Abstract
Transcriptional control of marginal zone (MZ) and follicular (FO) B cell development remains incompletely understood. The transcription factor, IFN regulatory factor (IRF)8, is known to play important roles in the differentiation of early B cells. In this article, we demonstrate that IRF8 is also required for normal development of MZ and FO B cells. Mice with a conventional knockout of Irf8 (IRF8–/–) or a point mutation in the IRF association domain of IRF8 had increased numbers of MZ B cells. To determine the B cell-intrinsic effects of IRF8 deficiency, we generated mice with a conditional allele of Irf8 crossed with CD19-Cre mice (designated IRF8-conditional knockout [CKO]). These mice had enlarged MZ and increased numbers of MZ and FO B cells compared with controls. The FO B cells of CKO mice exhibited reduced expression of CD23 and moderately increased expression of CD21. Gene-expression profiling showed that increased B cell production in IRF8-CKO mice was associated with changes in expression of genes involved in regulation of transcription, signaling, and inflammation. Functional studies showed that IRF8-CKO mice generated normal Ab responses to T-independent and T-dependent Ags. Thus, IRF8 controls the expansion and maturation of MZ and FO B cells but has little effect on B cell function.
B cell development follows ordered cellular stages characterized by expression of specific genes required for maturation and function. Rearrangements of Ig μH chain gene segments in pro-B cells and L chain gene loci in pre-B cells lead to expression of functional BCRs on the cell surface. BCR+ immature B cells are subject to positive and negative selection, resulting in survival or cell death through different mechanisms. Autoreactive B cells are deleted or arrested to undergo secondary Ig gene rearrangements at the small pre-BII cell stage. The latter mechanism, termed receptor editing, is thought to be used by up to 50% of all autoreactive B cells (1–4). Only those immature B cells bearing BCRs that are nonautoreactive or have reduced autoreactivity are allowed to leave the bone marrow (BM) and migrate to the spleen. There, they pass through defined transitional stages, termed T1, T2, and T3 by Allman et al. (5), or T1 and T2 by Loder et al. (6), to become fully mature B cells.
There are at least three mature B cell subsets in the spleen: follicular (FO) B2 cells, marginal zone (MZ) B cells, and CD5+ B1a cells. These subsets differ significantly in phenotype, function, and anatomical location. How these subsets are selected remains incompletely understood. Growing evidence has supported an important role for transcription factors in regulating MZ and FO B cell fate. This is largely based on the observations that genetically mutant mice deficient in a series of transcription factors exhibited imbalanced development of FO and MZ B cells. For example, mice deficient in Egr1 (7), Ets1 (8), Id2 (9), Aiolos (10, 11), and Relb (12) exhibit reduced MZ B cell compartments with or without altered FO B cell pools. Other mice bearing mutant alleles for E2a (13), Nfkb2 (14), and Fli1 (15) developed more MZ B cells than FO B cells.
In this study, we provide evidence that the transcription factor IRF8 restricts the expansion and maturation of MZ and FO B cells in the spleen. IRF8, also known as IFN consensus sequence binding protein, is one of nine members of the IRF family of transcription factors with functions involved in innate and adaptive immune responses. Stable expression of IRF8 has been found in B cells, macrophages, and CD11b– dendritic cells and is inducible in T cells. Expression is further increased by IFN-γ and TLR ligands (16). IRF8 regulates expression of target genes through its DNA-binding domain, forming heterodimers with other factors through its IRF association domain (IAD). IRF8-null mice (IRF8–/–) exhibit defects in development and function of macrophages and dendritic cells and produce excessive granulocytes causing splenomegaly (17–19). The mice also exhibit a cytokine imbalance because they are deficient in IFN-γ and IL-12 and overexpress IL-4 (17). BXH2 mice, bearing a near complete loss-of-function mutation in the IAD of IRF8, have phenotypes similar to those of IRF8–/– mice (20, 21).
Previous studies supported a role for IRF8 in regulation of B cell development at multiple stages. In the BM, there is a significant reduction in pre–pro-B cells in IRF8–/– mice compared with wild-type (WT) controls (22). This is associated with reduced expression of Pax5, Tcf3 (E2A), and Ebf1 and enhanced expression of Sfpi1 (PU.1) in common lymphoid progenitors and pre–pro-B cells. At the pre-B cell stage, IRF8 deficiency causes enhanced expression of the pre-BCR, leading to vigorous cycling (22). An even more severe deficit in pre-B cell development is seen in mice deficient in IRF8 and IRF4 (23), which exhibit a complete blockade of generation of small pre-B cells. IRF4 and IRF8 mutant mice also fail to initiate L chain gene rearrangement (24, 25). In the germinal center (GC) stage of development, IRF8 regulates expression of Aicda (encoding activation-induced cytidine deaminase), Bcl6 (26), and Mdm2 (27). Its absence causes development of irregular GCs (26, 28).
In addition to altered structures of the lymphoid organs in IRF8–/– mice due to excessive development of myeloid cells, these mice have a cytokine imbalance, which may alter normal mature B cell subset selection. To eliminate these complications for understanding IRF8 function in the B cell lineage, we generated mice with a B cell-specific deficiency in IRF8 using CD19-Cre and IRF8-flox systems. Our data indicated that IRF8 is involved in expansion and maturation of FO and MZ B cells.
Materials and Methods
Mice
IRF8 conventional knockout mice were described previously (17) and were kindly provided by Dr. Keiko Ozato (National Institute of Child Health and Human Development, National Institutes of Health). The mutation has been bred onto the C57BL/6J (B6) for >10 generations. Age-matched littermate mice (6–10 wk) were used as controls. B6 mice (8–10 wk old) were purchased from the Jackson Laboratory (Bar Harbor, ME). BXH2 mice bred to express ecotropic murine leukemia viruses (MuLV) at low levels (20) were also provided by Dr. Ozato. The use of mice in this study followed a protocol (LIP-4) approved by National Institute of Allergy and Infectious Diseases Animal Care and Use Committee.
Generation of IRF8 conditional knockout mice
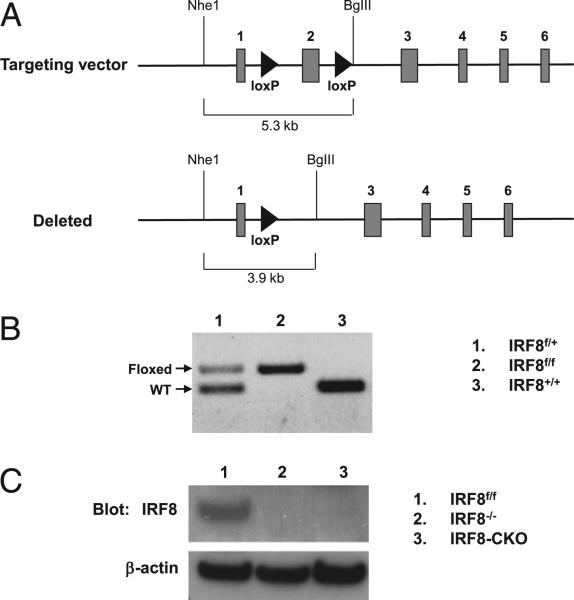
Mice bearing a conditional allele of Irf8 were generated at Ozgene using the strategy outlined in Fig. 3A. Exon 2 of Irf8 encoding the DNA-binding domain was flanked by loxP sites. A phosphoglycerine kinase-neo cassette flanked by Flp recombinase target sites (data not shown) was used for selection. Following homologous recombination of the vector in C57BL/6 embryonic stem cells and establishment of germline transmission, the PGK-neo cassette was excised using the FLP recombinase, leaving exon 2 flanked by loxP sites (Fig. 3A, top panel). Selective breeding to B6 mice was used to eliminate the FLP gene. Irf8f/f mice were crossed with CD19-Cre mice (Jackson Laboratory) to generate IRF8 conditional-deletion mice (designated IRF8-conditional knockout [CKO]) (Fig. 3A, bottom panel).
FIGURE 3.
Generation of IRF8-CKO mice. A, Targeting strategy to conditionally delete exon 2 of Irf8. B, PCR assay showed the floxed allele that was amplified from genomic DNA. C, Lack of IRF8 protein expression in splenic B cells of IRF8 CKO mice. Immunoblotting was performed with purified splenic B cells.
Immunization and Ab detection
For T-dependent immune responses, mice were immunized i.p. with 100 μg 4-hydroxy-3-nitrophenylacetyl–keyhole limpet hemocyanin (NP-KLH; Biosearch) in alum. Blood samples were taken before and 3 wk after immunization. For T-independent immune responses, mice were immunized i.p. with 50 μg NP-LPS (Biosearch) or 20 μg NP-FICOLL (Bio-search). Blood samples were taken before and 7 d after immunization.
Serum NP-specific Abs were tested by ELISA. In brief, 96-well plates were coated with NP(23)-BSA (Biosearch), followed by incubation with diluted serum samples and developed with HRP-conjugated mouse-specific anti-IgM, IgG1, and IgG3 Abs and substrate o-phenylenediamine dihydrochloride (Sigma-Aldrich). The reaction was read at 450 nm using an ELISA plate reader.
Flow cytometry (FACS)
Cells were prepared and stained as previously reported (29). mAbs specific for mouse B220 (RA3-6B2), IgM (II/41), CD43 (S7), CD5 (53-7.3), CD19 (1D3), CD23 (B3B4), CD24 (M1/69), CD21/CD35 (7G6), CD11b (M1/70), CD3 (145-2C11), CD86 (GL1), CD80 (16-10A1), CD40 (HM40-3), and MHC class II (M5/114.15.2) were obtained from BD-PharMingen. The anti–Gr-1 (RB6-8C5) Ab was purchased from eBioscience. Abs were directly conjugated to FITC, R-PE, PE-Cy7, allophycocyanin, allophycocyanin-Cy7, PerCP, Pacific blue, Alexa Fluor 700, or biotin. Biotinylated Abs were revealed with streptavidin-conjugated PerCP (BD-PharMingen) or Qdot655 (Invitrogen). Cells were analyzed using a FACSCalibur or an LSR II Analyzer (Becton Dickinson).
Confocal microscopy and immunohistochemistry
Immunofluorescence histological staining was performed on frozen sections of mice spleens using a standard protocol (28).
Microarray gene-profiling analysis
A total of 1 × 104 MZ and FO B cells were sort purified from individual mice in six independent experiments. A combination of CD23, CD21, and CD19 markers was included in the sorting scheme. PE-labeled anti-CD23 Ab was used to separate MZ and FO B cells (Supplemental Fig. 3). RNA extraction, labeling, and hybridization using Nugen Ribo-SPIA technology (Nugen Technologies, San Carlos, CA) and GeneChip Mouse gene 1.0 ST Array (Affymetrix, Santa Clara, CA) were processed by Research Technology Branch, National Institute of Allergy and Infectious Diseases. Data were collected using GeneChip Operating Software (GCOS v1.4) and imported into Partek Genomics Suite. The robust multiarray averaging and quantile normalization were performed. The t test was carried out to identify differentially expressed genes in MZ and FO B cells of IRF8-CKO mice versus control mice. The microarray data were deposited in the Gene Expression Omnibus database under accession number GSE24972 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24972).
Quantitative real-time RT-PCR
MZ and FO B cells were sort purified from three mice per group. Total RNA was isolated using the RNeasy mini Kit coupled with DNase set (both from QIAGEN). Reverse transcription was performed using random hexamer primers (Invitrogen) and a Superscript II kit (Invitrogen). cDNAs were amplified using regular SYBR-Green reagents (Applied Biosystems) in 384-well plates containing primer pairs (30). The expression of the genes of interest relative to HPRT was determined using the 2(–ΔΔCt) method.
Results
Abnormal peripheral B cell development in BXH2 and IRF8–/– mice
BXH2 is a recombinant inbred mouse strain derived from a cross between strains C57BL/6J and C3H/HeJ (25) that carries a point mutation resulting in an arginine to cystine substitution at aa 294 in the IAD of IRF8 (21). These mice exhibit a series of abnormalities in myeloid differentiation and function that are strikingly similar to those of IRF8–/– mice (21) but are distinguishable in terms of dendritic cell development and cytokine expression (20).
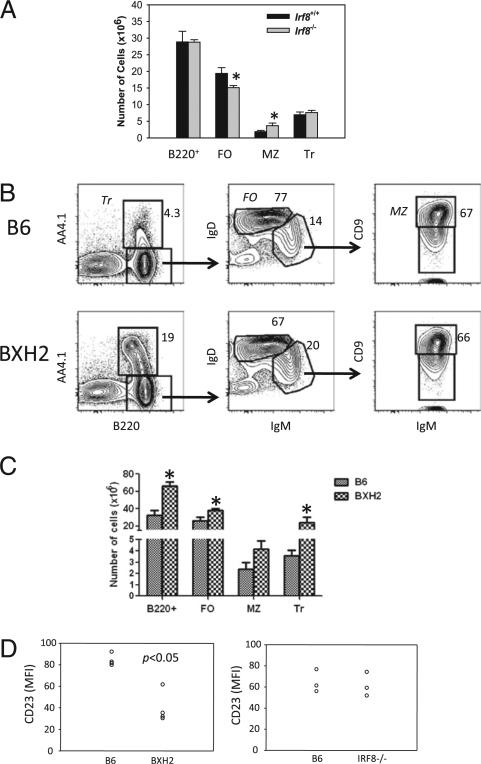
We recently reported that BXH2 and IRF8–/– mice exhibit defective development of pre–pro-B cells (22, 31). Interestingly, this deficiency was mostly corrected at the pre-B cell stage, a finding we attributed to abnormally increased expression of the pre-BCR and vigorous clonal expansion of pre-B cells (22, 23). Although the total B cell numbers in spleens of IRF8–/– and WT mice were similar, careful analysis of B cell subpopulations revealed that IRF8–/– mice had an expanded MZ B cell compartment (p < 0.05) and a moderately decreased FO B cell pool (p < 0.05), whereas the population of transitional B cells was unchanged (28) (Fig. 1A).
FIGURE 1.
Enhanced MZ B cell development in IRF8-deficient mice. A, Splenic B cell subsets of IRF8–/– and IRF8+/+ mice were assessed by FACS. FO (B220+IgM+CD23+CD21lo), MZ (B220+IgM+CD232CD21hi), and transitional (Tr) (B220+IgM+CD232CD212) B cells were enumerated from eight mice per group. B and C, Splenocytes of BXH2 and B6 mice were analyzed by FACS. Cells were gated on lymphocytes. Numbers indicate the percentage of cells falling in each gate. Data are representative of four experiments. D, The levels of CD23 expression on FO B cells (B220+IgM+CD21int) of BXH2, IRF8–/–, and control mice. Each symbol represents a mouse. *p < 0.05.
BXH2 mice express high levels of endogenous B-tropic ecotropic MuLV that are passed to offspring through the milk and contribute to the development of myeloid leukemias by proviral insertional mutagenesis of proto-oncogenes (reviewed in Ref. 21). To avoid the potential effects of high viral load on B cell development, we crossed male BXH2 mice with female B6 mice to generate progeny that, like B6 mice, only express low levels of MuLV late in life. In 8-wk-old BXH2 mice, the characteristics of B cell development in the BM were quite similar to those seen in IRF8–/– mice (31). In spleens of BXH2 mice, the numbers of total B cells, as well as FO and transitional B cell subsets, were increased (p < 0.05; Fig. 1B, 1C). The numbers of MZ B cells were also increased, but not significantly. These analyses suggested that mutation of the IRF8 IAD domain, which was shown to abrogate or markedly reduce the ability of IRF8 to complex with its partners (28), induced abnormal development of MZ B cells to a degree similar to that seen in IRF8–/– mice. However, the marked increase in transitional B cells and moderate increase in FO B cells seen in BXH2 mice, but not in IRF8–/– mice, indicate that a distinct mechanism may cause overproduction or accumulation of transitional B cells in the BXH2 strain. Moreover, it is noteworthy that the levels of CD23 expressed by FO B cells of BXH2 mice, but not of IRF8–/– mice, were significantly lower than for FO B cells of B6 mice (Fig. 1D). The reason for the differing patterns of B cell distribution between BXH2 and IRF8–/– mice warrants further investigation.
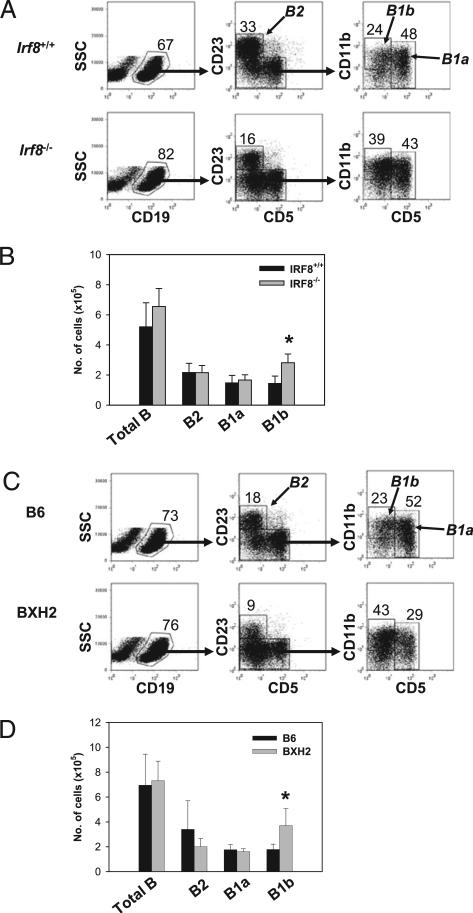
In the peritoneum, the total numbers of B, B2, and B1a cells were comparable between BXH2 and B6 mice (Fig. 2C, 2D), a pattern similar to that seen in comparisons of IRF8–/– and WT mice (Fig. 2A, 2B). The numbers of B1b cells were consistently increased in IRF8–/– mice (Fig. 2A, 2B) and BXH2 mice (Fig. 2C, 2D). These data indicated that the selection and development of peripheral B cell subsets, including MZ, FO, and B1b, were affected in mice bearing a null allele of IRF8 or a point mutation in the IAD domain.
FIGURE 2.
Enlarged B1b cell compartment in the peritoneum of IRF8-deficient mice. A and C, Peritoneal cells from indicated mice were analyzed by FACS. Numbers indicate the percentage of cells falling in each gate. B and D, Absolute cell numbers in the peritoneum of IRF8-deficent and control mice. The data are means ± SEM of five mice per group (B) and three mice per group (D). *p < 0.05.
B cell-specific deletion of IRF8 alters FO and MZ B cell fate
B cell-specific IRF8 conditional deletion mice were generated to permit the identification of B cell-intrinsic effects of IRF8 on B cell differentiation and function. Fig. 3A illustrates the strategy for conditional deletion of exon 2 of Irf8. The floxed and WT alleles were detected by PCR in genomic DNA (Fig. 3B). After crossing with CD19-Cre mice, the homozygous Irf8f/f mice bearing the Cre gene exhibited a complete depletion of IRF8 protein in splenic B cells (Fig. 3C).
In the BM of IRF8-CKO mice, the sizes of pro-B (B220+IgM2 CD43+), pre-B (B220+IgM2CD432), immature B (B220loIgM+), and mature B (B220hiIgM+) compartments were comparable to controls (Supplemental Fig. 1). This differs from conventional IRF8–/– mice, in which early B cell compartments were signifi cantly reduced (22). This could be due to incomplete deletion of IRF8 in early B cells of IRF8-CKO mice, an outcome similar to that found in other models involving deletion of a floxed allele when crossed with the CD19-Cre strain (32).
The sizes of spleens and lymph nodes were essentially normal in IRF8-CKO mice (data not shown). This again differs from IRF8–/– mice, which have enlarged spleens and lymph nodes due to excessive production of myeloid cells, particularly granulocytes (17).
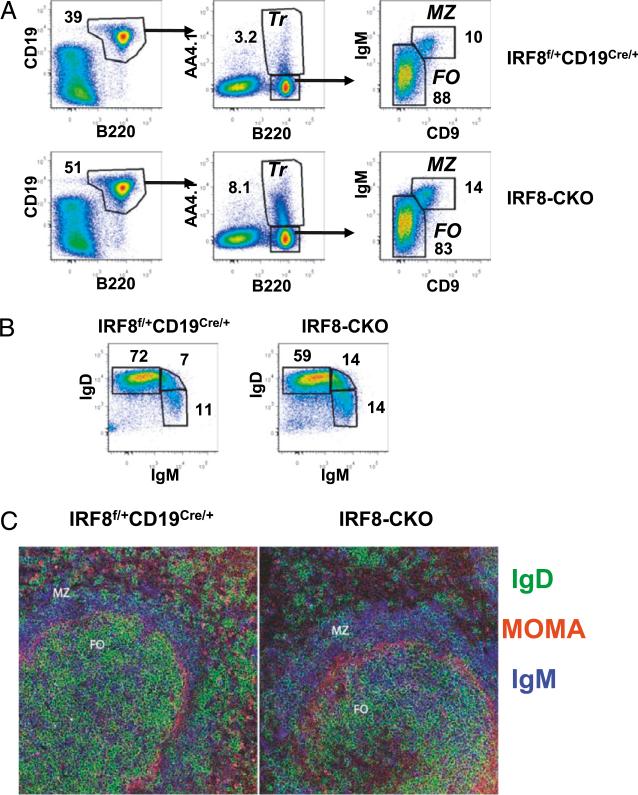
The distribution of B cell subsets in the spleen of IRF8-CKO mice was analyzed by FACS. The frequency of CD19+B220+ B cells was higher in IRF8-CKO mice than in controls (Fig. 4A). The frequency of B220+AA4+ transitional B cells was also increased in IRF8-CKO mice. Because the expression levels of CD23 were 50% lower in IRF8-CKO B cells (Fig. 5A), we used the combination of CD9 and IgM to define MZ B cells as CD9+IgMhi. The frequency of AA42 B220+IgMhiCD9+ MZ B cells was increased in IRF8-CKO mice, whereas the proportion of AA42B220+IgMloCD92 FO B cells was decreased (Fig. 4A). This distribution of B cell subsets was consistent with that defined using IgD plus IgM (Fig. 4B). In the latter case, FO B cells were regarded as IgD+IgMlo, transitional B cells were regarded as IgD+IgM+, and MZ/transitional 1 B cells were regarded as IgD2 IgMhi. Although the frequency of FO B cells was lower in IRF8-CKO mice than in control mice, the absolute numbers of FO B cells, as well as MZ and transitional B cells, were significantly higher in IRF8-CKO mice than in controls (Table I). Given the consistent observation of increased MZ B cell numbers in IRF8–/–, BXH2, and CKO mice, we further examined the organization of MZ B cells in the spleen by immunohistochemistry. The sections were stained with Abs to IgM, IgD, and MOMA1. MOMA1 is expressed by metallophilic macrophages that lie immediately adjacent to the white pulp and the inner border of the MZ. The staining intensities of IgM and IgD distinguish IgMhiIgDlo/2 MZ B cells from IgDhiIgMlo FO B cells, as shown for a control spleen in the left panel of Fig. 4C. Parallel studies of sections from spleens of IRF8-CKO mice revealed significantly enlarged MZs surrounding white pulp follicles, consistent with the data obtained by FACS.
FIGURE 4.
Enlarged MZ B cell compartment in IRF8-CKO mice. A, Splenocytes of IRF8-CKO and control mice were stained with Abs against B220, CD19, AA4.1, IgM, and CD9 and were analyzed by FACS. Dead cells were excluded by propidium iodide staining. The numbers represent the percentage of cells falling in each gate. B, Splenocytes were stained and analyzed by FACS. Cells were gated on B220+ PI2 cells. C, Immunofluorescence staining of frozen sections from spleens of IRF8-CKO and control mice. IgD (green) showed FO cells, MOMA (red) showed MZ macrophages, and IgM (blue) showed MZ B cells. Original magnification ×20.
FIGURE 5.
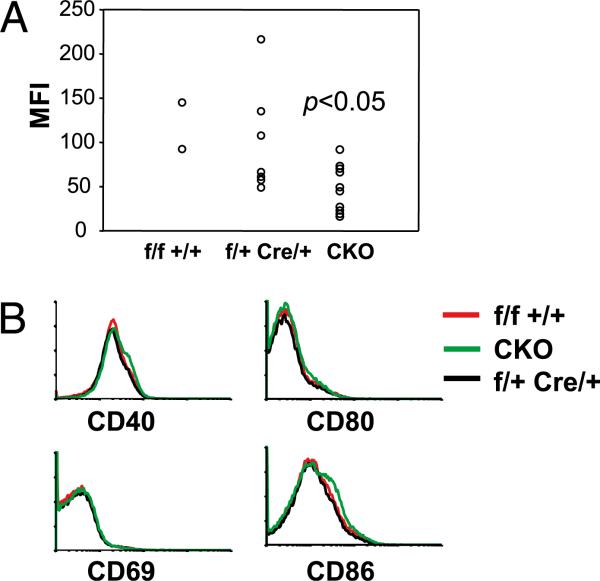
Decreased expression of CD23 but normal expression of activation markers in B cells of IRF8-CKO mice. A, Splenocytes were analyzed by FACS. The expression levels of CD23 on FO B cells (B220+ IgM+CD21int) were measured as mean fluorescence intensity (MFI). Each symbol represents a mouse. B, Expression levels of activation markers on splenic B cells (B220+IgM+) from the indicated mice were measured by FACS.
Table I.
B cell development in the spleen of IRF8-CKO mice
| Mice | n | Number of Cells (×106) |
|||
|---|---|---|---|---|---|
| Total B | Transitional | MZ | FO | ||
| f/+ Cre/+ | 6 | 6.65 ± 2.2 | 0.73 ± 0.1 | 0.87 ± 0.1 | 3.94 ± 0.6 |
| IRF8-CKO | 6 | 11.75 ± 3.3* | 1.31 ± 0.1** | 1.98 ± 0.3* | 6.77 ± 1.0* |
Splenocytes were stained and analyzed by FACS. Absolute cell numbers of each population were emulsified from a gating scheme described in Fig. 4A. Data are means ± SEM.
p < 0.05
p < 0.001, compared with controls.
Having identified enlarged MZ and FO B cell compartments in IRF8-CKO mice, we examined expression of other cell surface markers to determine whether IRF8-deficent B cells were phenotypically normal. For this analysis, a simple gating scheme was used to define MZ and FO B cells based on expression of B220, IgM, CD21, and CD23 (Supplemental Fig. 2). FO B cells of IRF8-CKO mice expressed modestly higher levels of CD21 than did FO B cells from controls (Supplemental Fig. 2), indicating that IRF8 is required for expression of a normal FO B cell phenotype. CD21 levels were comparable on MZ B cells from mice of both genotypes. Moreover, the levels of activation markers, including CD40, CD69, CD80, and CD86, were unchanged from normal in B cell subsets of IRF8-CKO mice (Fig. 5B).
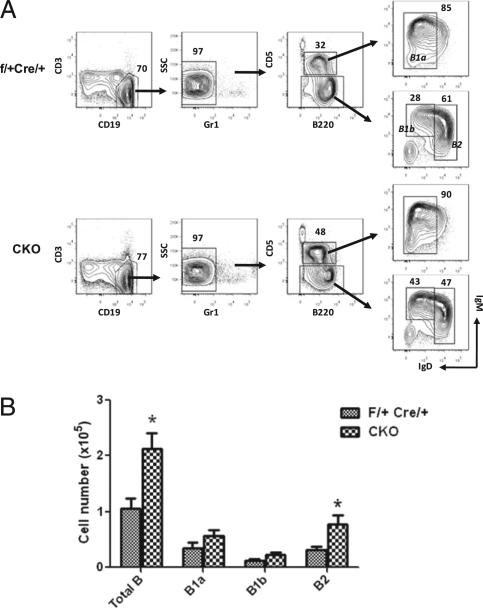
Peritoneal B cell subsets of IRF8-CKO mice were also analyzed by flow cytometry using Abs against CD3, Gr-1, CD19, CD5, B220, IgM, and IgD. B2, B1a, and B1b cells were readily distinguished using the gating scheme indicated in Fig. 6A. Although the frequencies of each B cell subset varied among individual mice of the IRF8-CKO and control groups, the numbers of total B cells and of B2 cells were significantly greater in CKO mice than in controls (Fig. 6B; p < 0.05; n = 6). The trend to increased B1b cells seen in IRF8–/– and BXH2 mice was also evident in the CKO mice. Interestingly, the increased numbers of B2 cells seen in CKO mice was not seen in IRF8–/– or BXH2 mice. These results suggested that IRF8 is involved in regulating the development of peritoneal B2 and B1b cells.
FIGURE 6.
Increased B cell subsets in the peritoneum of IRF8-CKO mice. Peritoneal cells were stained and analyzed by FACS. A, Numbers indicate percentage of cells falling in each gate. Cells were gated on 7AAD2 cells. B, Absolute cell numbers of peritoneal B cell subpopulations from indicated groups of mice. The data are means ± SEM of six mice per group. *p < 0.05; compared with controls.
IRF8-CKO mice develop normal Ab responses to T-independent and T-dependent Ags
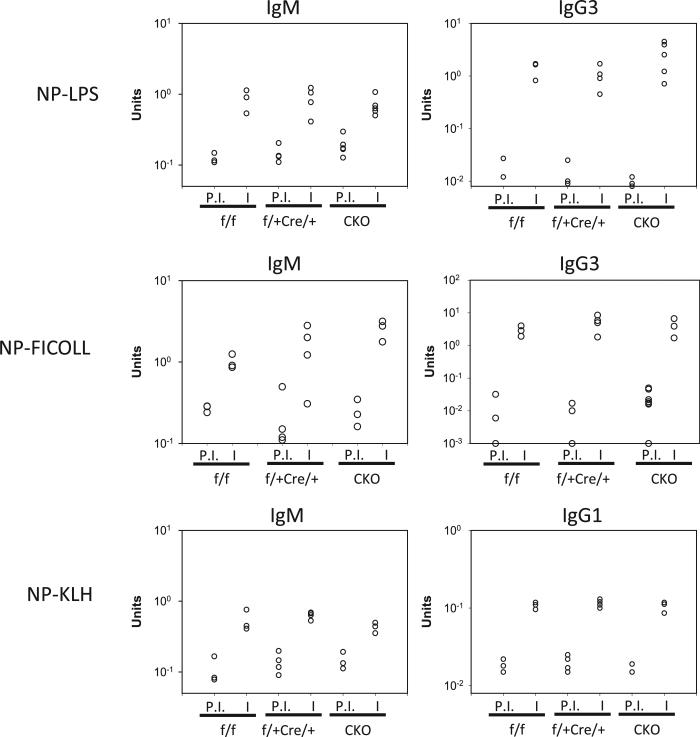
To determine whether a cell-intrinsic IRF8 deficiency in B cells would affect humoral immune responses in vivo, we immunized mice with T-independent (TI)-I (NP-LPS), TI-II (NP-FICOLL), and T-dependent (TD) (NP-KLH) Ags. In all cases, IRF8-CKO mice generated equivalent levels of NP-specific Abs compared with control mice (Fig. 7). The titers of high-affinity anti-NP Abs were also comparable between CKO and control mice (data not shown). Confocal microscopic studies of spleen sections revealed normal structures of GCs (data not shown), although the proliferation rate of GC B cells was slightly less in CKO mice than in control mice (28). This finding differs from that found in IRF8–/– mice in which the GCs were irregular in shape, a finding that we attributed to inefficient regulation of expression of Aicda (encoding activation-induced cytidine deaminase [AID]) and Bcl6 by IRF8 (28). To further determine whether IRF8 deletion in B cells has an effect on expression of AID and Bcl6 during a GC response, we measured the expression levels of Aicda and Bcl6 transcripts in sorted GC cells from IRF8-CKO and control mice that had been immunized with NP-KLH in alum. As shown in Supplemental Fig. 4, the levels of Aicda and Bcl6 transcripts in GC B cells of IRF8-CKO mice were slightly lower than seen with cells from control mice. We conclude that the abnormal development of GCs in IRF8–/– mice was likely due to B cell-extrinsic IRF8-dependent changes.
FIGURE 7.
Ab production in mice immunized with NP-LPS, NP-FICOLL, or NP-KLH. The concentrations of NP-specific Abs (IgM, IgG1, and IgG3) in sera were measured by ELISA. Each symbol represents a single mouse. I, immune; P.I., preimmune.
Gene expression-profiling analyses
The observation that B cells in spleens of CKO mice were composed of increased numbers of MZ and FO B cells prompted us to use Affymetrix microarrays to evaluate gene-expression patterns that distinguished FO or MZ B cells derived from CKO and control mice (IRF8f/+CD19Cre/+). These studies identified 25 genes that differed significantly (p < 0.01) with regard to transcript levels in sort-purified FO B cells from mice of the two genotypes (Table II). Of these, 7 (28%) had previously been identified as direct targets of IRF8 (26) or in chromatin immunoprecipitation (ChIP)-on-chip analyses of three GC-derived mouse B cell lymphoma cell lines (C.-H. Lee and H.C. Morse, III, unpublished observations). Prominent among the genes expressed at higher levels among the normal FO B cells were those involved in metabolic processes, transcriptional regulation, and signaling.
Table II.
List of differentially expressed genes in FO and MZ B cells of IRF8-CKO versus control mice
| FO |
MZ |
||||
|---|---|---|---|---|---|
| Gene Symbol | RefSeq | Fold | Gene Symbol | RefSeq | Fold |
| Trim12a | NM_023835 | –3.4 | Kynua | NM_027552 | –2.1 |
| Kynua | NM_027552 | –2.5 | Gml | –1.6 | |
| Rnf146a | NM_001110197 | –2.1 | Olfr99 | NM_146515 | –1.6 |
| Syne2a | NM_001005510 | –2.1 | Sema5a | NM_009154 | –1.6 |
| Gm1966 | –1.7 | 4933427I18Rik | –1.5 | ||
| Spic | NM_011461 | –1.7 | Lrrc16a | NM_026825 | –1.5 |
| 9230105E10Rik | NM_175677 | –1.6 | Nab1 | NM_008667 | –1.5 |
| Zfp294 | NM_001081068 | –1.6 | Rasgef1b | NM_145839 | –1.5 |
| BC023892 | –1.5 | Stk3a | NM_019635 | –1.5 | |
| Ceacam1 | NM_001039185 | –1.5 | Gmfg | BC011488 | 1.5 |
| ENSMUSG00000073981 | AK051661 | –1.5 | Sgk3 | NM_133220 | 1.5 |
| Kmoa | NM_133809 | –1.5 | ENSMUSG00000068293 | AY512926 | 1.6 |
| Stard3nl | NM_024270 | –1.5 | Hs3st1 | NM_010474 | 1.6 |
| LOC100046496 | 1.5 | LOC641050 | M11024 | 1.6 | |
| Sesn1 | NM_001013370 | 1.5 | Pgap1 | 1.6 | |
| St6galnac2 | NM_009180 | 1.5 | St6galnac2 | NM_009180 | 1.6 |
| Zfp1 | NM_001037665 | 1.5 | Ptpn22a | NM_008979 | 1.7 |
| Cd69a | NM_001033122 | 1.6 | Sesn1 | NM_001013370 | 1.8 |
| Gprc5b | NM_022420 | 1.7 | Cxcr4 | NM_009911 | 1.9 |
| Ptpn22a | NM_008979 | 1.9 | Gprc5b | NM_022420 | 1.9 |
| St6galnac1 | NM_011371 | 1.9 | BC005685 | BC005685 | 2.0 |
| ENSMUSG00000056897 | BC057193 | 2.0 | LOC100046764 | XR_032907 | 2.1 |
| LOC641050 | M11024 | 2.1 | BC018473 | NR_003364 | 6.4 |
| BC005685 | BC005685 | 2.2 | |||
| BC018473 | NR_003364 | 5.2 | |||
IRF8 targets identified by ChIP-on-Chip.
Parallel studies of sort-purified MZ B cells from control and CKO mice identified 23 genes whose expression differed significantly (p < 0.01) in the two groups of mice (Table II). Three genes (13%) in this group had been identified as direct targets of IRF8 by the ChIP-on-chip studies described above. The genes expressed at higher levels in normal MZ B cells included a number expressed at the cell surface or affecting signaling at the plasma membrane, as well as transcriptional regulation.
Thus, of the 48 genes found to distinguish IRF8-deficient and -competent MZ or FO B cells, 10 (21%) were previously identified as IRF8 targets. Interestingly, only 5 of these genes commonly distinguished the two FO and the two MZ B cell populations (St6galnac2, Kynu, Ptpn22, Sesn1, and Gprc5b), and each of the genes was similarly up- or downregulated in both comparisons. Quantitative real-time RT-PCR (qPCR) was used to validate array analyses of selected genes (Cd69, Kynu, Ptpn22, Sens1, Cxcr4) and showed changes in expression levels ~2-fold (data not shown), consistent with the microarray data. Taken together, these results indicated that a significant proportion of the genes that distinguish the FO and MZ compartments of IRF8+ and IRF82 B cells are known IRF8 targets and that some of these targets, as well as genes not known to be IRF8 targets, are similarly regulated in both mature B cell compartments.
Discussion
Peripheral B cell maturation is a dynamic biological process involving constant cellular interactions with environmental cues, including Ags, the cytokine BAFF, and ligands for B cell accessory receptors. How these signals influence MZ versus FO lineage choice remains elusive. There are several mechanisms that may explain peripheral B cell lineage determination. First, BCR signaling strength is reported to play a critical role in mature B cell fate decision. In the absence of ligand stimulation, BCR basal signaling is essential for survival (33) and differentiation (34). Monroe and colleagues showed that the cytoplasmic tails of Iga and Igβ in the absence of Ag binding are sufficient to drive FO B2 but not MZ and B1 cell differentiation (34, 35). Consistent with this finding, Hayakawa et al. (36) showed that immature B cells expressing a transgenic BCR specific for the self Thy-1 glycoprotein develop into FO B2 cells in the absence of Thy-1 expression. However, the same B cells acquire an MZ B cell phenotype in the presence of low-dose self-Ags and a B1a cell phenotype in the highest dose of self-Ags (36, 37). In line with these findings, an increase in expression of a BCR transgene often results in an enlarged MZ B cell compartment (38, 39). These studies were consistent with a correlation between BCR signaling strength and peripheral B cell maturation, with the dependence of BCR signaling strength for lineage specification being B1a > MZ > FO.
An alternative view of the relationship between BCR signaling strength and B cell lineage choice holds that FO B cell development is dependent on strong BCR signaling, whereas signals driving MZ B cell development are weak (40). These contrasting conclusions are not readily reconciled, but they can probably be ascribed to differences in the systems under study. In addition, the subsets have different environmental signaling requirements for their development and maintenance, including NOTCH2 for MZ B cells (see later discussion) and BAFF for FO B cells, which integrate with BCR signal strength in fate determination.
Second, the NOTCH signaling pathway is essential for mature B cell fate determination. Deletion of Notch2 (41), Rbpj (42), Dll1 (43), or Maml1 (44) selectively impaired MZ B cell development, whereas inhibition of NOTCH signaling by MINT biased toward FO B2 cell differentiation (45). Moreover, expression of an active form of NOTCH2 blocks development of B2 cells at the pre-B cell stage but promotes the development of B1a cells (46). More recently, Moran et al. (47) provided evidence for cross-talk between NOTCH and BCR signaling pathways in showing that NF-κB1/p50, a downstream effector of the BCR, synergizes with NOTCH2 to regulate MZ B cell fate.
Third, a transcription-factor network governs MZ versus FO B cell lineage determination. BCR signaling, growth factor stimulation, NOTCH signaling, and other unknown signals generated from the cell membrane are differentially interpreted in the nucleus by transcription factors. The studies from mutant mice with disrupted expression of Id2, Aiolos, Fli1, E2a, and Nfkb2 revealed that one group of transcription factors, including Id2 and Aiolos (9, 10), may counteract the activities of a second subset comprising Fli1, E2a, and Nfkb2 (13–15) to promote development of one lineage over the other. IRF8 seemed to be distinct in its effects from the other transcription factors in that it normally restricts expansion of the MZ and FO B cell pools.
Because the downstream target genes of a given transcription factor can number in the thousands, it is difficult to pinpoint the exact pathways that are responsible for MZ and FO B cell lineage determination. Our microarray gene expression-profiling analysis of IRF8 CKO MZ and FO cells versus cells from WT mice revealed significant changes in expression for only 48 of 28,853 genes evaluated. This means that the IRF8-determined gene expression differences between CKO and WT cells were surprisingly limited. In addition, because the degree of changes in gene expression, even when reassessed by qPCR, was not profound for most of these genes (≤2-fold), it reinforces the idea that the effect of IRF8 deficiency on development of MZ and FO B cells is combinatorial, affecting different signaling pathways involved in many aspects of cellular function. It is worthwhile noting that although the expression levels of CD23 and CD21 were altered in IRF8-CKO FO B cells at the protein level, we did not detect significant changes at the mRNA level. This argues that IRF8 regulation of B cell maturation may be evidenced at the transcriptional and posttranscriptional levels.
Although the development of MZ and FO B cells was affected in the absence of IRF8, there was no evidence that the IRF8-deficient mature B cells of either subset were functionally deficient to any significant extent. B cells purified from IRF8-CKO and IRF8–/– mice exhibited normal levels of proliferation in response to LPS, CpG, and anti-IgM in vitro (data not shown). IRF8-CKO mice could mount normal Ab responses to TD and TI Ags in vivo. Although the proliferation of GC B cells from IRF8–/– mice was slightly less than for normal controls, as assessed by in vivo BrdU labeling (27), the histologic features of GCs were indistinguishable between the two groups (data not shown). Given the fact that IRF8 regulates several key GC factors, including BCL6 and AID (26), the relative normality of GC development in IRF8-CKO mice raises possibilities that IRF8 partners may compensate for the functions of IRF8 in its absence. This hypothesis is supported by the demonstration that mice deficient in PU.1, a well-established partner of IRF8, have no prominent mature B cell abnormalities (48). We are testing whether mice bearing conditional alleles of both genes exhibit aberrant development of GCs or other peripheral B cell subsets.
In conclusion, this study demonstrated that IRF8 deficiency was associated with increased production of MZ and FO B cells. B cell-specific disruption of IRF8 in conditional deletion mice resulted in expansion of the MZ and FO B cell compartments, demonstrating conclusively that the effect of IRF8 on the development of MZ and FO B cells is B cell autonomous. Moreover, IRF8 is required for maintaining the identity of FO B cells by regulating the expression of CD23. Although IRF8 may modulate the expansion and maturation of transitional B cells, it is not required for subsequent cellular responses to antigenic stimulation.
Acknowledgments
We thank Dr. Keiko Ozato and Dr. Prafullakumar Tailor for providing mice and Mehrnoosh Abshari for technical assistance. We also thank members of the Rocky Mountain Laboratories, Research Technologies Section of the Research Technology Branch, National Institute of Allergy and Infectious Diseases, for technical assistance.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Abbreviations used in this article
- AID
activation-induced cytidine deaminase
- BM
bone marrow
- ChIP
chromatin immunoprecipitation
- CKO
conditional knockout
- FO
follicular
- GC
germinal center
- IAD
IRF association domain
- IRF
IFN regulatory factor
- MuLV
murine leukemia virus
- MZ
marginal zone
- NP-KLH
4-hydroxy-3-nitrophenylacetyl–keyhole limpet hemocyanin
- qPCR
quantitative real-time RTPCR
- TD
T-dependent
- TI
T-independent
- WT
wild-type
Footnotes
The sequences presented in this article have been submitted to the Gene Expression Omnibus database under accession number GSE24972.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Casellas R, Shih TA, Kleinewietfeld M, Rakonjac J, Nemazee D, Rajewsky K, Nussenzweig MC. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 2.Halverson R, Torres RM, Pelanda R. Receptor editing is the main mechanism of B cell tolerance toward membrane antigens. Nat. Immunol. 2004;5:645–650. doi: 10.1038/ni1076. [DOI] [PubMed] [Google Scholar]
- 3.Retter MW, Nemazee D. Receptor editing occurs frequently during normal B cell development. J. Exp. Med. 1998;188:1231–1238. doi: 10.1084/jem.188.7.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wardemann H, Hammersen J, Nussenzweig MC. Human auto-antibody silencing by immunoglobulin light chains. J. Exp. Med. 2004;200:191–199. doi: 10.1084/jem.20040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allman D, Lindsley RC, DeMuth W, Rudd K, Shinton SA, Hardy RR. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J. Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- 6.Loder F, Mutschler B, Ray RJ, Paige CJ, Sideras P, Torres R, Lamers MC, Carsetti R. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J. Exp. Med. 1999;190:75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gururajan M, Simmons A, Dasu T, Spear BT, Calulot C, Robertson DA, Wiest DL, Monroe JG, Bondada S. Early growth response genes regulate B cell development, proliferation, and immune response. J. Immunol. 2008;181:4590–4602. doi: 10.4049/jimmunol.181.7.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, John SA, Clements JL, Percy DH, Barton KP, Garrett-Sinha LA. Ets-1 deficiency leads to altered B cell differentiation, hyper-responsiveness to TLR9 and autoimmune disease. Int. Immunol. 2005;17:1179–1191. doi: 10.1093/intimm/dxh295. [DOI] [PubMed] [Google Scholar]
- 9.Becker-Herman S, Lantner F, Shachar I. Id2 negatively regulates B cell differentiation in the spleen. J. Immunol. 2002;168:5507–5513. doi: 10.4049/jimmunol.168.11.5507. [DOI] [PubMed] [Google Scholar]
- 10.Cariappa A, Tang M, Parng C, Nebelitskiy E, Carroll M, Georgopoulos K, Pillai S. The follicular versus marginal zone B lymphocyte cell fate decision is regulated by Aiolos, Btk, and CD21. Immunity. 2001;14:603–615. doi: 10.1016/s1074-7613(01)00135-2. [DOI] [PubMed] [Google Scholar]
- 11.Wang JH, Avitahl N, Cariappa A, Friedrich C, Ikeda T, Renold A, Andrikopoulos K, Liang L, Pillai S, Morgan BA, Georgopoulos K. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- 12.Weih DS, Yilmaz ZB, Weih F. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J. Immunol. 2001;167:1909–1919. doi: 10.4049/jimmunol.167.4.1909. [DOI] [PubMed] [Google Scholar]
- 13.Quong MW, Martensson A, Langerak AW, Rivera RR, Nemazee D, Murre C. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J. Exp. Med. 2004;199:1101–1112. doi: 10.1084/jem.20031180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo F, Weih D, Meier E, Weih F. Constitutive alternative NF-kappaB signaling promotes marginal zone B-cell development but disrupts the marginal sinus and induces HEV-like structures in the spleen. Blood. 2007;110:2381–2389. doi: 10.1182/blood-2007-02-075143. [DOI] [PubMed] [Google Scholar]
- 15.Zhang XK, Moussa O, LaRue A, Bradshaw S, Molano I, Spyropoulos DD, Gilkeson GS, Watson DK. The transcription factor Fli-1 modulates marginal zone and follicular B cell development in mice. J. Immunol. 2008;181:1644–1654. doi: 10.4049/jimmunol.181.3.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 17.Holtschke T, Löhler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 18.Tailor P, Tamura T, Kong HJ, Kubota T, Kubota M, Borghi P, Gabriele L, Ozato K. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007;27:228–239. doi: 10.1016/j.immuni.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 20.Tailor P, Tamura T, Morse HC, III, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turcotte K, Gauthier S, Tuite A, Mullick A, Malo D, Gros P. A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia-like syndrome in BXH-2 mice. J. Exp. Med. 2005;201:881–890. doi: 10.1084/jem.20042170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Lee CH, Qi C, Tailor P, Feng J, Abbasi S, Atsumi T, Morse HC., III IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–4038. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S, Pathak S, Trinh L, Lu R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood. 2008;111:1396–1403. doi: 10.1182/blood-2007-08-110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma S, Turetsky A, Trinh L, Lu R. IFN regulatory factor 4 and 8 promote Ig light chain kappa locus activation in pre-B cell development. J. Immunol. 2006;177:7898–7904. doi: 10.4049/jimmunol.177.11.7898. [DOI] [PubMed] [Google Scholar]
- 26.Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou JX, Lee CH, Qi CF, Wang H, Naghashfar Z, Abbasi S, Morse HC., III IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J. Immunol. 2009;183:3188–3194. doi: 10.4049/jimmunol.0803693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi CF, Li Z, Raffeld M, Wang H, Kovalchuk AL, Morse HC., III Differential expression of IRF8 in subsets of macrophages and dendritic cells and effects of IRF8 deficiency on splenic B cell and macrophage compartments. Immunol. Res. 2009;45:62–74. doi: 10.1007/s12026-008-8032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Ye J, Arnold LW, McCray SK, Clarke SH. A VH12 transgenic mouse exhibits defects in pre-B cell development and is unable to make IgM+ B cells. J. Immunol. 2001;167:1254–1262. doi: 10.4049/jimmunol.167.3.1254. [DOI] [PubMed] [Google Scholar]
- 30.Shin DM, Shaffer DJ, Wang H, Roopenian DC, Morse HC., III NOTCH is part of the transcriptional network regulating cell growth and survival in mouse plasmacytomas. Cancer Res. 2008;68:9202–9211. doi: 10.1158/0008-5472.CAN-07-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Morse HC., III IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol. Res. 2009;43:109–117. doi: 10.1007/s12026-008-8055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 34.Bannish G, Fuentes-Pananá EM, Cambier JC, Pear WS, Monroe JG. Ligand-independent signaling functions for the B lymphocyte antigen receptor and their role in positive selection during B lymphopoiesis. J. Exp. Med. 2001;194:1583–1596. doi: 10.1084/jem.194.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuentes-Pananá EM, Bannish G, Karnell FG, Treml JF, Monroe JG. Analysis of the individual contributions of Igalpha (CD79a)- and Igbeta (CD79b)-mediated tonic signaling for bone marrow B cell development and peripheral B cell maturation. J. Immunol. 2006;177:7913–7922. doi: 10.4049/jimmunol.177.11.7913. [DOI] [PubMed] [Google Scholar]
- 36.Hayakawa K, Asano M, Shinton SA, Gui M, Wen LJ, Dashoff J, Hardy RR. Positive selection of anti-thy-1 autoreactive B-1 cells and natural serum autoantibody production independent from bone marrow B cell development. J. Exp. Med. 2003;197:87–99. doi: 10.1084/jem.20021459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayakawa K, Asano M, Shinton SA, Gui M, Allman D, Stewart CL, Silver J, Hardy RR. Positive selection of natural autoreactive B cells. Science. 1999;285:113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- 38.Heltemes LM, Manser T. Level of B cell antigen receptor surface expression influences both positive and negative selection of B cells during primary development. J. Immunol. 2002;169:1283–1292. doi: 10.4049/jimmunol.169.3.1283. [DOI] [PubMed] [Google Scholar]
- 39.Martin F, Kearney JF. Marginal-zone B cells. Nat. Rev. Immunol. 2002;2:323–335. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- 40.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat. Rev. Immunol. 2009;9:767–777. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- 41.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 42.Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 43.Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S, Owen MJ. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 44.Oyama T, Harigaya K, Muradil A, Hozumi K, Habu S, Oguro H, Iwama A, Matsuno K, Sakamoto R, Sato M, et al. Mastermind-1 is required for Notch signal-dependent steps in lymphocyte development in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:9764–9769. doi: 10.1073/pnas.0700240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, Honjo T. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- 46.Witt CM, Hurez V, Swindle CS, Hamada Y, Klug CA. Activated Notch2 potentiates CD8 lineage maturation and promotes the selective development of B1 B cells. Mol. Cell. Biol. 2003;23:8637–8650. doi: 10.1128/MCB.23.23.8637-8650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran ST, Cariappa A, Liu H, Muir B, Sgroi D, Boboila C, Pillai S. Synergism between NF-kappa B1/p50 and Notch2 during the development of marginal zone B lymphocytes. J. Immunol. 2007;179:195–200. doi: 10.4049/jimmunol.179.1.195. [DOI] [PubMed] [Google Scholar]
- 48.Polli M, Dakic A, Light A, Wu L, Tarlinton DM, Nutt SL. The development of functional B lymphocytes in conditional PU.1 knock-out mice. Blood. 2005;106:2083–2090. doi: 10.1182/blood-2005-01-0283. [DOI] [PubMed] [Google Scholar]