Abstract
The molecular pathology of thymic epithelial tumors (TETs) is largely unknown. Using array comparative genomic hybridization (CGH), we evaluated 59 TETs and identified recurrent patterns of copy number (CN) aberrations in different histotypes. GISTIC algorithm revealed the presence of 126 significant peaks of CN aberration, which included 13 cancer-related genes. Among these peaks, CN gain of BCL2 and CN loss of CDKN2A/B were the only genes in the respective regions of CN aberration and were associated with poor outcome. TET cell lines were sensitive to siRNA knockdown of the anti-apoptotic molecules BCL2 and MCL1. Gx15-070, a pan-BCL2 inhibitor, induced autophagy-dependent necroptosis in TET cells via a mechanism involving mTOR pathways, and inhibited TET xenograft growth. ABT263, an inhibitor of BCL2/BCL-XL/BCL-W, reduced proliferation in TET cells when administered in combination with sorafenib, a tyrosine kinase inhibitor able to downregulate MCL1. Immunohistochemistry on 132 TETs demonstrated that CN loss of CDKN2A correlated with lack of expression of its related protein p16INK4 and identified tumors with poor prognosis. The molecular markers BCL2 and CDKN2A may be of potential value in diagnosis and prognosis of TETs. Our study provides the first preclinical evidence that deregulated anti-apoptotic BCL2 family proteins may represent suitable targets for TET treatment.
Keywords: comparative genomic hybridization, thymic epithelial tumors, BCL2 anti-apoptotic family members, CDKN2A, target therapy
Thymic epithelial tumors (TETs) are rare with an estimated incidence of 0.32/100 000 persons/year.1 According to the 2004 WHO classification, there are two major categories of TETs: thymomas and thymic carcinomas (TCs). Thymomas are further classified into A, AB, B1, B2, and B3 subtypes according to cancer cell features, degree of atypia and number of intratumoral thymocytes.2 Histology of TCs is similar to carcinomas originating in organs other than the thymus and displays the characteristics of aggressive epithelial tumors.2 Surgery represents the mainstay of TET treatment, and survival is strongly influenced by the stage of the disease and the completeness of tumor resection.2 Although chemotherapy is able to induce substantial tumor shrinkage of variable duration in metastatic and non-resectable TETs, it is not curative in patients with metastatic disease. Exploration of molecularly targeted drugs, which have become available for treatment of several cancer types, has so far been limited by the lack of understanding of the molecular changes of TETs.
Results
Correlation between chromosome arm-level aberrations and TET histotypes
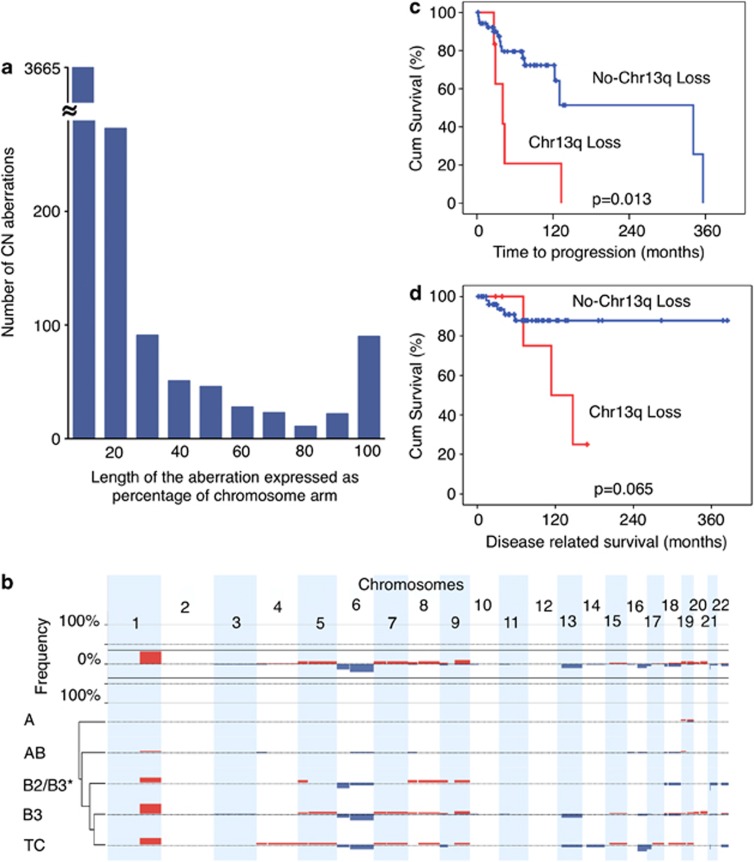
A total of 59 tumors rich in cancer cells (>80%) were selected for array comparative genomic hybridization (CGH) evaluation from 132 formalin-fixed paraffin-embedded (FFPE) TET samples (Table 1). CGH reveled a total of 4300 copy number (CN) aberrations, ranging from large events involving an entire chromosome arm to only few kilobases. The length of each CN aberration was expressed as the percentage of a relative chromosome arm, and CN aberrations were divided into 10 groups according to the proportion of the chromosome affected (Figure 1a). Small CN aberrations (0–10% of a chromosome arm length) were the most common (Figure 1a; n=3665). The number of CN aberrations progressively decreased in inverse proportion to their length up to 80% of their respective chromosome arms, and then rose again reaching a second peak at 90–100%. The number of expected 90–100% arm-level aberrations was estimated interpolating the values up to 80% by the formula y=6.5636x−2.546 and it was significantly lower than that observed (90 instead of 7; χ2 P<0.001). On the basis of this observation, we defined arm-level CN aberrations those with an extension longer than 80% of the respective chromosome arm. These data suggest that small (focal) and long arm-level aberrations might arise through different mechanisms during cancer development.
Table 1. Patient characteristics.
| Total (132) | CGH (59) | |
|---|---|---|
| Median age (range), years | 55 (20–86) | 57 (20–86) |
| Sex | ||
| Male: Female | 67: 65 | 28: 31 |
| Tumor sample | ||
| Primary tumor | 108 (82%) | 46 (78%) |
| Relapsed | 24 (18%) | 13 (22%) |
| Stage | ||
| I | 35/114 (31%) | 15/49 (31%) |
| II A | 26/114 (23%) | 9/49 (18%) |
| II B | 17/114 (15%) | 5/49 (10%) |
| III A | 16/114 (14%) | 7/49 (14%) |
| III B | 3/114 (3%) | 2/49 (4%) |
| IV A | 5/114 (4%) | 3/49 (6%) |
| IV B | 12/114 (11%) | 8/49 (16%) |
| Na* | 18 | 10 |
| Completeness of resection | ||
| R0 | 79/112 (71%) | 33/47 (70%) |
| R1 | 23/112 (21%) | 7/47 (14%) |
| R2 | 10/112 (9%) | 7/47 (14%) |
| Na* | 20 | 12 |
| WHO histotype | ||
| A | 15/132 (11%) | 12/59 (20%) |
| AB | 28/132 (21%) | 14/59 (24%) |
| B1 | 24/132 (18%) | 0 |
| B1/B2 | 6/132 (5%) | 0 |
| B2 | 8/132 (6%) | 1/59 (2%) |
| B2,B3 | 11/132 (8%) | 5/59 (8%) |
| B3 | 24/132 (18%) | 20/59 (34%) |
| C | 14/132 (11%) | 7/59 (12%) |
| Other# | 2/132 (2%) | 0 |
| Paraneoplastic syndromes | ||
| MG | 32/125 (26%) | 17/58 (29%) |
| Other+ | 2/125 (2%) | 1/58 (2%) |
| 10-Year DRS | 87% | 83% |
| 10-Year TTP | 69% | 65% |
Abbreviations: CGH, comparative genomic hybridization; DRS, disease-related survival; MG, myasthenia gravis; Na*, patients for whom data were not available at diagnosis; other#, one micronodular and one cystic thymoma; other+, one autoimmune glomerulonephritis (in the CGH analysis) and one autoimmune encephalopathy; TTP, Time to progression; WHO, World Health Organization
Figure 1.
Arm-level CN aberrations in TETs identified by CGH. (a) Arm-level CN aberrations were divided into 10 categories according to their length expressed as the percentage of the relative chromosome arm affect by the aberration. The number of CN aberrations covering <10% of the chromosome arm length were the most abundant. The power function y=6.5636x−2.546 summarizes the distribution of CN aberrations based on chromosome arm length from 0 to 80%, and based on this equation the expected number of 90–100% arm-length CN aberrations was 7 instead of 90 observed (χ2, P<0.001). (b) Arm-level CN gains (red) and losses (blue) of autosomal chromosomes. Arm-level CN aberrations were defined by the sum of all the CN gain or all the CN loss affecting one chromosome arm in each tumor. If this sum was >80% of the chromosome arm length it was considered an arm-level CN aberration. The top panel depicts the overall summary of frequency of arm-level CN aberrations. The arm-level CN aberrations grouped by histotype (A, AB, B2+B2/B3, B3 and TCs) are summarized in the bottom panel. * indicates B2 and B2/B3. Thymic carcinomas and B3 thymomas exhibit similar patterns of arm-level CN aberrations. Only few arm-level CN aberrations were observed in type A thymomas. (c) Time to progression and (d) disease-related survival curves in relation to 13q CN loss
Figure 1b depicts the frequency of arm-level CN aberrations and their distribution according to TET histotypes (see also Supplementary Table S1). Hierarchical cluster analysis of frequency of CN aberrations observed in different histotypes indicated a close relatedness between B3 and TCs. B2+B2/B3, AB and A types were incrementally dissimilar to TCs (Figure 1b). Type A thymoma showed occasional chromosome arm-level CN aberrations (Supplementary Table S1). On the contrary, types B3 and TCs shared frequent arm-level CN gains of 1q, as well as losses of chromosomes 6 and 13q. In addition, TCs presented frequent losses of 16p and 17q. Although the potential continuum with A subtypes is impossible to evaluate due to excess of non-neoplastic thymocytes in B1 and B2 histotypes, which were not evaluated in our study, our results support the presence of a continuum of genomic aberrations between B2/B3, B3 and TC histotypes (Figure 1b).
Chromosome arm-level CN loss of 13q occurs in more aggressive TETs
In univariate analyses, loss of 13q was associated with a poorer time to progression (TTP; log-rank test, P=0.013; Figure 1c) and disease-related survival (DRS; P=0.065; Figure 1d). Arm-level CN loss of 13q was found only in more aggressive histotypes (B3 and TCs; P=0.005). Conventional factors (sex, age, stage or extent of resection) were not associated with DRS in the 59 patients. Also, as there was no death in patients in a lower risk WHO category (A/AB/B1), it was not possible to construct a Cox model including the markers relative to conventional factors for this outcome in this subset of patients, because it would have an infinite hazard ratio (HR) and be unusable in a model. As such, it was not possible to show whether CN loss of 13q is an independent prognostic factor for DRS. CN loss of 13q was not an independent prognostic factor (P=0.17; HR: 2.11; 95% CI on HR: 0.73–6.07) after adjustment for WHO category (P=0.029; HR: 9.72; 95% CI on HR: 1.26–75.20 in a Cox multivariable analysis for TTP (Supplementary Table S2). Deletions of the whole or parts of chromosome 13 have been shown to have survival implications in other tumors, such as multiple myeloma (whole chromosome 13)3 and breast cancer (13q12-13).4
Identification of significant CN aberrations by GISTIC
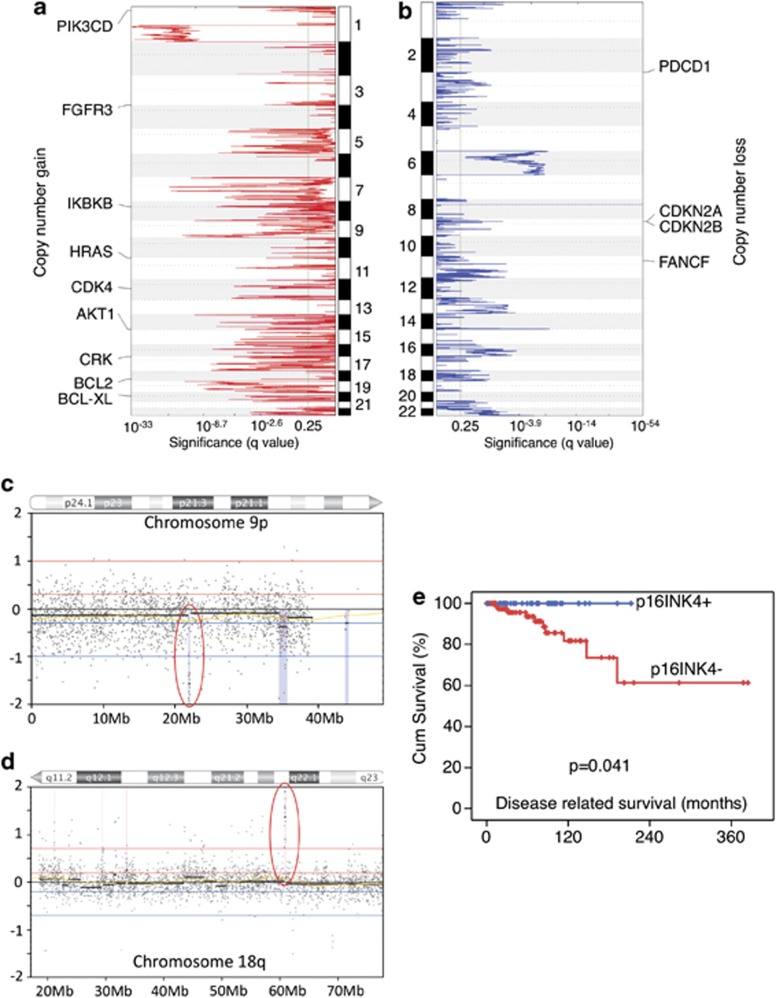
To identify the potential importance of CN aberrations in the biology of TETs, we applied the GISTIC algorithm5 to TET CGH data and identified 72 peaks of CN gain (817 genes+23 miRNAs) and 54 of CN loss (155 genes+3 miRNAs) (Figures 2a and b, Supplementary Table S3). Of the genes mapped into GISTIC peaks, known cancer-related genes were selected and depicted in Figures 2a and b based on the following criteria: (1) in focal GISTIC peaks (<5 Mb), (2) recurrent (in at least two tumors) and (3) <15 genes in a peak. CN aberrations of CDK4, CDKN2A/B and IKBKB have been reported previously in other tumors.6 Other known cancer-related genes identified in our analysis include HRAS and AKT/mTOR pathway signal transduction genes (PIK3CD and ATK1), or belong to FGF receptor family (FGFR3). Interestingly many genes regulating programmed cell death were also found in GISTIC peaks (BCL2, BCL-XL, PDCD1, CRK), suggesting an important role of apoptosis in TET pathogenesis. Moreover, CN loss of a DNA damage repair gene FANCF suggests a link with BRCA/ATM pathway and uncontrolled cell cycle progression.7
Figure 2.
Identification of significant CN aberration peaks with survival implications. (a) Peaks of CN gain and (b) CN loss identified by GISTIC algorithm. GISTIC q-values (x axis) are plotted across the genome (y axis). q-value of<0.25 was considered significant. Thirteen cancer-related genes were identified in the GISTIC peaks. (c) Representative samples with focal CN loss of CDKN2A/B locus and (d) focal CN gain of BCL2 locus on chromosomes 9p and 18q, respectively. x axis indicates genome map position, and y axis the log2 ratio of red and green signals from the array. Blue dots represent the array probes. Red circles indicate CDKN2A/B and BCL2 loci. (e) Disease-related survival in relation to CDKN2A expression evaluated by immunohistochemistry
Among these peaks, the CN gain of BCL2 and the CN loss of CDKN2A and CDKN2B loci were selected for further characterization. The role of BCL2 has been well characterized in cancer and an amplification of this gene may result in a block of apoptosis with consequent accumulation of tumor cells. The deletion of CDKN2A and CDKN2B, two known tumor suppressor genes acting on the control of cell cycle, may be related to uncontrolled tumor cell proliferation. The CN gain peak of 18q21.33 affected only the BCL2 gene locus, and the amplitude of the gain suggested the presence of several copies of the gene (Figure 2c). The CN loss peak of 9p21.3 included only CDKN2A/B loci and was found in four tumors. The amplitude of the deletion suggested the presence of a homozygous CDKN2A/B deletion (Figure 2d). Moreover, BCL2 CN gains or CDKN2A/B CN losses were both associated with poorer prognosis (DRS and TTP log-rank test, P<0.05). In the multivariate analysis for TTP, there was a trend toward a significant gain by adding BCL2 CN (P=0.069; HR 2.92; 95% CI on HR: 0.92–9.27) to WHO category, the only significant independent prognostic factor (details in Supplementary Results).
CDKN2A CN loss correlates with low p16INK4 expression and poor prognosis
Tumors carrying homozygous 9p21.3 CN loss (two B3 thymomas and two TCs) (Figure 2c) had a significantly worse DRS (log-rank test, P=0.021) and TTP (log-rank test, P=0.019) in univariate analysis but not in the TTP multivariate model (details in Supplementary Methods). Focal deletion of 9p21.3 is a frequent event in cancer (40% overall and 16% focal CN loss),6 and poor outcome for patients carrying 9p21.3 deletion has also been described in lymphoblastic leukemia.8 This region contains two known tumor suppressor genes, CDKN2A and CDKN2B. CDKN2A encodes p16INK4 and p14ARF by alternative splicing. We confirmed the CN loss of CDKN2A identified by CGH using CN-PCR analysis in all the four tumors evaluated but not in five TETs without CDKN2A deletion assessed by CGH (Fisher exact test, P=0.008).
We evaluated p16INK4 expression in TET samples using a tissue microarray containing 132 TET tumors, which include those evaluated by CGH. None of the five tumors carrying 9p21.3 CN loss expressed p16INK4 (four with CN loss log2 ratio less than −0.7 and one with CN loss −0.3<x<−0.7). Out of 119 evaluable cases, 34 were positive for p16INK4 (Supplementary Table S4), which was associated with better DRS (Figure 2e; log-rank test, P=0.041) but not with TTP. As no events were observed for DRS in p16-positive tumors, the multivariate model could not be built. Interestingly, 61% of the primary TETs (34/56) and the three TET cell lines showed no detectable p16INK4 expression in the absence of CDKN2A CN loss (Supplementary Figure S1), indicating that negative p16INK4 expression was not exclusively due to CDKN2A CN loss. The loss of p16INK4 expression has been shown to be possibly related to p16INK4 promoter methylation9, 10 or miR-24 deregulation.11
Deregulation of BCL2 family genes in TETs
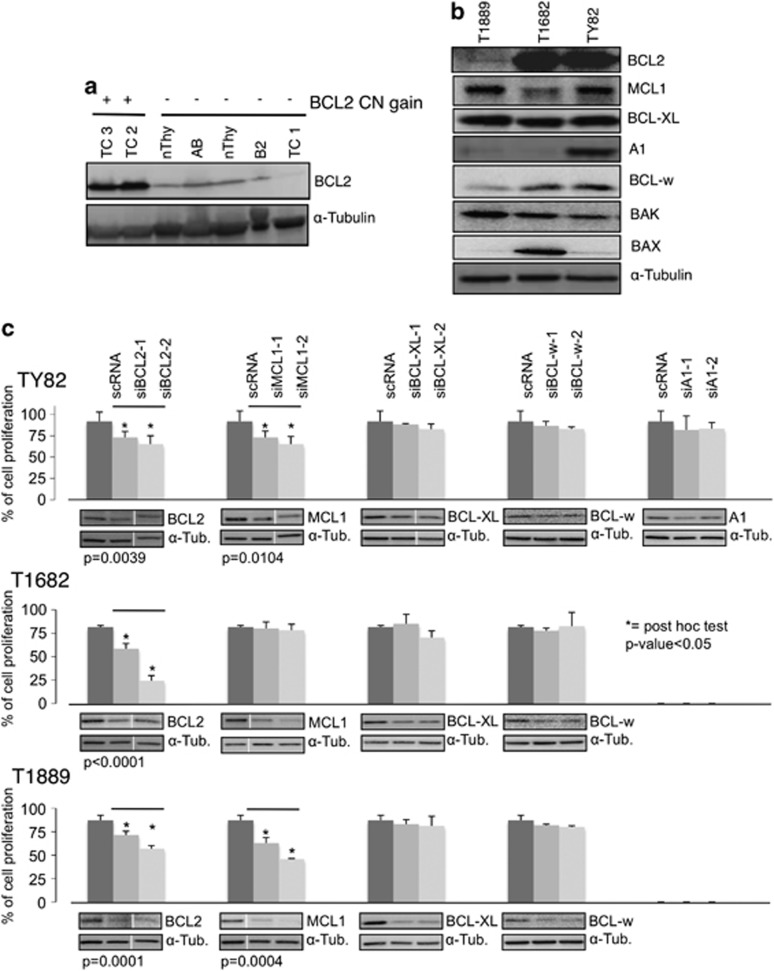
BCL2 locus presented CN gain in 10% (6 out of 59) of the TET samples, including one type A, two B3 thymomas and three TCs. Moreover, focal BCL2 amplification (Figure 2d) was also confirmed in five TCs of an independent series of 12 frozen TETs (42%). For two TCs, which showed BCL2 CN gain, there was enough material to test BCL2 expression by western blot, which demonstrated an increased expression of BCL2 protein in comparison with normal thymus, thymomas (AB and B2) and a TC without BCL2 CN gain (Figure 3a). Previous study showed that BCL2 is expressed in about 60% of type A and the A component of type AB thymomas. More than 90% of TCs express BCL2, whereas only few type B thymomas are positive for BCL2.12, 13, 14, 15 Consistent with previous reports that MCL1 and BCL2 were frequently coexpressed in TCs16 and MCL1 CN gain is a frequent event in several cancers,6 we observed MCL1 CN gain in 51% of all TET cases, and higher in B3 (70%) and TCs (57%). However, this CN gain was mainly the result of the whole 1q gain rather than focal MCL1 CN amplification. Moreover, BCL-XL locus was identified in a significant peak of CN gain by GISTIC analysis (Supplementary Table S3).
Figure 3.
Deregulation of BCL2 family genes in TETs. (a) BCL2 protein is more expressed in thymic carcinomas with BCL2 CN gain (TC2 and TC3) than in samples without CN gain. TC1, TC2, TC3: thymic carcinomas; B2 and AB: thymomas; nThy: normal thymus. (b) Expression of BCL2 anti-apoptotic and pro-apoptotic molecules in three TET cell lines by western blot analysis. (c) siRNA knockdown of anti-apoptotic BCL2 family proteins in the three TET cell lines. Cell proliferation was measured using MTS assay 72 h after siRNA transfection. Data were normalized to those of untransfected cells. BCL2 and MCL1 siRNAs induced a significant reduction of cell proliferation in all three TET cell lines, and in T1889 and TY82, respectively (*P<0.05). Western blots showed siRNA knockdown in each experiment. Note longer exposure revealed that BCL2 expression was detectable in T1889 cells (b and c)
CGH evaluation of TET cell lines revealed only MCL1 CN gain and BCL2 CN loss in T1889 (Supplementary Figures S1C–F). BCL2 expression was higher in TY82 and T1682 than in T1889 (Figure 3b). Although all TET cell lines expressed BCL-XL, MCL1, BCL-w, A1 expression was prominent only in Ty82 cells (Figure 3b). To figure out which of these anti-apoptotic proteins was necessary for TET proliferation, siRNA knockdowns were performed for each anti-apoptotic molecule (BCL2, MCL1, BCL-XL, BCL-W and A1) in all cell lines (Figure 3c). BCL2 knockdown induced a significant reduction of cell proliferation in all three TET cell lines whereas MCL1 knockdown affected exclusively proliferation of T1889 and Ty82. These results suggest that these cells may be addicted to BCL2 and MCL1 for growth.
TET cell lines are resistant to ABT263 but sensitive to ABT263/sorafenib combination treatment
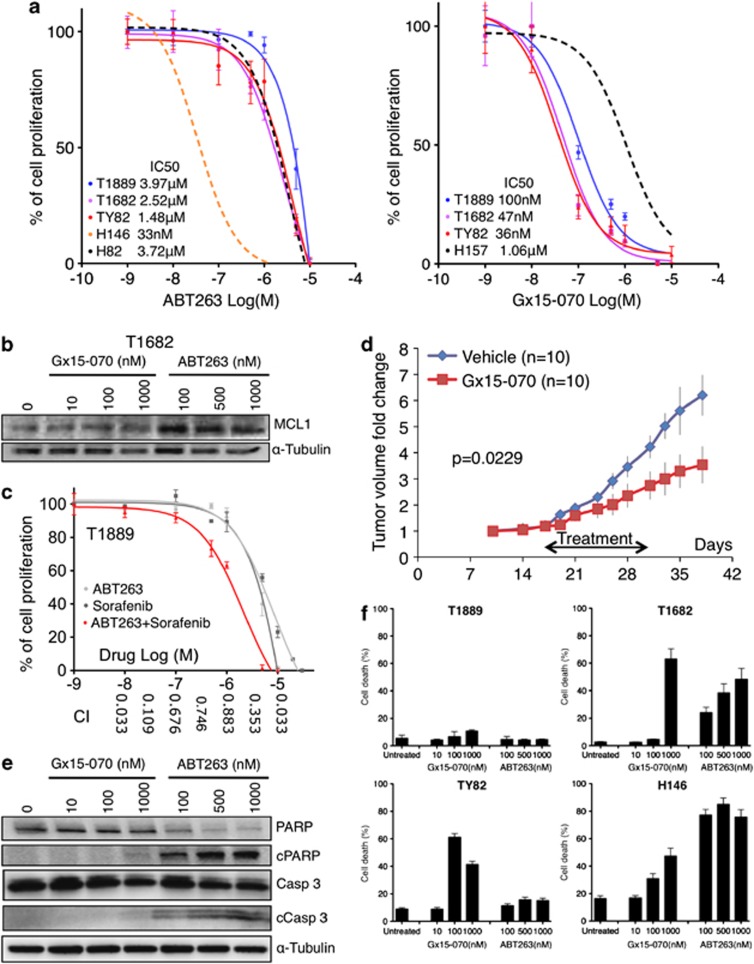
All TET cell lines were relatively resistant to ABT263, a small-molecule BH3 mimetic that inhibits the anti-apoptotic proteins BCL2, BCL-XL and BCL-W17 (Figure 4a, left panel), similar to H82, a small cell lung cancer cell line that does not express BCL2.17 Resistance to ABT263 treatment has been associated to MCL1 expression in multiple cell line models.17 Although T1682 expressed MCL1 at relatively low level, T1682 cells were highly resistant to ABT263 (IC50=2.52 μM) as compared with H146 small cell lung cancer cells (IC50=33 nM) (Figure 4a), which may be attributed to the MCL1 overexpression induced by ABT263 in T1682 cells (Figure 4b).
Figure 4.
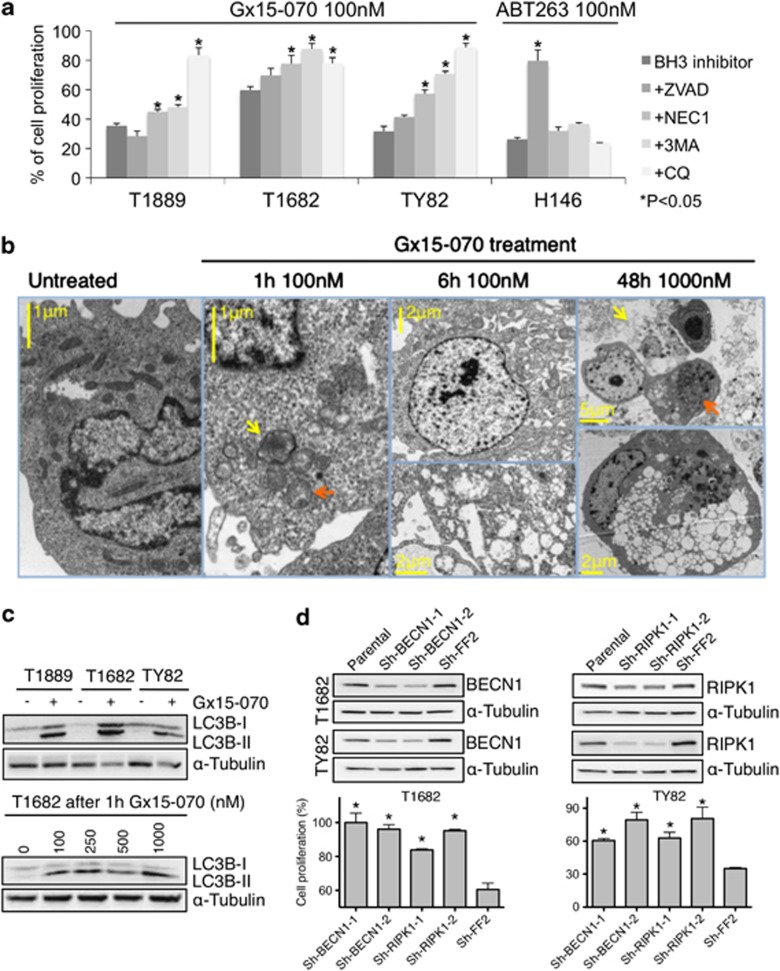
Effect of two BCL2 anti-apopotic family inhibitors, ABT263 and Gx15-070, in TET cell lines. (a) MTS assay of ABT263 and Gx15-070 in TET and control cell lines. H146, H82 small cell lung cancer cell lines were used as references in ABT263 and H157 non-small cell lung cancer cell line in Gx15-070 experiments.27 Experiments were repeated four times. (b) In T1682 cells, ABT263 treatment increases MCL1 level compared with untreated cells or Gx15-070-treated cells. (c) T1889 was more sensitive to ABT263 and Sorafenib combination than to each single drug treatment as shown by MTS assay. Combination index (CI) values, reported on x axis, were calculated using Calcusync and demonstrated a synergistic effect for the drug combination: values <1 denotes synergism. Results for T1889 and TY82 are summarized in Supplementary Table S5. Experiments repeated three times. (d) Gx15-070 suppressed the growth of TY82 xenografts. Average fold change of tumor volume of 10 mice treated by Gx15-070 and 10 by vehicle alone are reported. The black arrow indicates the timing of treatment. (e) Cleavage of PARP and caspase-3 was assessed by western blot after 48-h treatment with various concentrations of Gx15-070 and ABT263 in T1682 cells. (f) Cell death induced by Gx15-070 and ABT263 was expressed as percentage of dead cells over total number of cells (cell death %). Cells were treated with the indicated concentrations of Gx15-070 and ABT263 for 48 h, stained with TOPRO3 and followed by flow cytometry analysis. Note that the highest effect of Gx15-070 in Ty82 cells was detected at 100 nM and was less pronounced at 1000 nM. This could be due to the off-target effect of Gx15-070 at high concentration that antagonized on-target effect of the drug (experiments were repeated two times)
Sorafenib, a tyrosine kinase inhibitor, known to attenuate MCL1 expression18 (Supplementary Figure S2A), was used in ABT263 combination studies. Combination of sorafenib and ABT263 resulted in an impressive synergistic effect for most concentrations tested (Figure 4c; Supplementary Table S5). These data reinforce the evidence that TET cell lines are addicted to MCL1 and BCL2 and suggest that these proteins can be targets for therapy.
Gx15-070 inhibits TET cells growth through autophagy-dependent necroptosis
Obatoclax (Gx15-070), a small-molecule BH3 mimetic, bona fide inhibitor of all the anti-apoptotic BCL2 family proteins, including MCL1,19 reduced the proliferation of TET cell lines with an IC50 ranging from 36–100 nM, (Figure 4a, right panel). Tumor xenografts in mice treated with Gx15-070 grew significantly slower than in those treated with placebo (P=0.0229; Figure 4d).
Interestingly, Gx15-070 treatment did not enhance caspase-3 activity in TET cell lines, whereas induction of caspase-3 activity was clearly observed in ABT263-treated BCL2-positive TET cells (Supplementary Figure S2C). Cleavage of PARP and caspase-3 was predominantly observed in T1682 cells treated with ABT263 but not with Gx15-070 (Figure 4e), which was in sharp contrast with the finding that 60% of T1682 cells underwent cell death at 1000 nM Gx15-070, whereas only 25% of the cells were dead at 100 nM ABT263 (Figure 4f). T1689 cells were resistant to ABT263 as determined by MTS assay (Figure 4a), which measures the proliferation rate based on both the cell growth and survival. However, TOPRO3 viability assay demonstrated that ABT263 induced cell death in T1689 cells (Figure 4f). Moreover, the pan-caspase inhibitor ZVAD-FMK was able to restore cell proliferation of ABT263 – but not of Gx15-070-treated TET cell lines (Figure 5a). Together these findings demonstrate that Gx15-070 and ABT263 induce caspase-3-independent and -dependent cell death, respectively. As Gx15-070 only elicited minimal cell death in T1889 cells (Figure 4f), we examined the effect of Gx15-070 on cell proliferation. Though no sign of cell cycle arrest was found (data not shown), a dramatic increase of cell doubling time was observed in all Gx15-070-treated TET cells (Supplementary Figures S3A–C). These data suggest that Gx15-070 treatment of TET cells leads to both growth inhibition and cell death. As recent studies suggested that Gx15-070 induces autophagy-dependent necroptosis in acute lymphoblastic leukemia cells,20 we performed electron microscopy (EM) in TET cells, which demonstrated changes in mitochondrial structure as early signs of autophagy after 1 h Gx15-070 treatment (Figure 5b). After 6 h treatment, cytoplasmic vacuolization and mitochondrial swelling with early signs of necroptotic cell death were evident. Intriguingly, cytoplasmic vacuolization, late-stage necroptotic or autophagic cell death were the main features 48 h after Gx15-070 (1 μM) treatment. EM showed no increase of apoptotic cell death in Gx15-070-treated cells, being the apoptotic cells less than 1% in untreated and treated cells; consistent with the observation that caspase-3 was not involved in Gx15-070-induced cell death (Figures 4E and 5A; Supplementary Figure S2C). Out of 100 cells evaluated by EM for each condition, necroptotic cells were only 1% of untreated T1682 cells but increased to 9.5% after 1 h and to 12.8% after 6 h treatment with 250 nM Gx15-070. After 48 h, 77% of T1682 cells treated with 100 nM Gx15-070 were necroptotic. As shown in Figure 5c, accumulation of the autophagy marker LC3B-II was detected in all Gx15-070-treated TET cell lines and occurred as early as 1 h after Gx15-070 treatment, suggesting that Gx15-070 activates the autophagy pathway in TET cells. To confirm that Gx15-070 induces autophagy-dependent necroptotic cell death, we evaluated the effect of necrostatin 1 (Nec1; necroptosis inhibitor), 3MA (autophagosome formation inhibitor) and chloroquine (CQ; autophagosome maturation inhibitor) in Gx15-070-treated TET cells, and demonstrated that Gx15-070-induced growth inhibition (Figure 5a) and cell death (Supplementary Figure S4) could be effectively rescued by necroptosis and autophagy inhibition. Depletion of Beclin 1 (BECN1), a key regulator of autophagy pathway, and RIPK1, a key regulator of necroptotic pathway, (Figure 5d) recapitulated the results obtained with Nec1 and CQ (Figure 5a). In line with previous reports, our results suggest that Gx15-070-induced necroptosis is autophagy-dependent.20
Figure 5.
Gx15-070 induces autophagy and necroptosis in TET cells. (a) Inhibitors of pan-caspase (50 μM ZVAD-FMK), necroptosis (30 μM NEC1), autophagosome formation (5 μM 3MA) and autophagosome maturation (25 μM CQ) rescued Gx15-070-induced growth inhibition. Percentage of reduction in cell growth induced by 100 nM Gx15-070 was calculated from the MTS data in the presence or absence of the indicated inhibitors for the three TET cell lines and for the small cell lung cancer cell line H146 as control. (*one-way ANOVA post hoc P<0.05; experiments were repeated three times). (b) Time-course electron microscopic morphology of T1682 cells treated with Gx15-070 at the indicated concentrations. After 1-h treatment, lamellar bodies (yellow arrow) consistent with early signs of autophagy and mitochondrial structural changes with loss of cristae (orange arrow) were evident. After 6-h treatment, early signs of necroptosis with intact nuclear envelope (top panel), and mitochondrial swelling with loss of mitochondrial matrix and cytoplasm vacuolization were observed (bottom panel). After 48-h treatment, late stages of necroptotic cell death (yellow arrow) and advanced stages of autophagy (orange arrow) were evident. The bottom panel depicts advanced stage of autophagic cell death with cytoplasmic vacuolization and lamellar bound structures consistent with autophagosomes. (c) Gx15-070 (1 μM) induced LC3BII accumulation after 48-h treatments in TET cells (top blot). LC3BII accumulation was observed in T1682 cells 1-h after Gx15-070 treatment at a concentration as low as 100 nM (bottom blot). (d) BECN1 and RIPK1 knockdown rescues Gx15-070-induced growth inhibition. Western blot shows shRNA knockdown of Beclin1 (sh-BECN1-1 and sh-BECN1-2) and RIPK1 (sh-RIPK1-1 and sh-RIPK1-2) in T1682 and TY82 cell lines. Sh-FF2: shRNA control vector carrying anti-exogenous luciferase shRNA FF2 (top). MTS assay of BECN1 and RIPK1 knockdown cells treated with 250 nM Gx15-070 (bottom). *denotes statistically significant differences compared with control (P<0.05); experiments were repeated three times
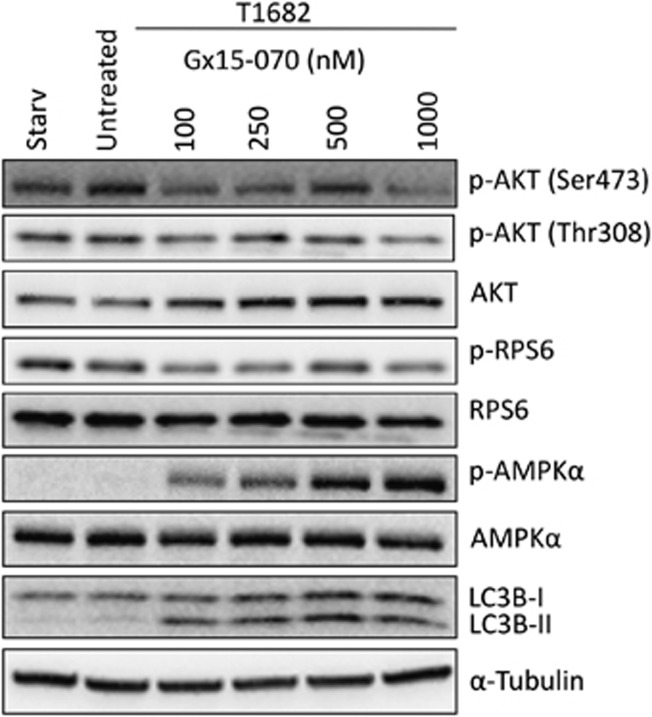
Gx15-070 also induced a progressive reduction of AKT Ser473 and Thr308 phosphorylation and a reduction of p-RPS6 phosphorylation, suggesting inhibition of mTOR pathway, in line with previous reports 21 (Figure 6). Moreover, we noticed that Gx15-070 induced phosphorylation of AMPKα, a Ser/Thr kinase normally activated when ATP level is reduced.22 AMPK phosphorylation and LC3B cleavage appeared within 1 h after Gx15-070 treatment (Figure 6). Together, these data suggest that Gx15-070 may trigger autophagy either through direct inhibition of the mTOR pathway21 or activation of AMPK via mitochondria damage as revealed by our EM study.
Figure 6.
Effect of Gx15-070 of mTOR pathway and on AMPKα. Effect of 6h Gx15-070 treatment on T1682 cells compared with untreated cells and cells starved for 6 h. The treatment induced a progressive reduction of p-AKT (Ser473 and Thr308) and p-RPS6 with the exception of 500 nM concentration. Progressive increase of LC3B-II and AMPKα phosphorylation was observed in Gx15-070 concentration-dependent manner
Discussion
We studied the genomic CN aberrations of TETs to identify potential prognostic factors and targets suitable for therapy. High-resolution array-CGH allowed to identify CN aberrations that cosegregate with WHO histotypes. A total of 126 significant CN aberrations including focal/recurrent CN aberrations of 13 cancer-related genes were found by GISTIC algorithm analysis. Combining GISTIC analysis and patients' survival data, we selected BCL2 CN gain and CDKN2A/B CN loss for further studies, because they were associated with a poor DRS and TTP, and because they are well-known cancer genes in other tumors. Moreover, loss of p16INK4 expression was associated with worse prognosis. Using RNAi and small molecule inhibitor approaches, we showed that TET cells underwent autophagy-dependent necroptosis after Gx15-070 treatment and that ABT263-sorafenib combination inhibited TET cell growth, providing the first preclinical evidence that deregulated BCL2 family genes may serve as suitable targets for TET therapies.
Large arm-level aberrations identified in our study are in general agreement with genetic alterations reported by conventional CGH.23, 24, 25 Only few CN aberrations and LOHs have been reported in type A thymomas, which have a very indolent behavior and excellent survival.25 The B2/B3 histotypes showed frequent arm-level CN gain of 1q and loss of chromosome 6, together with other CN aberrations that are present in less than 20% of the cases. B3 thymomas and TCs displayed a pattern of CN aberrations similar to B2/B3 tumors except for the losses of 13q, 16q and 17p, which were not observed in B2/B3 tumors. Consistent with previous reports,23, 26 hierarchical cluster analysis of arm-level CN aberrations revealed that type A thymomas were very distantly related to aggressive tumors (B3 and C), suggesting that arm-level CN aberrations determine the aggressiveness of the disease. In addition, loss of 13q, which has been previously described in B3 and TCs by conventional CGH,24 correlated with worse prognosis. RB1, a well-characterized tumor suppressor gene, has been mapped to 13q. We found only one TET with concomitant loss of 13q and CDKN2A locus. The latter encodes p16INK4 and p14ARF via alterative splicing. As p16INK4, an inhibitor of CDK4-cyclin D, acts upstream of RB, inactivation of either gene will be in theory sufficient to compromise the RB pathway. The near mutual exclusive loss of 13q and CDKN2A, which together represented 15% (9/59) of TETs in our series, and the tight association of poor DRS with either loss of 13q CN (Figures 1c and d) or p16INK4 expression (Figure 2e) suggest that the RB pathway has an important role in TET pathogenesis.
Using GISTIC algorithm we identified several cancer related genes involved in cell cycle control (CDKN2A/B and CDK4), or intracellular signal transduction (AKT1, PIK3CD and HRAS). Interestingly, many of these genes (BCL2, BCL-XL, PDCD1, CRK) were related to apoptosis control, and BCL2 CN gain was associated with poorer TTP and DRS. CN gain of MCL1, another member of the BCL2 family gene, was found in 51% of TETs and mainly in aggressive histotypes (83% B2/B3, 70% B3 and 57%TCs). However, MCL1 CN gain was mostly the result of the whole 1q gain rather than focal MCL1 amplification.
The oncogenic role of BCL2 is well-known in follicular lymphomas in which constitutive BCL2 activation due to IgH-BLC2 juxtaposition is thought to result in lymphomagenesis by interfering with the normal apoptosis of B lymphocytes.27 BCL2 overexpression was observed in other types of lympho-proliferative disorders and small cell lung cancer.17 In addition to their involvement in regulation of apoptosis, BCL2 family members are also involved in the regulation of autophagy and necroptosis pathways. We validated the functional significance of BCL2 family genes in TET cells via siRNA approaches, and demonstrated that all TET cell lines (T1889, TY82 and T1682) were addicted to BCL2, whereas T1889 and TY82 required also MCL1 for growth and survival. More importantly, we showed that sorafenib (a multi-tyrosine kinase inhibitor), as shown in other models,18 could reduce MCL1 expression induced by ABT263 in all the TET cell lines tested, and that ABT263/sorafenib combination was highly synergistic in inhibiting TET cell proliferation. We also showed that Gx15-070, a pan inhibitor of BH3 BCL2 anti-apoptotic family members, potently inhibited TET cell proliferation and induced cell death. Interestingly, cell death could be rescued by Beclin1, RIPK1 knockdown or by autophagy and necroptosis inhibition, suggesting that Gx15-070 induced cell death is autophagy-dependent. Taken together, our data strongly indicate that BCL2 family genes are deregulated in TET cells and are potential targets for TET treatment.
In conclusion, we identified CN losses of CDKN2A and 13q as potentially poor prognostic markers in TETs, implying that RB pathway deregulation may be important in TET pathogenesis. Targeting anti-apoptotic BCL2 family members by Gx15-070 or by ABT263-sorafenib combination may represent a novel treatment strategy for TET patients.
Materials and Methods
This study was conducted in agreement with the Declaration of Helsinki and was approved by the involved institutional ethical review boards (ClinicalTrials.gov ID: NCT00965627).
Patients
FFPE samples were collected from a series of 132 consecutive patients who underwent surgery for TET. This series has been described previously in detail elsewhere.28 Patients were staged according to the Masaoka staging system.29 Completeness of resection was defined as R0 when complete, R1 when residual disease was microscopic and R2 when macroscopic.30 After resection, tumor diagnosis was confirmed by two independent pathologists (HSL and MR) and histology was according to the 2004 WHO classification.2 Patient characteristics are reported in Table 1. Median follow-up was 84.5 months (95% CI 72.1–96.8). Frozen tumors were collected from an additional 12 patients who underwent tumor biopsy or radical resection within the past 2 years at the National Cancer Institute (Bethesda, MD, USA).
DNA extraction and array-CGH
Of the 132 samples, 59 only were selected for CGH, based on presence of >80% cancer cells in the sample, as assessed by Hematoxylin- and eosin-stained slides. DNA was extracted using DNeasy kit (Qiagen, Inc., Valencia, CA, USA) according to Genomic DNA ULS labeling kit protocol (Agilent Technologies, Inc., Palo Alto, CA, USA). The labeling procedure was conducted using Genomic DNA ULS labeling kit (Agilent), according to the vendor instructions. Twenty samples were hybridized on Human Genome CGH Array 105A (Agilent) and the remaining on SurePrint G3 Human CGH Array 180K (Agilent). The arrays were scanned on a laser-based microarray scanner (Agilent), and the data were extracted and normalized using Feature Extraction 10.5 (Agilent). Row data were centered to the mode using R script. CN profile was inferred using Rank Segmentation algorithm in Nexus 5.1 (Biodiscovery Inc., El Segundo, CA, USA). CN call thresholds were defined: high gain +0.7, gain +0.3, loss −0.3 and homozygous loss −0.7. Sex chromosomes were excluded from the analysis. GISTIC algorithm was applied using the online version on Gene Pattern server 2.0.31 GISTIC algorithm scores the frequency and amplitude of CN changes (number of abnormal copies) across the sample set. To assess chromosomal regions that significantly associate with CN aberrations, GISTIC identifies regions with scores above the random aberrations simulated by null distribution. The generated P-values were corrected for multiple hypotheses testing by Benjamini–Hochberg FDR procedure to obtain q-values.5 Q-value<0.25 were considered significant.
Immunohistochemistry
Immunohistochemistry was performed on a tissue microarray that included samples from 132 TET patients as previously described.28, 32 The staining with anti-p16INK4 antibody (BD PharMingen, San Diego, CA, USA) was graded according to the percentage of positive cells and intensity of the staining. Percentage of positive cancer cells was ranked 0 (0–5%), 1 (6–25%), 2 (26–50%), 3 (51–75%) or 4 (76–100%). Signal intensity (1–3) was multiplied by rank of positive cells in order to get a positivity score. P16INK4, scores 0-3 were considered negative (G0), 4-6 were G1, 7-9 were G2, and 10-12 were G3.
Western blot
Protein extraction, western blot and immunoprecipitation were performed as previously described.33, 34 Anti-BCL2 antibody was purchased from DAKO (Carpinteria, CA, USA), anti-MCL1 and anti-p16INK4 from BD PharMingen and anti-A1 from Abcam (Cambridge, MA, USA). Antibodies against BCL-XL, BCL-W, BAK, BAX, α-tubulin, PARP, caspase-3, cleaved caspase-3, LC3B, Beclin1 and RIPK1 were purchased from Cell Signaling (Danvers, MA, USA).
Real-time PCR
TaqMan CN assay was used to confirm the deletion of CDKN2A locus (Applied Biosystems, Foster City, CA, USA). Primers for CDKN2A (Applied Biosystems) and the reported locus RPPH1 (Applied Biosystems), the endogenous control, were combined to estimate the CN status. Real-time PCR was performed on ABI 7900HT fast real-time PCR system (Applied Biosystems). Gene CN was determined by CopyCaller software v1.0 (Applied Biosystems). Normal male diploid genomic DNA (Promega, Madison, WI, USA) fragmented at 99 °C for 10 min was used as diploid reference.
Cell lines and drug treatment
TC T1889 and B1 thymoma T1682 cell lines were kindly provided by Dr. Marco Breinig.35 TY82 TC cell line was purchased from Japan Health Science Foundation (Tokyo, Japan), whereas HEK-293, H157, NCI-H146 and NCI-H82 were from ATCC (Manassas, VA, USA). All cell lines were cultured in RPMI 1640 containing 200 mM ℒ-Glutamine (Invitrogen, Grand Island, NY, USA), 50 U/ml penicillin, 50 U/ml streptomycin (Invitrogen) and 10% heat-inactivated calf serum (Invitrogen) and grown in a 37 °C incubator with humidified 5% CO2 atmosphere. Media was supplemented with 25 nM Hepes for T1889 and T1682.
Activity of ABT263 and Gx15-070 (Selleck Chemicals, Houston, TX, USA) was tested using a proliferation assay (CellTiter 96 Non-Radioactive Cell Proliferation Assay, Promega),36 a viability assay (CytoTox 96 Non-Radioactive Cytotoxicity Assay, Promega) and a membrane permeability assay using TOPRO3 (Invitrogen) staining for flow cytometry (FACScalibur and Cellquest, Becton, Dickinson and Company, Franklin Lakes, IL, USA),37 because Gx15-070 generates auto-fluorescence that interferes with propidium iodide and 7AAD. Caspase-3 activity was calculated using a colorimetric caspase-3 Activity Detection Kit (Millipore, Billerica, MA, USA), as previously described.38
In vivo studies
Athymic NCr-nu/nu mice, purchased from NCI-Frederick (Frederick, MD, USA) were inoculated subcutaneously with 106 TY82 cells together with Matrigel (Becton, Dickinson and Company). Seventeen days after transplantation, 10 mice received 3 mg/kg/day of Gx15-070 and 10 mice the drug diluents as control by intramuscular injection for 5 consecutive days/week for 2 weeks. Tumor volume was calculated by the formula volume=1/2 × length (mm) × (width (mm))2 and assessed three times per week.
ShRNA, siRNA and transfection experiments
BECN1 shRNAs (1: V2HS_23694 and 2: V2HS_241693) and RIPK1 shRNAs (1: V2HS_17422 and 2: V2HS_241668) were purchased from Open-Biosystems (Thermo Scientific, Huntsville, AL, USA) and subcloned into pMSCV-PM-pheS retroviral expression vector (Clontech, Mountain View, CA, USA). High titer shRNA viral particles were produced in HEK-293T packaging cells and infection of TET cell lines was carried out in the presence of 4 μg/ml polybrene (Millipore).
Anti-apoptotic BCL2 family member ON-TARGETplus siRNAs were purchased from Dharmacon (Lafayette, CO, USA). Transfection was performed using PepMute siRNA Transfection reagent (SignaGen Laboratories, Rockville, MD, USA) and 10 nM of each siRNA. Two ON-TARGETplus siRNA (Dharmacon) against each gene were used to knockdown protein expression; a scrambled siRNA was used as control. Protein expression was evaluated by western blot after 48 h from the transfection and cell proliferation tested after 72 h.
Statistical analysis
Clinical and biological characteristics were compared using the Fisher's exact test, χ2 test or one-way ANOVA with Dunn test for post-hoc comparisons, when appropriate. Correlations were considered significant at the<0.05 level (two tailed). Survival curves were generated using the Kaplan–Meier method. DRS was calculated from the date of surgery to the date of death due to tumor progression. TTP was calculated from date of surgery to relapse or progression. For all survival calculations, the date of surgery for the primary tumor was considered as start date. The statistical difference between the survival curves was determined by a log-rank test. Subsequently, those factors, that appeared to be at least modestly associated with outcome (P<0.10) in univariate analyses were evaluated for their joint impact on TTP or DRS using a Cox proportional hazard model. All P-values are two tailed and have not been adjusted for multiple comparisons. All tests were performed using the SPSS version 17 (SPSS, Inc., Chicago, IL, USA) and SAS version 8.2 (SAS Institute, Inc., Cary, NC, USA). Synergism of drug combination was calculated using CalcuSyn 2.1 (Biosoft, Cambridge, UK).
Acknowledgments
The NIH/NCI intramural program funded this project. We thank Dr. Abu-Asab M for technical assistance for the electron microscopy pictures.
Glossary
- TET
thymic epithelial tumors
- CGH
comparative genomic hybridization
- TC
thymic carcinoma
- CN
copy number
- GISTIC
genomic identification of significant targets in cancer
- WHO
World Health Organization
- FFPE
formalin fixed paraffin embedded
- MG
Myasthenia gravis
- TTP
Time To Progression
- DRS
disease related survival
- HR
hazard ratio
- CI
confidence interval
- siRNA
small interfering RNA
- MTS
CellTiter 96 AQueous non-radioactive cell proliferation assay
- Gx15-070
Obatoclax
- BH3
BCL2 homology domain 3
- IC50
inhibitory concentration 50
- EM
electron microscopy
- Nec1
Necrostatin 1
- 3MA
3-Methyladenine
- CQ
chloroquine
- shRNA
short hairpin RNA
- LOH
loss of heterozygosity
- RNAi
inhibitory RNA
- FDR
false discovery rate
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- de Jong WK, Blaauwgeers JL, Schaapveld M, Timens W, Klinkenberg TJ, Groen HJ. Thymic epithelial tumours: a population-based study of the incidence, diagnostic procedures and therapy. Eur J Cancer. 2008;44:123–130. doi: 10.1016/j.ejca.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. Pathology and Genetics: Tumors of the Lung, Pleura, Thymus And Heart. IARC Press: Lyon, France; 2004. [Google Scholar]
- Chiecchio L, Protheroe RK, Ibrahim AH, Cheung KL, Rudduck C, Dagrada GP, et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia. 2006;20:1610–1617. doi: 10.1038/sj.leu.2404304. [DOI] [PubMed] [Google Scholar]
- van den Berg J, Johannsson O, Hakansson S, Olsson H, Borg A. Allelic loss at chromosome 13q12-q13 is associated with poor prognosis in familial and sporadic breast cancer. Br J Cancer. 1996;74:1615–1619. doi: 10.1038/bjc.1996.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Kim M, Yim SH, Cho NS, Kang SH, Ko DH, Oh B, et al. Homozygous deletion of CDKN2A (p16, p14) and CDKN2B (p15) genes is a poor prognostic factor in adult but not in childhood B-lineage acute lymphoblastic leukemia: a comparative deletion and hypermethylation study. Cancer Genet Cytogenet. 2009;195:59–65. doi: 10.1016/j.cancergencyto.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Kondo K, Takizawa H, Nagao T, Nakagawa Y, Fujino H, et al. Aberrant methylation of tumour-related genes in thymic epithelial tumours. Lung Cancer. 2009;64:155–159. doi: 10.1016/j.lungcan.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Hirabayashi H, Fujii Y, Sakaguchi M, Tanaka H, Yoon HE, Komoto Y, et al. p16INK4, pRB, p53 and cyclin D1 expression and hypermethylation of CDKN2 gene in thymoma and thymic carcinoma. Int J Cancer. 1997;73:639–644. doi: 10.1002/(sici)1097-0215(19971127)73:5<639::aid-ijc5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann JrR, Srikantan S, et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanaki K, Rontogianni D, Kouvidou CH, Bolioti S, Delides G, Pantelidaki A, et al. Expression of p53, mdm2, p21/waf1 and bcl-2 proteins in thymomas. Histopathology. 1997;30:549–555. doi: 10.1046/j.1365-2559.1997.5730805.x. [DOI] [PubMed] [Google Scholar]
- Pan CC, Chen PC, Wang LS, Lee JY, Chiang H. Expression of apoptosis-related markers and HER-2/neu in thymic epithelial tumours. Histopathology. 2003;43:165–172. doi: 10.1046/j.1365-2559.2003.01663.x. [DOI] [PubMed] [Google Scholar]
- Khoury T, Arshad A, Bogner P, Ramnath N, Zhang S, Chandrasekhar R, et al. Apoptosis-related (survivin, Bcl-2), tumor suppressor gene (p53), proliferation (Ki-67), and non-receptor tyrosine kinase (Src) markers expression and correlation with clinicopathologic variables in 60 thymic neoplasms. Chest. 2009;136:220–228. doi: 10.1378/chest.08-2482. [DOI] [PubMed] [Google Scholar]
- Hiroshima K, Iyoda A, Toyozaki T, Supriatna Y, Shibuya K, Shimamura F, et al. Proliferative activity and apoptosis in thymic epithelial neoplasms. Mod Pathol. 2002;15:1326–1332. doi: 10.1097/01.MP.0000038463.67854.84. [DOI] [PubMed] [Google Scholar]
- Chen FF, Yan JJ, Chang KC, Lai WW, Chen RM, Jin YT. Immunohistochemical localization of Mcl-1 and bcl-2 proteins in thymic epithelial tumours. Histopathology. 1996;29:541–547. doi: 10.1046/j.1365-2559.1996.d01-540.x. [DOI] [PubMed] [Google Scholar]
- Shoemaker AR, Mitten MJ, Adickes J, Ackler S, Refici M, Ferguson D, et al. Activity of the Bcl-2 family inhibitor ABT-263 in a panel of small cell lung cancer xenograft models. Clin Cancer Res. 2008;14:3268–3277. doi: 10.1158/1078-0432.CCR-07-4622. [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Murthy Madiraju SR, et al. Small molecule obatoclax (Gx15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci USA. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace L, Bornhauser BC, Schmitz M, Cario G, Ziegler U, Niggli FK, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest. 120:1310–1323. doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espona-Fiedler M, Soto-Cerrato V, Hosseini A, Lizcano JM, Guallar V, Quesada R, et al. Identification of dual mTORC1 and mTORC2 inhibitors in melanoma cells: prodigiosin vs. obatoclax. Biochem Pharmacol. 83:489–496. doi: 10.1016/j.bcp.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzel R, Hoegel J, Schmitz W, Blaeker H, Morresi-Hauf A, Aulmann S, et al. Clusters of chromosomal imbalances in thymic epithelial tumours are associated with the WHO classification and the staging system according to Masaoka. Int J Cancer. 2003;105:494–498. doi: 10.1002/ijc.11101. [DOI] [PubMed] [Google Scholar]
- Zettl A, Strobel P, Wagner K, Katzenberger T, Ott G, Rosenwald A, et al. Recurrent genetic aberrations in thymoma and thymic carcinoma. Am J Pathol. 2000;157:257–266. doi: 10.1016/S0002-9440(10)64536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Starostik P, Zettl A, Strobel P, Schwarz S, Scaravilli F, et al. Correlating genetic aberrations with World Health Organization-defined histology and stage across the spectrum of thymomas. Cancer Res. 2003;63:3708–3715. [PubMed] [Google Scholar]
- Girard N, Shen R, Guo T, Zakowski MF, Heguy A, Riely GJ, et al. Comprehensive genomic analysis reveals clinically relevant molecular distinctions between thymic carcinomas and thymomas. Clin Cancer Res. 2009;15:6790–6799. doi: 10.1158/1078-0432.CCR-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockenbery D, Nunez G, Milliman C, Schreiber RD, Korsmeyer SJ. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348:334–336. doi: 10.1038/348334a0. [DOI] [PubMed] [Google Scholar]
- Zucali PA, Petrini I, Lorenzi E, Merino M, Cao L, Di Tommaso L, et al. Insulin-like growth factor-1 receptor and phosphorylated AKT-serine 473 expression in 132 resected thymomas and thymic carcinomas. Cancer. 2010;116:4686–4695. doi: 10.1002/cncr.25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detterbeck FC, Nicholson AG, Kondo K, Van Schil P, Moran C. The Masaoka-Koga stage classification for thymic malignancies: clarification and definition of terms. J Thorac Oncol. 6 (7 Suppl 3:S1710–S1716. doi: 10.1097/JTO.0b013e31821e8cff. [DOI] [PubMed] [Google Scholar]
- Strobel P, Hartmann M, Jakob A, Mikesch K, Brink I, Dirnhofer S, et al. Thymic carcinoma with overexpression of mutated KIT and the response to imatinib. N Engl J Med. 2004;350:2625–2626. doi: 10.1056/NEJM200406173502523. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Petrini I, Zucali PA, Lee HS, Pineda MA, Meltzer PS, Walter-Rodriguez B, et al. Expression and mutational status of c-kit in thymic epithelial tumors. J Thorac Oncol. 5 (9:1447–1453. doi: 10.1097/JTO.0b013e3181e96e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarenza A, Lazarovici P, Lempereur L, Cantarella G, Bianchi A, Bernardini R. Tamoxifen inhibits nerve growth factor-induced proliferation of the human breast cancerous cell line MCF-7. Cancer Res. 2001;61:3002–3008. [PubMed] [Google Scholar]
- Harada T, Lopez-Chavez A, Xi L, Raffeld M, Wang Y, Giaccone G. Characterization of epidermal growth factor receptor mutations in non-small-cell lung cancer patients of African-American ancestry. Oncogene. 2011;30:1744–1745. doi: 10.1038/onc.2010.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehemann V, Kern MA, Breinig M, Schnabel PA, Gunawan B, Schulten HJ, et al. Establishment, characterization and drug sensitivity testing in primary cultures of human thymoma and thymic carcinoma. Int J Cancer. 2008;122:2719–2725. doi: 10.1002/ijc.23335. [DOI] [PubMed] [Google Scholar]
- Lee JH, Voortman J, Dingemans AM, Voeller DM, Pham T, Wang Y, et al. MicroRNA expression and clinical outcome of small cell lung cancer. PLoS One. 6:e21300. doi: 10.1371/journal.pone.0021300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavecchio M, Simone M, Bernasconi S, Tognon G, Mazzini G, Erba E. Multi-parametric flow cytometric cell cycle analysis using TO-PRO-3 iodide (TP3): detailed protocols. Acta Histochem. 2008;110:232–244. doi: 10.1016/j.acthis.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Palumbo S, Toscano CD, Parente L, Weigert R, Bosetti F. The cyclooxygenase-2 pathway via the PGE(2) EP2 receptor contributes to oligodendrocytes apoptosis in cuprizone-induced demyelination. J Neurochem. 2010;121:418–427. doi: 10.1111/j.1471-4159.2011.07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.