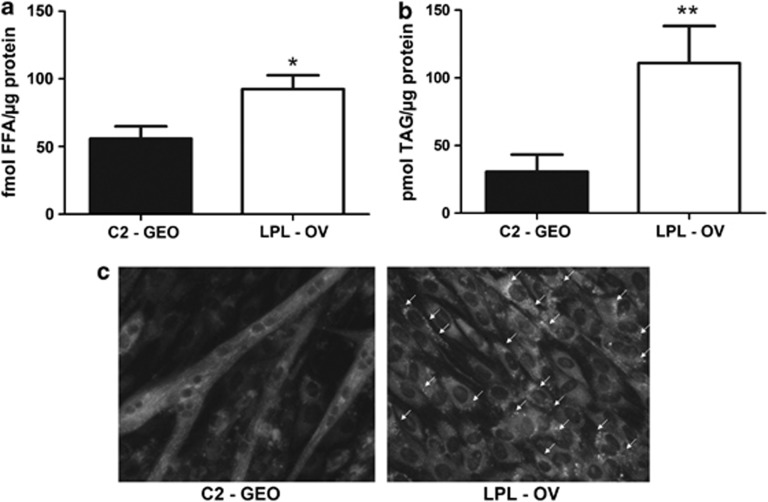
As a second model system to test effects of hLPL overexpression on myogenesis we used cultured C2C12 murine myoblasts, which are regarded as an excellent model system to study myogenesis by several groups.13, 14 C2-GEO (vector control) as well as hLPL-overexpressing LPL-OV cells proliferated at a comparable rate (Supplementary Figure 7B). Only after each culture had reached confluence, cells were induced for myogenic differentiation using 2% HS. At the time of induction both cultures showed comparable numbers of viable cells (Supplementary Figure 7C). Functional hLPL overexpression was confirmed in LPL-OV cells (Supplementary Figure 5). Biochemical analysis revealed that intracellular FFA (Figure 5a) were significantly (*P<0.05) increased, on average by 65.7% in LPL-OV compared with C2-GEO. Moreover, there was a highly significant (**P<0.005) increase in TAG levels (Figure 5b) on average by 176.4% in the LPL-OV line. Increased intracellular neutral lipids in hLPL-overexpressing cells were also evidenced by nile red (Figure 5c and Supplementary Figure 9) and bodipy (Supplementary Figure 6) stainings.
Figure 5.
(a) FFA levels were determined in 6 days myogenically differentiated LPL-OV and C2-GEO cells that had been serum starved for 6 h and are presented ±S.D. (n=3). LPL-OV contained significantly (*P<0.05) more FFA than wt (92.49±10.24 fmol/μg versus 55.81±9.16 fmol FFA/μg protein). (b) TAG was measured in the same samples and shows a significant (**P<0.005) increase in LPL-OV compared with C2-GEO (111.0±27.34 pmol TAG/μg protein versus 30.68±12.65 pmol/μg). (c) Cells were grown under identical culture conditions but on coverslips for microscopic analysis. Areas with Nile red-positive lipid droplets were seen exclusively in LPL-OV cells and are indicated by white arrows. Representative images are shown
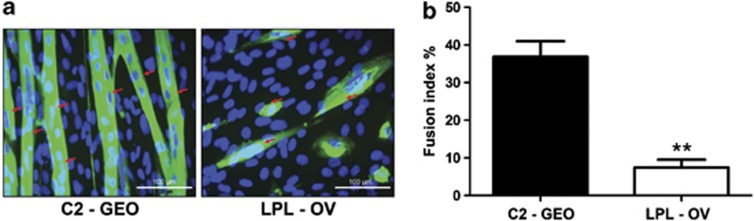
To assess the influence of LPL-triggered lipid accumulation on the myogenic capacity of C2C12 myoblasts we measured the fusion index of LPL-OV and C2-GEO cells. Remarkably, it was significantly (**P<0.005) decreased on average by 79.8% after 6 days of differentiation in hLPL-overexpressing C2C12 cells (Figures 6a and b), indicating a reduced differentiation potential. Supporting this finding, Supplementary Figure 7 shows representative images from differentiating cultures from both genotypes at d0, d3 and d6 after induction with 2% HS, indicating reduced myogenic capacity of hLPL-overexpressing C2C12 cells also at d3. Moreover, we detected a significant (**P<0.005) decrease in the satellite cell marker Pax7 mRNA expression by 88.1±13% in LPL-OV compared with C2-GEO 1 day after induction of myogenic differentiation. mRNA expression of myogenic transcription factors MyoD (myogenic differentiation antigen 1) and Myogenin was decreased by 88.8±27% and 55.6±27.8%, respectively, after 6 days of differentiation. Representative amplification blots and melting curves are shown in Supplementary Figure 1.
Figure 6.
(a) LPL-OV as well as C2-GEO cells (n=3), were induced for differentiation by medium containing 2% HS after reaching confluence and differentiated for 6 days. Cells were fixed and decorated with antiMyHC antibody followed by immunofluorescence detection to visualize myotubes (green channel) and nuclei (stained with DAPI; blue channel). Representative images are shown. Red arrows indicate myotubes. (b) Fusion indices were calculated from five images of each culture and represent mean values of nuclei in MyHC-positive stained areas versus total number of nuclei per image in percent ±SD. The fusion index of LPL-OV is significantly (**P<0.005) lower than the one of control cells (7.44±2.07% versus 36.86±4.12%)
Discussion
Excessive supply of FFA may have an important role in the pathogenesis of myopathy. In obese individuals increased FFA supply is often associated with the metabolic syndrome, whereas in cancer cachexia enhanced amounts of FFA are released from fat depots, for example by cytokines released from malignant tumors.19 Especially in the latter, severe myopathy may develop, drastically reducing the quality of life of affected individuals.20 In order to develop rational concepts for therapeutic intervention, it is necessary to understand the pathogenic mechanisms that cause lipotoxicity in skeletal muscle.
To directly investigate lipotoxicity in skeletal muscle we employed hLPL overexpression in two models. First, the MCK(m)-hLPL murine model system distinguished by a human LPL minigene ectopically expressed under the control of the muscle-specific MCK promoter.8 Although no systemically elevated lipid levels were found in these mice, they do show lipid overload in muscle when kept on standard chow.8, 9, 10, 11, 21 This allows direct study of lipid overload in muscle.1, 4, 6 As a second model system we used C2C12 murine myoblast cells overexpressing human LPL to investigate lipid-associated changes in myogenic potential. First we confirmed that muscle-specific hLPL expression leads to enhanced muscle lipid content in mice (Figures 3a and b). We also found a gross reduction in M. gast. and M. quad. muscle mass (Figures 1b and c) and impaired physical performance (Figure 1a) in hLPL transgenic animals. This phenotype was apparently aggravated by impaired muscle function due to structural changes reflected by ultrastructural morphology as illustrated by the irregularities of a representative sarcomere unit from M. gast. (Figure 2). The lack of phenotype in M. sol. in hLPL transgenic mice (Figure 1d) was probably due to a less pronounced hLPL transgene expression resulting in only slightly elevated FFA and TAG levels (compare Figures 3c and d with Figures 3a and b). Elevated caspase expression was detected immunohistochemically in single cells of M. gast. of hLPL transgenic mice and caspase 3 and/or 7 activites were elevated in muscle homogenates of M. gast. of hLPL transgenic mice (Supplementary Figure 8). Caspases are useful markers of apoptosis, however, they are also involved in muscle differentiation and repair.22, 23 Tunel staining substantiated increased apoptotic activity as it revealed a significantly (***P<0.0001) higher number of tunel-positive nuclei in M. gast. from MCK(m)-hLPL mice as compared with wt (Figures 3e and f). Enhanced protein degradation was evidenced by increased proteasomal activity in MCK(m)-hLPL M. gast. (Figure 3g). Both apoptosis and enhanced proteasomal activity might contribute to loss of M. gast. muscle mass in MCK(m)-hLPL mice (Figures 3a). Differences in the distribution of type IIa and IIb fibers in gastrocnemius and quadriceps muscles of LPL-overexpressing mice and controls have been reported recently.24 On staining type I and type II fibers (Supplementary Figure 3) we found no differences between hLPL transgenic and wt mice. In our study, we did not discriminate between type IIa and IIb fibers. However, we do not think that it is likely that the differences reported for type IIa and IIb fibers are sufficient to explain the severe myopathy observed in these animals.
Skeletal muscle is a highly regenerative tissue and it remains somewhat surprising that the observed rate of apoptosis and proteasome activation would be sufficient to cause this rather severe phenotype. An alternative or additional explanation for this observation might relate to the fact that the regenerative potential of muscle is mainly based on the proper function of satellite cells, which are activated after muscle damage.12 To investigate whether muscle regeneration was also affected by hLPL overexpression-induced lipotoxicity, we studied skeletal muscle regeneration in vivo in M. gast. after CTX-induced injury. Indeed, we established that the regenerative potential was severely impaired in MCK(m)-hLPL mice (Figures 4a and b).
To investigate cellular differentiation mechanisms we used hLPL overexpression in C2C12 murine myoblast cells. Staining for myotubes revealed that these cells had almost completely lost their myogenic potential (Figures 6a and b and Supplementary Figure 7). In addition, the classic satellite cell marker Pax7 was lost in confluent cultures, whereas expression of myogenic factors Myogenin and MyoD was suppressed in differentiated cultures of hLPL-overexpressing C2C12 cells. Therefore, we speculate that lipid accumulation might lead to a very early inhibition of myogenesis, possibly by changing the fate of myogenic precursor cells. Our results are supported by a recent study reporting that the trans-10, cis-12-conjugated linoleic acid also inhibits C2C12 cell differentiation.25
In summary, we propose that lipid overload causes impaired skeletal muscle function due to a reduction of muscle mass and ultrastructural damage. This is caused by a combination of activation of the proteosomal system, increased apoptosis and impairment of muscle regeneration potential, as evidenced in vivo and in cultured cells. The nature of the lipid species exerting its effects in these processes has yet to be clarified. This, however, is beyond the scope of the current study. Our results should provide a basis to better understand lipotoxic effects in pathological conditions as diverse as the metabolic syndrome and cachexia.1, 26
Material and Methods
Animal housing
C57BI/6J × CB wt mice and otherwise isogenic MCK(m)-hLPL mice8 were maintained on a regular light–dark cycle (12 h light,12 h dark) and kept on standard chow (4.5% w/w fat). Mice were genotyped by PCR (primer-sequences, see Supplementary Table 1; representative genotyping PCR DNA gel image, Supplementary Figure 10). All experiments were performed in accordance with animal protocol BMWF-66.010/0110-II/3b/2010, as approved by the Austrian government.
Treadmill experiments
Physical endurance of mice was assessed on a motorized treadmill (IITC Inc./Life Sciences, Woodland Hills, CA, USA) at v=0.23 m/s and slope=0°. A mild electrical stimulus (100–120 V; 2 mA) was applied to mice that reached the rear end of the treadmill lane. The time until a mouse received 10 stimuli was recorded.
Muscle isolation
Mice were anaesthetized using isoflurane and killed by cervical dislocation. Muscles were carefully anatomically isolated and utmost care was taken to excise only the muscle of interest free of contaminating other muscles or connective tissue. See Figure 1e for representative muscle preparations.
Quantitative reverse transcription real-time PCR
Total RNA was extracted from whole transverse sections of the respective muscles using Trizol LS from Invitrogen (Vienna, Austria) following the manufacturers protocol. cDNA was synthesized with High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Vienna, Austria). For qPCR, SYBR Green PCR Master Mix from Invitrogen and the Applied Biosystems 7900 HT Sequence Detection System were used (primers, see Supplementary Table 1). For computation of relative RNA contents we used the comparative CT method.27
Muscle weights
Whole freshly isolated muscles were weighed. Both respective muscles from each animal were weighed and the average value was used as a single data point for statistical analyses.
EM of muscle sections
Excised muscle tissue was prepared and EM was performed using a Philips CM 100 electron microscope (Philips, Eindhoven, The Netherlands) essentially as described earlier.28
FFA and TAG content
Lipids were either extracted from whole transverse sections of carefully isolated whole muscles or from C2C12 cell homogenates by the hexane isoproponal method.29 FFA content was measured using the NEFA-HR(2) kit from Wako Chemicals GmbH (Neuss, Germany) and the TAG content measured by the Triglyceride FS Kit from DiaSys Diagnostic Systems (Holzheim, Germany) following the manufacturer's instructions.
Muscle regeneration
Injury was triggered in M. gast. of 6-month-old MCK(m)-hLPL and wt mice by careful direct injection of 25 μl 10 μM CTX (Sigma-Aldrich, Vienna, Austria). Animals from both groups were also mock-injected with PBS (Sham) for control purposes. Whole M. gast. was isolated on the 4th day after injection to inspect the wound coagulum. From a second set of CTX- and Sham-injected animals from both genotypes, M. gast., M. sol. and M. edl. were carefully isolated at 10th day after injection for histological analysis of injury and muscle regeneration.30, 31 Whole muscles were fixed using 4% neutral buffered formaldehyde (formalin) and whole transversial sections were prepared and stained with hematoxylin&eosin (H&E).32 Images of H&E-stained myofibers were captured and whole transverse sections of each muscle were analyzed by microscopy. No injured areas were found in Sham or in M. sol. and M. edl. from CTX-injected mice confirming that CTX injection was confined to M. gast. The region of regeneration in CTX-injected M. gast. was defined by centrally nucleated fibers. XSA of 100–260 myofibers within a fixed area of regeneration (289 319 μm2) was measured.
Tunel staining
Whole muscle cross sections were stained using the Fluorescein In Situ Cell Death Detection Kit #11684795001 from Roche-Applied-Science (Vienna, Austria). Staining was performed according to the supplier's protocol. Slides were counterstained with Alexa Fluor 594 Phalloidin #A12381 for labeling of actin filaments (1 : 1000) and DAPI fluoroPure grade #D21490 (1 : 1000), both from Life Technologies (Vienna, Austria). Slides were visualized with an Olympus BX51 fluorescent microscope equipped with digital camera DP71 and images of the whole transverse sections analyzed with cell D software, Olympus (Vienna, Austria).
Immunohistochemistry
Skeletal muscles of killed mice were carefully isolated, fixed in formalin and embedded in paraffin. IHC was performed using active caspase3 antibody and appropriate secondary antibody (Supplementary Table 2). Muscle fiber-type staining was performed using monoclonal anti-Myosin fast or slow antibodies M4276 and M8421, respectively, both from Sigma-Aldrich.
Cell-culture
The C2C12-derived LPL-OV and C2-GEO cells were a generous gift from Dr. Eckel's research group33 and stably express geomycin or a human LPL minigene, respectively. Cells were propagated at 37 °C, 5% CO2 in growth medium RPMI 1640 high glucose (PAA, Vienna, Austria), 10% FBS (PAA). Cells were passaged at pre-confluence, essentially as described.16 Myogenic differentiation was induced in cultures that had reached confluence by switch to DMEM high glucose, 2% Donor Horse Serum from PAA.16, 34
C2C12 cell fusion index
Fixed (2% formaldehyde) and permeabilized (0.1% Triton-X100, Sigma-Aldrich) cells were analyzed for myosin heavy chain (MyHC) expression (see Supplementary Table 1 for antibodies used) and nuclei were counterstained by DAPI. Fusion index (%) was calculated from five representative images from each culture, dividing the number of nuclei in MyHC-positive stained areas by the total number of nuclei.
LPL activity
LPL was released from cells using Heparin (EBEWE-Pharma, Holzkirchen, Germany) and activity assays were performed as described earlier35 using scintillation cocktail (Opti-Phase SuperMix, PerkinElmer Life and Analytical Sciences, Inc., Santa Clara, CA, USA) in a Beckman Coulter Inc. (Vienna, Austria) type LS 6500 Multi-Purpose Scintillation Counter. Protein content of cells was determined for normalization.
Lipid staining
Nile Red staining and Bodipy staining were performed as described earlier (Greenspan et al.36 and Zhang et al.,37 respectively). Images were taken with a Nikon Eclipse TE 2000U; x-cite 120 Fluores Exfo (NIKON, Vienna, Austria).
Statistical analysis
Results are expressed as±S.D. Statistical significance was tested using unpaired t-test with Welch's correction. P<0.05 was considered significant (***P<0.0001, **P<0.005 and *P<0.05).
Biochemical caspase and 26S proteasome activity assays
Transversal muscle cross sections were homogenized and caspase activity determined using the CaspaseGLO 3/7 Kit from Promega (Madison, WI, USA). ATP-depended 26S proteasome activity was determined using homogenized transversal muscle cross sections in 5 mM ATP-containing buffer, essentially as described earlier.26, 38
Acknowledgments
We are grateful to Dr. Eckel, Division of Endocrinology, Metabolism and Diabetes, and Cardiology, University of Colorado, for providing C2C12 cells stably transduced with hLPL. We thank Michael D Huber from the Scripps Research Institute, La Jolla, CA for valuable discussions of the manuscript. This study was supported by the FWF (Austrian Science Foundation) SFB LIPOTOX grant no. F30 (to GH). TKP and SKD are supported by the doctoral program Molecular Medicine of the Medical University of Graz and WZ by the DK-Plus ‘Metabolic and Cardiovascular Disease' of the FWF grant no. W1226. PWV was supported by the FWF Erwin Schroedinger grant J 2922 and is currently supported by the AHA (American Heart Association) grant #11POST7590202.
KPT, HT, SKD and WZ planned and performed experiments and wrote parts of the manuscript. SS performed experiments. GH and PWV planned the experiments and wrote the manuscript.
Glossary
- C2-GEO
C2C12 cells stably transduced with a fusion gene of β-galactosidase and neomycin phosphotransferase (served as vector control)
- CTX
cardiotoxin
- EM
electron microscopy
- FBS
fetal bovine serum
- FFA
free fatty acid
- hLPL
human lipoprotein lipase
- LPL
lipoprotein lipase
- LPL-OV
C2C12 cells stably transduced with the human lipoprotein lipase gene
- M. gast.
Musculus gastrocnemius
- M. quad.
Musculus quadriceps
- M. sol.
Musculus soleus
- M. edl.
Musculus extensor digitorum longus
- MCK(m)-hLPL
human lipoprotein lipase minigene (hLPL) overexpressed under the control of the promoter from muscle creatin kinase (MCK) (the level of overexpression was medium, m)
- MyHC
myosin heavy chain
- MyoD
myogenic differentiation antigen 1
- NEFA
non-esterified fatty acid
- Pax7
paired box 7
- TAG
triacyl glycerol
- XSA
cross section area
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Unger RH, Clark GO, Scherer PE, Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim Biophys Acta. 2010;1801:209–214. doi: 10.1016/j.bbalip.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Pelleymounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, Boone T, et al. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Hammer RE, Ikemoto S, Brown MS, Goldstein JL. Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature. 1999;401:73–76. doi: 10.1038/43448. [DOI] [PubMed] [Google Scholar]
- Unger RH. Minireview: weapons of lean body mass destruction: the role of ectopic lipids in the metabolic syndrome. Endocrinology. 2003;144:5159–5165. doi: 10.1210/en.2003-0870. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hirose H, Ohneda M, Johnson JH, McGarry JD, Unger RH. Beta-cell lipotoxicity in the pathogenesis of non-insulin-dependent diabetes mellitus of obese rats: impairment in adipocyte-beta-cell relationships. Proc Natl Acad Sci USA. 1994;91:10878–10882. doi: 10.1073/pnas.91.23.10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herpen NA, Schrauwen-Hinderling VB. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol Behav. 2008;94:231–241. doi: 10.1016/j.physbeh.2007.11.049. [DOI] [PubMed] [Google Scholar]
- Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. J Mol Med. 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- Levak-Frank S, Radner H, Walsh A, Stollberger R, Knipping G, Hoefler G, et al. Muscle-specific overexpression of lipoprotein lipase causes a severe myopathy characterized by proliferation of mitochondria and peroxisomes in transgenic mice. J Clin Invest. 1995;96:976–986. doi: 10.1172/JCI118145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler W, Levak-Frank S, Radner H, Kostner GM, Zechner R. Muscle-specific overexpression of lipoprotein lipase in transgenic mice results in increased alpha-tocopherol levels in skeletal muscle. Biochem J. 1996;318 (Part 1:15–19. doi: 10.1042/bj3180015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefler G, Noehammer C, Levak-Frank S, el-Shabrawi Y, Schauer S, Zechner R, et al. Muscle-specific overexpression of human lipoprotein lipase in mice causes increased intracellular free fatty acids and induction of peroxisomal enzymes. Biochimie. 1997;79:163–168. doi: 10.1016/s0300-9084(97)81509-x. [DOI] [PubMed] [Google Scholar]
- Voshol PJ, Jong MC, Dahlmans VE, Kratky D, Levak-Frank S, Zechner R, et al. In muscle-specific lipoprotein lipase-overexpressing mice, muscle triglyceride content is increased without inhibition of insulin-stimulated whole-body and muscle-specific glucose uptake. Diabetes. 2001;50:2585–2590. doi: 10.2337/diabetes.50.11.2585. [DOI] [PubMed] [Google Scholar]
- Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20:1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- McNally EM, Pytel P. Muscle diseases: the muscular dystrophies. Annu Rev Pathol. 2007;2:87–109. doi: 10.1146/annurev.pathol.2.010506.091936. [DOI] [PubMed] [Google Scholar]
- Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Shimabukuro M, Higa M, Zhou YT, Wang MY, Newgard CB, Unger RH. Lipoapoptosis in beta-cells of obese prediabetic fa/fa rats. Role of serine palmitoyltransferase overexpression. J Biol Chem. 1998;273:32487–32490. doi: 10.1074/jbc.273.49.32487. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon KC. Cancer cachexia and fat-muscle physiology. N Engl J Med. 2011;365:565–567. doi: 10.1056/NEJMcibr1106880. [DOI] [PubMed] [Google Scholar]
- Tisdale MJ. Cachexia in cancer patients. Nat Rev Cancer. 2002;2:862–871. doi: 10.1038/nrc927. [DOI] [PubMed] [Google Scholar]
- Levak-Frank S, Weinstock PH, Hayek T, Verdery R, Hofmann W, Ramakrishnan R, et al. Induced mutant mice expressing lipoprotein lipase exclusively in muscle have subnormal triglycerides yet reduced high density lipoprotein cholesterol levels in plasma. J Biol Chem. 1997;272:17182–17190. doi: 10.1074/jbc.272.27.17182. [DOI] [PubMed] [Google Scholar]
- Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci USA. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray TV, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A, et al. A non-apoptotic role for caspase-9 in muscle differentiation. J Cell Sci. 2008;121 (Part 22:3786–3793. doi: 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- Jensen DR, Knaub LA, Konhilas JP, Leinwand LA, MacLean PS, Eckel RH. Increased thermoregulation in cold-exposed transgenic mice overexpressing lipoprotein lipase in skeletal muscle: an avian phenotype. J Lipid Res. 2008;49:870–879. doi: 10.1194/jlr.M700519-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tachibana H, Morinaga Y, Fujimura Y, Yamada K. Modulation of proliferation and differentiation of C2C12 skeletal muscle cells by fatty acids. Life Sci. 2009;84:415–420. doi: 10.1016/j.lfs.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Das SK, Eder S, Schauer S, Diwoky C, Temmel H, Guertl B, et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science. 2011;333:233–238. doi: 10.1126/science.1198973. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Venable JH, Coggeshall R. A simplified lead citrate stain for use in electron microscopy. J Cell Biol. 1965;25:407–408. doi: 10.1083/jcb.25.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara A, Radin NS. Lipid extraction of tissues with a low-toxicity solvent. Anal Biochem. 1978;90:420–426. doi: 10.1016/0003-2697(78)90046-5. [DOI] [PubMed] [Google Scholar]
- d'Albis A, Couteaux R, Janmot C, Roulet A, Mira JC. Regeneration after cardiotoxin injury of innervated and denervated slow and fast muscles of mammals. Myosin isoform analysis. Eur J Biochem. 1988;174:103–110. doi: 10.1111/j.1432-1033.1988.tb14068.x. [DOI] [PubMed] [Google Scholar]
- Horsley V, Pavlath GK. Forming a multinucleated cell: molecules that regulate myoblast fusion. Cells Tissues Organs. 2004;176:67–78. doi: 10.1159/000075028. [DOI] [PubMed] [Google Scholar]
- Staber PB, Vesely P, Haq N, Ott RG, Funato K, Bambach I, et al. The oncoprotein NPM-ALK of anaplastic large-cell lymphoma induces JUNB transcription via ERK1/2 and JunB translation via mTOR signaling. Blood. 2007;110:3374–3383. doi: 10.1182/blood-2007-02-071258. [DOI] [PubMed] [Google Scholar]
- Poirier P, Marcell T, Huey PU, Schlaepfer IR, Owens GC, Jensen DR, et al. Increased intracellular triglyceride in C(2)C(12) muscle cells transfected with human lipoprotein lipase. Biochem Biophys Res Commun. 2000;270:997–1001. doi: 10.1006/bbrc.2000.2528. [DOI] [PubMed] [Google Scholar]
- Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, et al. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- Nilsson-Ehle P. Human lipoprotein lipase activity: comparison of assay methods. Clin Chim Acta. 1974;54:283–291. doi: 10.1016/0009-8981(74)90247-2. [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SO, Trimble R, Guo F, Mak HY. Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol. 2010;11:96. doi: 10.1186/1471-2121-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky IG, Suzara D, Furman M, Goldspink P, Ford GC, Nair KS, et al. Proteasome production in human muscle during nutritional inhibition of myofibrillar protein degradation. Metabolism. 2004;53:340–347. doi: 10.1016/j.metabol.2003.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.