Background: Axonal SMN is a truncated product of the spinal muscular atrophy (SMA) disease gene SMN1.
Results: Forced expression of axonal SMN in motoneuron-like NSC34 cells modulates growth, axonogenesis, and motility.
Conclusion: Axonal SMN induces CCL2/CCL7 chemokines and the IGF-1 growth factor. CCL2 contributes to axonal SMN-induced motility and axonogenesis.
Significance: Insights into the function and underlying mechanisms with relevance for axonal SMN in SMA are provided.
Keywords: Axon, Cell Motility, Differentiation, Neurite Outgrowth, Neurons, CCL2, IGF1, SMN1, Motoneuron, Spinal Muscular Atrophy
Abstract
Spinal muscular atrophy is a fatal genetic disease of motoneurons due to loss of full-length survival of motor neuron protein, the main product of the disease gene SMN1. Axonal SMN (a-SMN) is an alternatively spliced isoform of SMN1, generated by retention of intron 3. To study a-SMN function, we generated cellular clones for the expression of the protein in mouse motoneuron-like NSC34 cells. The model was instrumental in providing evidence that a-SMN decreases cell growth and plays an important role in the processes of axon growth and cellular motility. In our conditions, low levels of a-SMN expression were sufficient to trigger the observed biological effects, which were not modified by further increasing the amounts of the expressed protein. Differential transcriptome analysis led to the identification of novel a-SMN-regulated factors, i.e. the transcripts coding for the two chemokines, C-C motif ligands 2 and 7 (CCL2 and CCL7), as well as the neuronal and myotrophic factor, insulin-like growth factor-1 (IGF1). a-SMN-dependent induction of CCL2 and IGF1 mRNAs resulted in increased intracellular levels and secretion of the respective protein products. Induction of CCL2 contributes to the a-SMN effects, mediating part of the action on axon growth and random cell motility, as indicated by chemokine knockdown and re-addition studies. Our results shed new light on a-SMN function and the underlying molecular mechanisms. The data provide a rational framework to understand the role of a-SMN deficiency in the etiopathogenesis of spinal muscular atrophy.
Introduction
Spinal muscular atrophy (SMA)3 is a disease causing selective motoneuron death. SMA affects the pediatric population, and it is the leading genetic cause of infant mortality (1). Loss of telomeric SMN1 (Survival of Motor Neuron 1), the pathogenetic gene, is at the basis of SMA, whereas SMN2, the duplicated centromeric gene, modulates the severity of this disease (2). SMN1 codes for a functional protein, full-length SMN (FL-SMN), and the primary product of SMN2 is Δ7-SMN, an unstable protein of uncertain significance (3). FL-SMN is a ubiquitous protein localizing to the nucleus and cytoplasm of many cell types (4). It is well established that FL-SMN acts as an assembly factor for small nuclear ribonucleoprotein particles or small nucleolar ribonucleoproteins involved in mRNA splicing (2, 5). However, it is unclear how reduced levels of a ubiquitous protein like FL-SMN lead to the selective degeneration of motoneurons in SMA.
We demonstrated that FL-SMN is not the sole product of the SMN1 gene, which generates a second and much less abundant protein via an alternative splicing event resulting in retention of intron 3 (6). The alternative protein product is shorter than FL-SMN because of the presence of an in-frame stop codon located in intron 3. The same splicing variant is observed in mice and rats, indicating evolutionary conservation of the transcript and corresponding protein (7). Unlike FL-SMN, expression of this novel gene product is relatively tissue-specific and temporally restricted. In fact, the a-SMN mRNA and protein are detectable in spinal cord motoneurons and some peripheral tissues, such as liver and heart, only during the late phases of embryogenesis and early postnatal life (6). In the motoneuron, the SMN splicing variant localizes to axons and is excluded from the nucleus. For this reason we named the protein a-SMN. It is conceivable that loss of function of human a-SMN contributes to the pathogenesis of this disease (8).
In this study, we describe the development of a cellular model for the expression of a-SMN in NSC34 motoneuron-like cells. The model was used to support the functional significance of a-SMN in axonogenesis and to establish an important role for the protein in the control of cell motility. In addition, whole-genome gene expression studies permitted the identification of IGF1 (insulin-like growth factor-1), CCL2, and CCL7 (C-C motif ligands 2 and 7) as factors associated with a-SMN expression. Functional studies performed on CCL2 indicate that the chemokine contributes to the axonogenic and pro-motility action of a-SMN.
EXPERIMENTAL PROCEDURES
Cell Cultures
The NCS34 cell line (9) was maintained in low glucose (1 g/liter) DMEM (Invitrogen) supplemented with 5% TET System-approved fetal calf serum (Clontech). To obtain stable deposition of neuronal axons, cells were grown in culture dishes pre-coated for 1 h with Matrigel matrix basement membrane (200 μg/ml, BD Biosciences) (10). The TR4 clone was grown in the presence of 10 μg/ml blasticidin S (Invitrogen), whereas a-SMN-expressing clones were cultured in the presence of 10 μg/ml blasticidin S and 50 μg/ml Zeocin (Invitrogen). Induction of a-SMN was performed in medium containing 1 μg/ml TET without addition of other antibiotics.
Plasmid Generation and Transfections
The cDNA coding for human a-SMN (6) was amplified using the following oligonucleotides: forward primer, 5′-ctaagcttatggcgatgagcagcggcggca-3′ (consisting of the underlined HindIII site, placed upstream of the sequence corresponding to nucleotides 164–185 of NM_000344); reverse primer, 5′-agtctagagcaggtttttgttataagggt-3′ (consisting of an XbaI site placed upstream of the sequence complementary to nucleotides 119–140 of SMN1 intron 3). The resultant cDNA fragment encoding a-SMN was digested with HindIII/XbaI and inserted into the pcDNA4/TO plasmid (Invitrogen) digested with the appropriate enzymes. The human FL-SMN cDNA has already been described (6). Transient transfection of NSC34 cells with the human a-SMN and FL-SMN cDNAs was performed as described (6). Transfections were performed with the FuGENE HD transfection reagent (Roche Applied Science), according to the manufacturer's instructions.
Generation of a-SMN-Expressing NSC34-derived Cellular Clones
For the forced expression of human a-SMN, we used a TET-dependent system and an approach involving a two-step selection strategy. The NSC34 cell line was stably transfected with a TET repressor plasmid construct (pcDNA6/TR, Invitrogen). We obtained a number of cell clones characterized by resistance to blasticidin S that were subsequently isolated by serial dilution. Four clones were evaluated for the expression of the TET repressor by transient transfection of an appropriate TET-dependent β-galactosidase reporter construct. The cell clone TR4 was chosen for further studies, given the low level of reporter expression in basal conditions and significant induction upon challenge with TET. TR4 was transfected with a construct containing the entire coding region of the human a-SMN cDNA under the control of a promoter bearing TET repressor-binding sites. After selection for Zeocin resistance, numerous clones were isolated by serial dilution.
Antibodies
The anti-peptide polyclonal antibody raised against the C-terminal region of the human a-SMN (number 910) was prepared in rabbits by NeoMPS (Strasbourg, France) and used at 1:500 dilution in all the experiments. The following antibodies were also used: mouse anti-SMN (clone 8) from BD Biosciences (diluted 1:25,000), directed against the N-terminal region of the SMN protein; mouse anti-actin from Chemicon (Temecula, CA; diluted 1:5000 for Western blot and 1:500 for IF experiments); mouse anti-α-tubulin (Sigma; diluted 1:500); mouse anti-JIP-1 from Santa Cruz Biotechnology (Santa Cruz, CA; diluted 1:200); mouse anti-CCL2 and mouse anti-IGF1 from R&D Systems (Minneapolis, MN; diluted 1:20).
Morphological Analysis and IF Experiments
For the evaluation of cellular morphology, cells were fixed with 4% paraformaldehyde, 4% sucrose or with 4% paraformaldehyde, 2% glutaraldehyde for 30 min at room temperature. For IF, NSC34 cells were rinsed three times in low salt (150 mm NaCl) 10 mm phosphate buffer, pH 7.4, and three times in high salt (500 mm NaCl) 20 mm phosphate buffer, pH 7.4, to avoid cross-linking with nonspecific epitopes. Subsequently, fixed cells were incubated in goat serum diluted in 20 mm phosphate buffer, pH 7.4, containing 3% goat serum, 0.1% Triton X-100, and 500 mm NaCl. The primary antibodies were incubated overnight at 4 °C. The secondary Alexa Fluor 546 and 488 antibodies were incubated for 1 h at room temperature. After all experiments, cells were repeatedly rinsed with FluorSave (Calbiochem) and examined with a confocal microscope (Radiance 2100, Bio-Rad; Olympus Fluoview, Olympus, Segrate, Italy). For fluorescence intensity analysis of JIP-1, 15 confocal nonoverlapping images were chosen at ×400 magnification from triplicate experiments. Background fluorescence intensity was subtracted. Fluorescence was quantified using Image-ProPlus software and expressed as mean ± S.D. To quantify the surface area occupied by growth cones, NSC34 and a-SMN81 cells were labeled with DNase Alexa Fluor® 488 (Invitrogen), binding and highlighting G-actin (11), for 15 min at room temperature. Confocal images at ×600 were taken from at least 30 growth cones in triplicate experiments and analyzed with the Image-ProPlus software. Data were statistically analyzed by one-way ANOVA followed by the Bonferroni post hoc comparison test. For the atomic force microscopy experiments, cells were grown on glass bottom dishes (WillCo-Wells, model GWSt 5040) for 72 h, fixed in 2% glutaraldehyde phosphate buffer at pH 7.4 (for 30 min), gently washed in bi-distilled water, and stored in 0.6% glutaraldehyde solution at 4 °C. Cells were analyzed using a BioscopeII atomic force microscope equipped with the Digital Instruments NanoScope III (Veeco, Santa Barbara, CA) and integrated with the inverted optical microscope Axio Observer A1 (Zeiss, Oberkochen, Germany). Atomic force microscopy images (512 × 512 pixels) were recorded at room temperature in contact mode (in both height and deflection modes) with nonconducive silicon nitride tips Model MLCT (Veeco, Santa Barbara, CA), with typical nominal spring constants ranging from 0.01 to 0.60 n/m and nominal resonance frequency ranging from 7 to 125 kHz. The scan range was 65 μm, and the scan rate was 0.5 μm/s. Randomly selected cells were scanned and qualitatively analyzed to evaluate morphological features of cell bodies, neurites, and growth cones.
Gene Expression Analysis and Validation Studies
Total RNA was prepared using miRNeasy mini kit (Qiagen, Valencia, CA). Transcriptome analysis was performed with the use of Whole Mouse Genome Oligo Microarray kit (Agilent, Palo Alto, CA), and the results were deposited in the GEO data base (accession: E-MEXP-3402). The levels of the a-SMN transcript were measured by SYBR Green-based real time PCR, using amplimers specific for the a-SMN mRNA as follows: 5′-ttacccagctaccattgcttcaat-3′ (nucleotides 487–509 in NM_000344) and 5′-tagtctctgcttccagaaattgaat-3′ (complementary to nucleotides 23–47 of intron 3, numbering the 5′-end nucleotide of intron 3 as 1). Validation of the microarray results was performed with the following TaqMan probes: Ccl2, Mm00441242_m1; Ccl7, Mm01308393_g1; Igf1, Mm00439560_m1; c-Myc, Mm00487804_m1; Actb, Mm01205647_g1 (Applied Biosystems, Carlsbad, CA).
Measurement of CCL2 and IGF1 Proteins
Cells were lysed, and the protein extracts were used for Western blot analysis according to a routine procedure detailed elsewhere (6). The levels of CCL2 and IGF1 proteins in the cell lysates and the conditioned medium were measured by ELISA using mouse CCL2/JE/MCP-1 and mouse/rat IGF1 immunoassay kits (R&D Systems), respectively.
Silencing Experiments
Synthetic anti-a-SMN (target sequence located in intron 3, 5′-tgtccattcctgaaagtgg-3′) and nontargeting siRNAs were purchased from Dharmacon (ThermoFisher Scientific, Lafayette, CO). Transfection (1 μm siRNAs) was performed following the manufacturer's instructions. Synthetic anti-CCL2 siRNAs (Ccl2MSS276974(3_RNAI) and Ccl2MSS276976(3_RNAI)) and the relative control siRNA (catalog no. 12935-400, high GC Duplex) were purchased from Applied Biosystems and transfected with Lipo2000 (Invitrogen) according to the instructions of the manufacturer. For the experiments involving cell motility with anti-CCL2 siRNAs, cells were co-transfected with the green fluorescent protein (GFP) expressing plasmid pEGFP-N1 (Clontech). After 72 h, the efficiency of RNA interference was monitored by RT/real time PCR using the TaqMan probes described above.
Single Cell Motility and Measurement of Axon Length
Single cell motility assays were performed on Matrigel (200 μg/ml)-coated substrate (12), as described (13), using the Imaging Station Cell[caret]R (Olympus, Segrate, Italy). Image sequences, converted to.tif format files, were analyzed with the image analysis software ImageJ (W. Rasband, National Institutes of Health, Bethesda). In particular, each cell in the field was tracked individually using the “Manual Tracking” plug-in distributed with the ImageJ software. At 24 and 60 h, the length of neurites was manually measured outlining each neurite with the free-hand line tool of ImageJ.
Statistical Analysis
Unless otherwise indicated, statistical evaluation of the quantitative results was performed according to one-way ANOVA followed by post-hoc test.
RESULTS
Characterization of NSC34-derived Cell Lines Expressing Human a-SMN
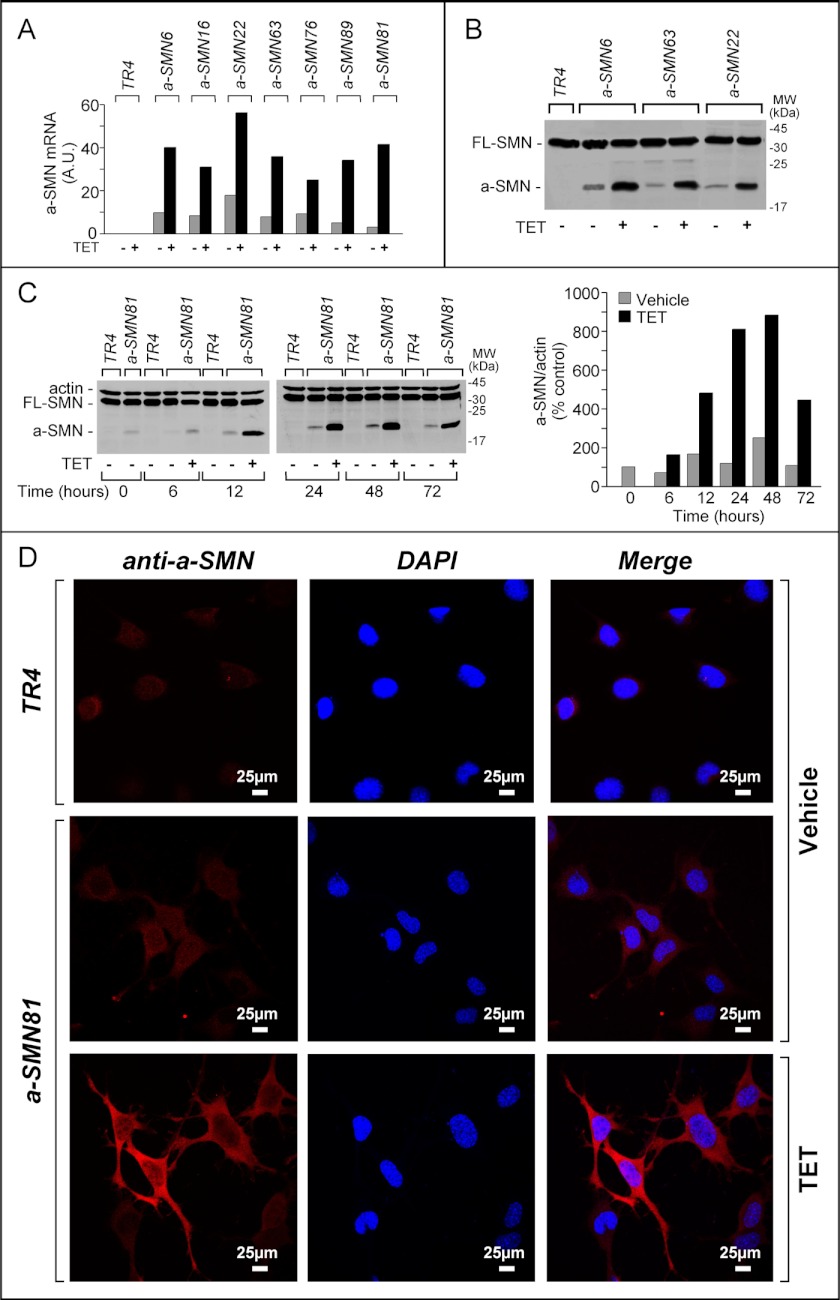
NSC34 is an immortalized murine hybrid neuroblastoma/spinal cord cell line with a motoneuron-like phenotype (9). NSC34 cells express readily measurable amounts of endogenous FL-SMN upon Western blot analysis (6, 8). However, in the same experimental conditions, we were unable to determine detectable levels of the a-SMN counterpart.4 For these reasons, we used NSC34 cells to obtain regulated expression of human a-SMN and to study its biological function. We established a cellular model showing basal expression and TET-dependent induction of the protein. First, we generated the NSC34-derived TR4 cell line, which is characterized by stable insertion of the TET repressor. Similar to NSC34, TR4 cells showed a motoneuron-like morphology (data not shown). Subsequently, a human a-SMN expression plasmid was introduced in TR4 cells, and several derived clones were isolated by serial dilution. We evaluated the expression of the a-SMN mRNA in basal conditions and after treatment with TET for 24 h in numerous clones. Seven representative clones are shown in Fig. 1A. Detectable amounts of human a-SMN mRNA were observed in all the clones analyzed even in the absence of TET. TET up-regulated the expression of the a-SMN transcript and protein in all the clones tested (Fig. 1, B and C). The kinetics of a-SMN induction were very similar in all clones, as illustrated for a-SMN81 cells (Fig. 1C). TET-dependent induction of a-SMN protein was evident after 12 h, peaked at 48 h, and diminished by 72 h. IF studies performed in the a-SMN81 cell line confirmed the Western blot data, revealing a-SMN labeling in basal conditions and TET-dependent induction of the a-SMN signal (Fig. 1D). The a-SMN protein showed the expected intracellular localization (6), which was limited to the soma/neuritis (Fig. 1D). This is at variance with what has been described for FL-SMN whose localization extends to the nucleus.
FIGURE 1.
TET-dependent induction of the a-SMN transcript and protein. A, indicated a-SMN-expressing clones and the parental cell line, TR4, were grown to confluency. Cells were subsequently treated with medium in the presence/absence of TET for 24 h. Total RNA was isolated and used for RT/real time PCR analysis. The relative quantity of a-SMN transcript is expressed in arbitrary units (A.U.) following normalization for the actin transcript. B, indicated a-SMN-expressing clones and TR4 cells were treated with or without TET for 24 h. Protein extracts were separated on 12% denatured PAGE and subjected to Western blot analysis using anti-SMN antibodies (clone 8). MW, molecular weight. C, a-SMN81 clone and the TR4 counterpart were treated with TET for the indicated amount of time, and protein extracts were analyzed by Western blot as in B. The densitometric quantification of the a-SMN bands following normalization for actin is illustrated by the bar graph on the right. D, a-SMN81 and TR4 clones were treated with or without TET for 24 h and were analyzed by means of confocal IF after staining with anti-human a-SMN antibodies (No. 910) (left panels) and counterstaining with the nucleus-specific dye DAPI (middle panels). Merged images are shown on the right.
a-SMN Causes Growth Inhibition, Stimulates Axonogenesis, and Increases Cell Motility
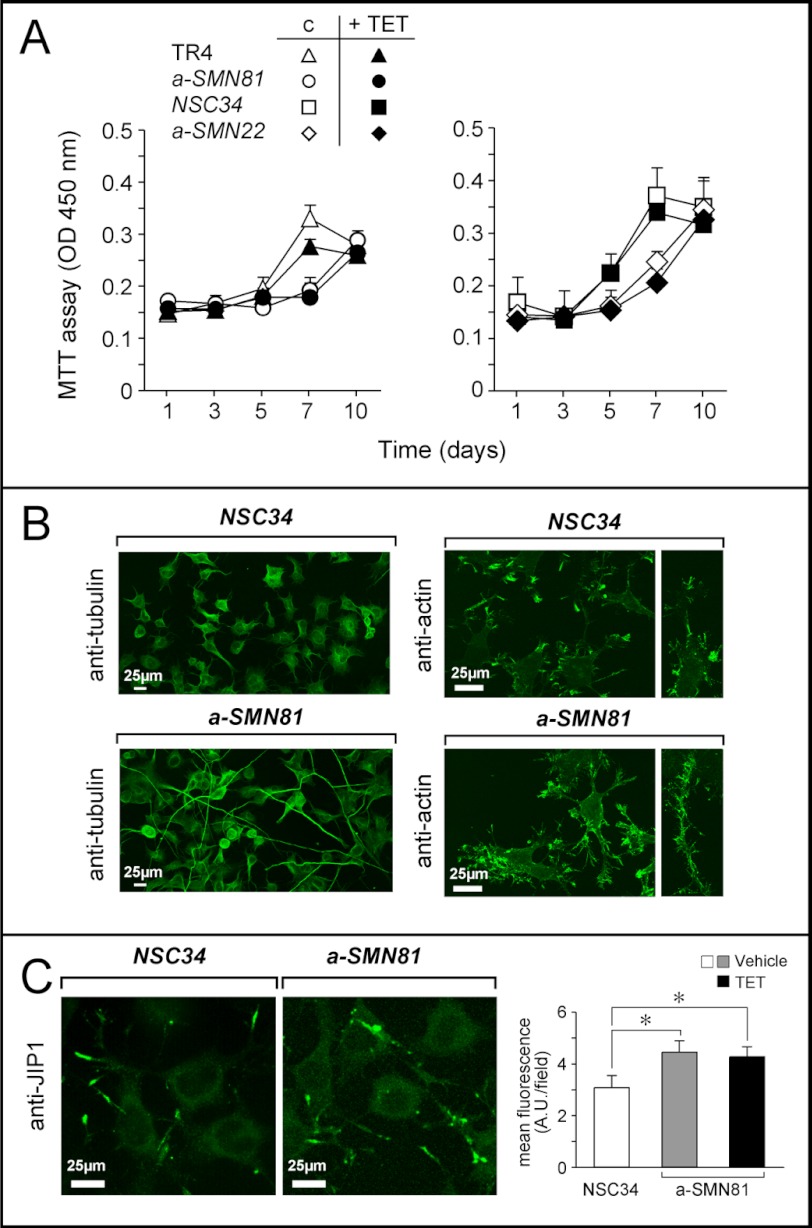
We evaluated the consequences of a-SMN expression on various aspects of cellular homeostasis. Initially, we compared the growth of a-SMN22 and a-SMN81 clones, two extremes in terms of basal a-SMN levels and TET-dependent induction, with that of the NSC34 and TR4 controls. We performed these experiments upon serum starvation (0.5%) and subsequent synchronization by serum re-addition (10%). After a lag time of ∼24–36 h, NSC34 and TR4 cells started to grow and reached confluency by 7 days (Fig. 2A). In contrast, the a-SMN22 and a-SMN81 cells were characterized by a slower growth rate and did not reach confluency even 10 days after re-addition of serum. The growth of the two clones and the parental cell lines was left largely unaffected by treatment with TET. This suggests that the amounts of a-SMN expressed in basal conditions are enough to cause maximal reduction of cell growth.
FIGURE 2.
Growth and morphology of the a-SMN expressing clones. A, NSC34, TR4, a-SMN81, and a-SMN22 cells were grown in medium containing 0.5% serum for 3 days. Subsequently, cells were trypsinized and seeded at 20,000 cells/ml in medium containing 5% serum. Cell growth, in the presence/absence of TET, was followed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Each value is the mean ± S.D. of three separate cultures. B and C, NSC34 and a-SMN81 cells were grown in basal conditions. The morphological features of the cells were analyzed by means of confocal IF with anti-α-tubulin (B, left panel), anti-actin (B, right panel), and anti-JIP-1 (C) antibodies. The bar graph on the right shows quantification of the JIP-1 fluorescence results. *, significantly different (p < 0.05, one-way ANOVA followed by the Bonferroni post hoc comparison test).
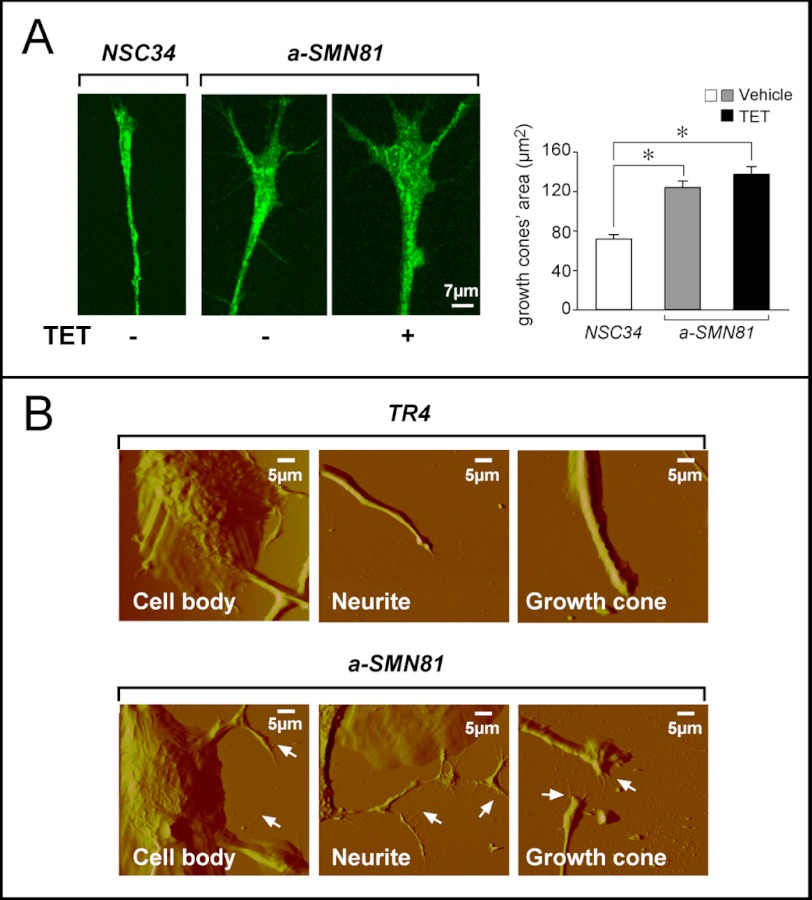
As growth inhibition is often accompanied by cell differentiation (13, 14), we evaluated whether basal expression of a-SMN was associated with increased neuronal maturation. All the a-SMN-expressing clones, including a-SMN81, presented a morphology characterized by signs of neuronal differentiation, with increases in axonal thickness, elongation, and branching relative to the NSC34 or TR4 controls. As illustrated for a-SMN81 cells grown in basal conditions (Fig. 2, B and C), these morphological changes were particularly evident upon IF. IF studies conducted with specific anti-tubulin and anti-actin antibodies demonstrated the presence of longer and more branched neurites in a-SMN81 than in NSC34 cells (Fig. 2B). Increased length and branching of axons was paralleled by increased complexity of growth cones, as indicated by the specific markers, JIP-1 (Fig. 2C, left panel) (15) and G-actin (Fig. 3A, left panel) (16, 17). JIP-1-associated fluorescence was significantly higher in growth cones of a-SMN81 cells relative to the NSC34 controls (Fig. 2C, right panel). Induction of a-SMN by TET did not affect JIP-1 levels. After staining for G-actin, quantitative determination of growth cone size revealed that the areas occupied by these structures were significantly larger in a-SMN81 relative to NSC34 cells (Fig. 3A, right panel). Addition of TET to the medium of growing a-SMN81 cells did not alter this parameter significantly. Sustained axonogenesis of a-SMN-expressing clones was confirmed by atomic force microscopy analysis, comparing a-SMN81 and TR4 cells (Fig. 3B). More numerous and thicker filopodia emerging from the cell body, as well as more complex neurites and growth cones, were observed in the a-SMN81 clone.
FIGURE 3.
Morphology of axons and growth cones in NSC34, TR4, and a-SMN81 cells. A, to determine the surface area occupied by growth cones, NSC34 and a-SMN81 cells grown in the absence or presence of TET, as indicated, were stained for G-actin for 15 min at room temperature. Confocal images were taken from at least 30 growth cones in triplicate experiments and analyzed with the Image-ProPlus software. Left panel, representative images. Right panel, quantification of the growth cone surface area. Data were analyzed by one-way ANOVA followed by the Bonferroni post hoc comparison test. *, p < 0.05. B, representative atomic force micrographs of TR4 and a-SMN81 cells illustrate the morphological features of cell bodies, neurites, and growth cones (arrows).
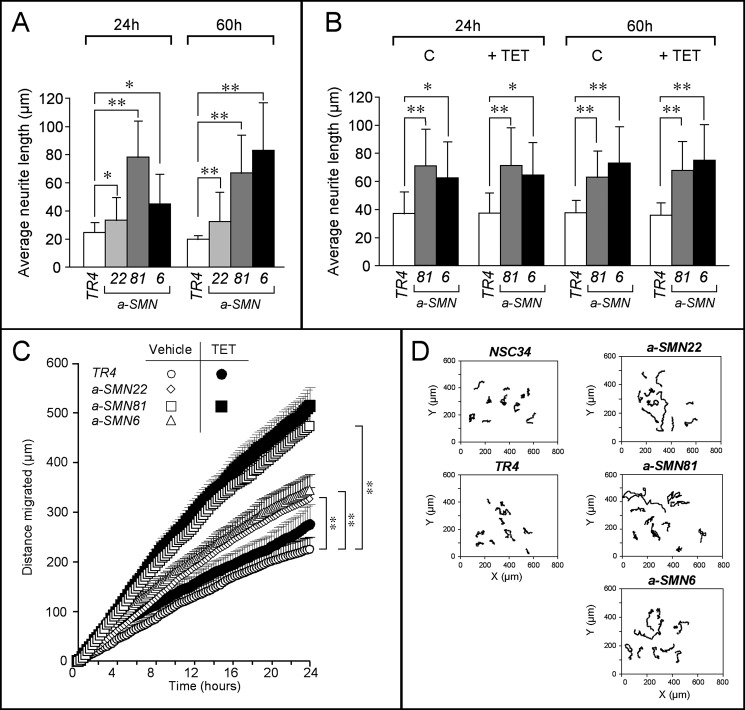
We quantified the axonogenic effect of a-SMN expression by image analysis. The average axon length determined in a-SMN22, a-SMN81, and a-SMN6 cells grown in basal conditions was superior to that determined in the TR4 controls (Fig. 4A). As exemplified in the case a-SMN81 and a-SMN6 cells, TET caused no further increase in average axon length at the two time points considered (Fig. 4B).
FIGURE 4.
Axonogenesis and cell motility of the a-SMN-expressing clones. TR4, a-SMN6, and a-SMN81 clones were seeded at 3000 cells/ml in the presence/absence of TET. Neurite length (after 24 and 60 h of treatment) and random cell motility were determined by time-lapse microscopy. A and B, average neurite length was calculated on at least 40 individual cells per each experimental point. C, random cell motility was also calculated on at least 40 individual cells per each experimental point. D, migration pathway of ten representative cells/each group is illustrated. **, statistically significant according to two-way ANOVA (p < 0.01); *, statistically significant according to two-way ANOVA (p < 0.05).
Axon outgrowth and cell motility are often associated processes (18), both involving reorganization of tubulin and actin, the two main components of the cytoskeleton (19, 20). Given the changes induced by a-SMN on tubulin and actin organization (see Fig. 2B), we determined the effect of a-SMN expression on cell motility. In basal conditions, the a-SMN22, a-SMN81, and a-SMN6 clones were all characterized by increased motility relative to the TR4 (Fig. 4, C and D) and the NSC34 controls (Fig. 4D). a-SMN81 was the clone characterized by the highest level of motility, although it was not the clone expressing the largest a-SMN amounts, probably as a result of other and as yet to be defined concomitant factors. Addition of TET to the medium did not lead to any further enhancement of cell motility, as illustrated for a-SMN81 cells and TR4 controls. The increase in cell motility is the consequence of an effect on the random component, as indicated by the lack of directionality observed in cell movement (Fig. 4D).
Taken together our data demonstrate that a-SMN expression exerts effects on cell growth, axonogenesis, and cell motility. These effects are already maximal in basal conditions and are not modified by TET addition. Within the range of the a-SMN expression levels determined in our clones, the results obtained are consistent with a threshold effect.
Human a-SMN Expression Perturbs the NSC34 Transcriptome
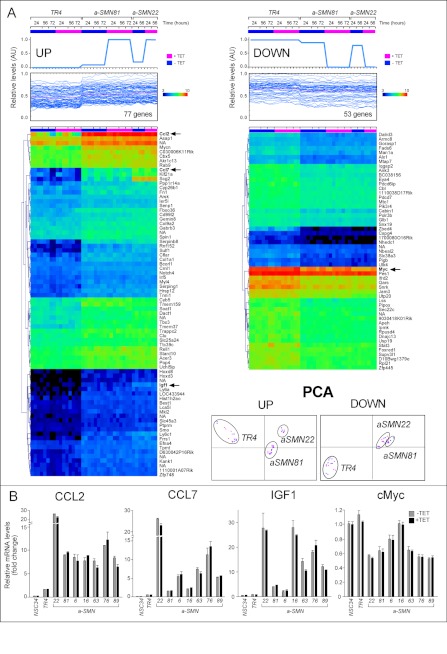
To identify genes of functional significance for the growth inhibitory, pro-axonogenic, and pro-motility action of a-SMN, we defined the transcriptomic profiles of TR4, a-SMN81, and a-SMN22 cells grown for different amounts of time (24, 56, and 72 h) in the absence/presence of TET, using whole-genome gene expression microarrays (Fig. 5A). Consistent with the functional data, we did not identify genes significantly regulated by TET in a-SMN81 and a-SMN22 cells. This is similar to what was observed in TR4 cells. Thus, we refined our analysis, looking for genes whose expression was quantitatively and directly or inversely correlated to a-SMN protein levels in the various conditions considered, using a template-matching algorithm (Pavlidis Template Matching). Pavlidis Template Matching resulted in the identification of genes whose expression was consistently higher (77 genes) or lower (53 genes) in a-SMN81 and a-SMN22 relative to the control TR4 cells at all the time points analyzed (Fig. 5A; up-regulated genes, left panel, and down-regulated genes, right panel). However, we did not find transcripts whose levels were quantitatively and strictly correlated to the amounts of the a-SMN transgene product expressed (Table 1 lists the top 20 up- and down-regulated genes). In fact, up-regulation or down-regulation of all these genes was already maximal in untreated a-SMN81/a-SMN22 cells, and expression was not influenced by TET or by time. The observations were confirmed by Principal Component Analysis, which showed remarkable differences in the transcriptomic profiles of TR4 and a-SMN81 or a-SMN22 cells, regardless of TET treatment (Fig. 5A, bottom right panel). This is similar to what was observed for the effects exerted by a-SMN on growth, axonogenesis, and motility. Taken together, our gene expression data are consistent with the idea that amounts of a-SMN above a threshold, which is overcome by basal expression of the transgene, are enough to produce the observed perturbations in the transcriptome.
FIGURE 5.
Transcriptome profiles of TR4, a-SMN22, and a-SMN81 cells and validation of the results. TR4, a-SMN22, and a-SMN81 cells were treated for 24, 56, and 72 h with or without TET. Total RNA was extracted, labeled, and used to hybridize whole-genome gene expression microarrays. A, upper panel, panels (left = dose-dependent up-regulation; right = dose-dependent down-regulation) show the templates used for Pavlidis template matching analysis that correspond to the a-SMN protein levels determined by Western blot (see Fig. 1C). Middle panel, expression profiles of the genes significantly matching the templates (p < 0.01) are shown. Lower panel, heat maps of the same genes up- or down-regulated in a-SMN81 and a-SMN22 clones are illustrated. Bottom right panel, principal component analysis (PCA) of the transcriptomes determined in TR4, a-SMN22 and a-SMN81 cells is presented (magenta, without TET; blue, with TET). AU, absorbance units. B, relative mRNA levels of CCL2, CCL7, IGF1, and c-Myc determined in the indicated a-SMN expressing clones, as well as NSC34 and TR4 control cells, were validated by RT/real time PCR. The data are expressed in fold change relative to the NSC34 control taken as 1. The results were obtained from triplicate cultures and are expressed as the means ± S.D.
TABLE 1.
Top over- and underexpressed genes in a-SMN expressing clones
Table contains the quantitative results obtained after microarray gene expression profiling of a-SMN81, a-SMN22, and TR4 cells. Data are expressed in log2 of the intensities and represent 8–16 samples for each cell line taken at different times in the absence and presence of TET (columns 3–8). The last two columns show the fold change in the two a-SMN expressing clones relative to the control TR4 clone. The top 20 genes are concordantly over- and underexpressed in a-SMN81 and a-SMN22 clones are shown.
| Symbol | Name |
TR4 |
a-SMN81 |
a-SMN22 |
a-SMN81 |
a-SMN22, fold versus TR4 | |||
|---|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | Mean | S.D. | fold vs TR4 | |||
| Scg2 | Secretogranin II | 5.7 | 0.4 | 8.9 | 0.7 | 12.2 | 0.1 | 9.2 | 94.7 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 6.0 | 0.8 | 9.4 | 0.5 | 10.7 | 0.5 | 10.7 | 26.0 |
| Kif21a | Kinesin family member 21A | 6.4 | 1.0 | 9.6 | 0.6 | 10.6 | 0.2 | 9.0 | 18.4 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 10.8 | 1.5 | 14.1 | 0.4 | 14.5 | 0.4 | 10.2 | 12.7 |
| Serpinb8 | Serine peptidase inhibitor, clade B- 8 | 5.7 | 0.8 | 8.7 | 0.3 | 9.0 | 0.2 | 7.6 | 9.6 |
| Sulf1 | Sulfatase 1 | 5.1 | 0.5 | 7.6 | 0.3 | 7.9 | 0.3 | 5.5 | 7.0 |
| Rnf152 | Ring finger protein 152 | 5.1 | 0.4 | 7.3 | 0.4 | 7.9 | 0.2 | 4.5 | 7.1 |
| Ly6c1 | Lymphocyte antigen 6 complex, C1 | 4.4 | 0.2 | 6.2 | 0.4 | 7.4 | 0.2 | 3.5 | 7.9 |
| Igf1 | Insulin-like growth factor 1 | 4.3 | 0.2 | 5.5 | 0.3 | 6.9 | 0.2 | 2.3 | 6.0 |
| Ppp1r14a | Protein phosphatase 1, regulatory subunit domain 14A | 6.8 | 0.6 | 8.2 | 0.2 | 8.9 | 0.4 | 2.5 | 4.3 |
| Ly6a | Lymphocyte antigen 6 complex, A | 4.3 | 0.3 | 5.7 | 0.4 | 6.3 | 0.3 | 2.7 | 4.1 |
| Tmem37 | Transmembrane protein 37 | 8.4 | 0.4 | 10.1 | 0.4 | 10.2 | 0.2 | 3.2 | 3.4 |
| Fn1 | Fibronectin 1 | 7.1 | 0.5 | 8.9 | 0.5 | 8.8 | 0.3 | 3.3 | 3.2 |
| Tmem159 | Transmembrane protein 159 | 9.0 | 0.4 | 10.9 | 0.3 | 10.4 | 0.1 | 3.7 | 2.7 |
| Frrs1 | Ferric chelate reductase 1 | 4.9 | 0.5 | 6.1 | 0.4 | 6.8 | 0.4 | 2.5 | 3.9 |
| Cflar | CASP8 and FADD-like apoptosis regulatory domain | 6.2 | 0.6 | 7.9 | 0.3 | 7.5 | 0.1 | 3.2 | 2.5 |
| Hoxd3 | Homeobox D3 | 4.0 | 0.3 | 5.3 | 0.3 | 5.6 | 0.3 | 2.5 | 3.0 |
| Cyb5 | Cytochrome b5 | 9.1 | 0.5 | 10.7 | 0.2 | 10.3 | 0.1 | 3.0 | 2.2 |
| LOC433944 | Hypothetical gene (AK083542) | 4.5 | 0.4 | 5.9 | 0.2 | 5.8 | 0.3 | 2.7 | 2.5 |
| Col1a1 | Collagen, type I, α1 | 6.5 | 0.5 | 8.0 | 0.2 | 7.7 | 0.2 | 2.9 | 2.3 |
| Cabin1 | Calcineurin-binding protein 1 | 9.1 | 0.2 | 8.3 | 0.4 | 8.1 | 0.2 | 0.6 | 0.5 |
| Unknown | Unknown (probe ID A_52_P309084) | 11.4 | 0.2 | 10.4 | 0.2 | 10.5 | 0.1 | 0.5 | 0.5 |
| Myc | Myelocytomatosis oncogene | 13.5 | 0.2 | 12.8 | 0.3 | 12.4 | 0.2 | 0.6 | 0.5 |
| Alx1 | ALX homeobox 1 | 7.8 | 0.1 | 6.7 | 0.2 | 7.0 | 0.2 | 0.5 | 0.6 |
| Dnajc13 | DnaJ (Hsp40) homolog, C-13 | 10.9 | 0.4 | 10.0 | 0.3 | 9.9 | 0.2 | 0.5 | 0.5 |
| Pdcd6ip | Programmed cell death 6 int protein | 10.2 | 0.3 | 9.3 | 0.3 | 9.1 | 0.3 | 0.6 | 0.5 |
| Stat3 | Signal transducer and activator of transcription 3 | 11.6 | 0.4 | 10.5 | 0.2 | 10.7 | 0.2 | 0.5 | 0.5 |
| Eya4 | Eyes absent 4 homolog (Dm) | 10.2 | 0.4 | 9.2 | 0.2 | 9.1 | 0.2 | 0.5 | 0.5 |
| Nhedc1 | Na+/H+ exchanger domain-containing 1 | 5.3 | 0.3 | 4.4 | 0.2 | 4.2 | 0.3 | 0.5 | 0.5 |
| Unknown | Unknown (probe ID A_52_P1101021) | 11.0 | 0.2 | 10.2 | 0.3 | 9.7 | 0.1 | 0.6 | 0.4 |
| Lss | Lanosterol synthase | 10.7 | 0.1 | 9.7 | 0.5 | 9.7 | 0.2 | 0.5 | 0.5 |
| Ulk4 | Unc-51-like kinase 4 (Caenorhabditis elegans) | 6.7 | 0.4 | 5.6 | 0.2 | 5.5 | 0.3 | 0.5 | 0.4 |
| Man1a | Mannosidase 1, α | 8.7 | 0.3 | 7.5 | 0.2 | 7.6 | 0.3 | 0.4 | 0.5 |
| Unknown | Unknown (probe ID A_52_P144286) | 10.0 | 0.2 | 8.9 | 0.2 | 8.8 | 0.1 | 0.5 | 0.4 |
| Iqgap2 | IQ motif containing GTPase act protein 2 | 10.2 | 0.3 | 9.0 | 0.4 | 9.0 | 0.2 | 0.4 | 0.4 |
| Mtap7 | Microtubule-associated protein 7 | 8.1 | 0.5 | 6.8 | 0.3 | 6.8 | 0.1 | 0.4 | 0.4 |
| Dalrd3 | DALR anticodon binding domain-containing | 8.5 | 0.2 | 7.1 | 0.3 | 7.3 | 0.2 | 0.4 | 0.4 |
| 1700080O16Rik | RIKEN cDNA 1700080O16 gene | 5.6 | 0.4 | 4.3 | 0.3 | 4.1 | 0.3 | 0.4 | 0.4 |
| Slc38a3 | Solute carrier family 38, member 3 | 7.3 | 0.3 | 6.0 | 0.4 | 5.7 | 0.2 | 0.4 | 0.3 |
| Unknown | Unknown (probe ID A_51_P453158) | 7.8 | 0.7 | 4.6 | 0.3 | 4.1 | 0.4 | 0.1 | 0.1 |
We focused our attention on four genes because of their potential significance for the a-SMN-dependent effects on cell growth, axonogenesis, and motility (Ccl2, Ccl7, Igf1, and c-Myc). The two chemokines, CCL2 and CCL7, possess chemoattractant properties (21) and control neuronal development as well as repair from injury (22, 23). IGF1 regulates the growth and survival of neuronal cells (24, 25). c-myc (v-Myc myelocytomatosis viral related oncogene) is a well known protein involved in cell proliferation (26). To verify the association between the expression of a-SMN and these genes, we used RT/real time PCR to compare the levels of the four transcripts in a larger panel of cell clones (Fig. 5B). Increased levels of CCL2, CCL7, and IGF1 were present in all the a-SMN-expressing clones considered, and reduced cMyc expression was found in six out of seven clones. In all cases, mRNA levels were not influenced by treatment with TET, further confirming the a-SMN threshold effect hypothesized on the basis of the microarray results.
a-SMN-induced Expression of CCL2, CCL7, and IGF1
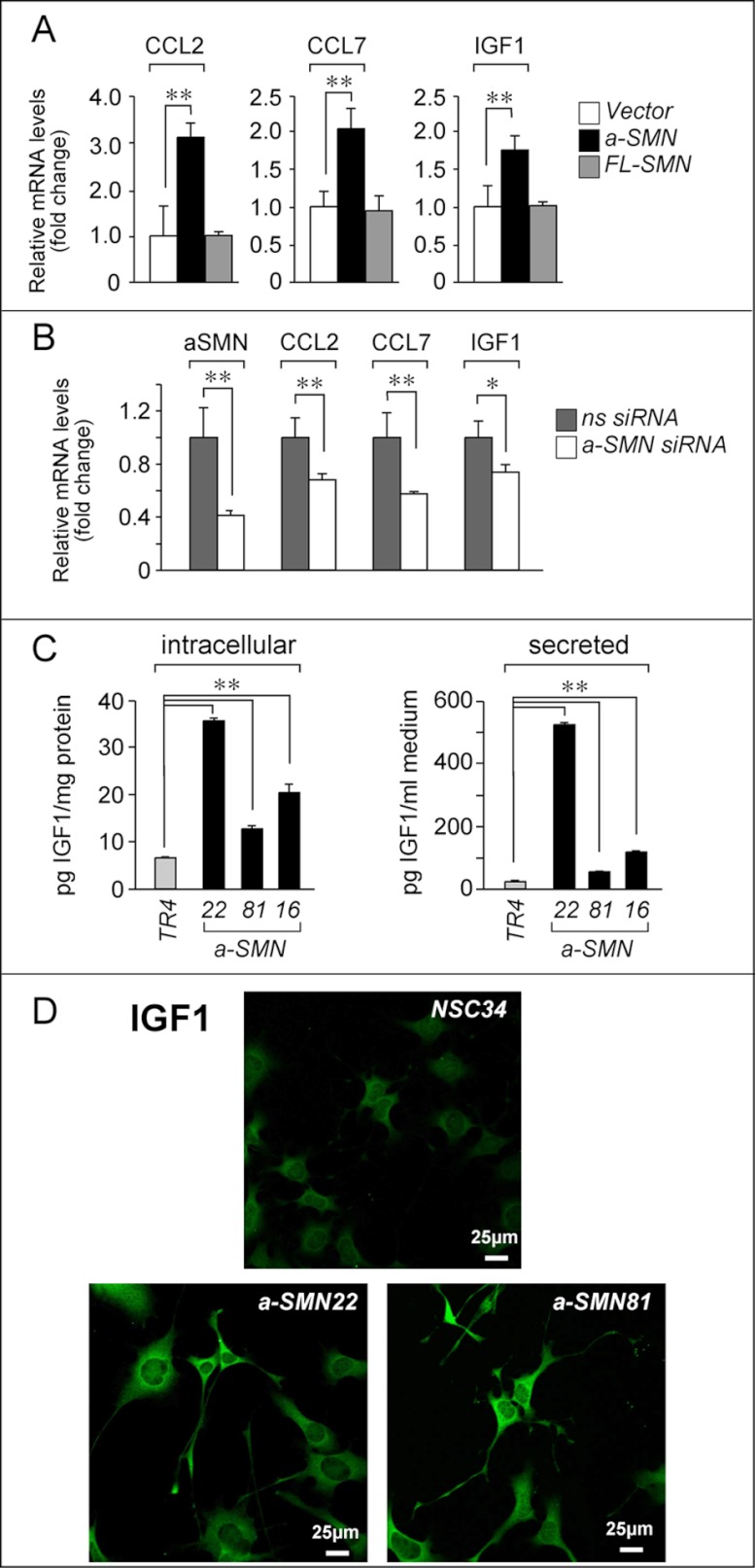
To strengthen the cause/effect relationship between a-SMN expression and CCL2, CCL7, or IGF1 up-regulation and to evaluate the specificity of these effects, NSC34 cells were transiently transfected with expression plasmids containing either the human a-SMN or the human FL-SMN cDNA (Fig. 6A) (6). Specific expression of the human a-SMN and human FL-SMN mRNAs was verified after transfection of the corresponding cDNAs, upon RT/real time PCR analysis (data not shown). Forced expression of human a-SMN resulted in increased levels of the mRNAs encoding CCL2, CCL7, and IGF1. In contrast, no significant change of the three transcripts was observed after transfection of the FL-SMN cDNA. This indicates that induction of CCL2, CCL7, and IGF1 is specifically associated with a-SMN expression. To support the results, we conducted mirror experiments in the representative a-SMN6 clone, knocking down a-SMN with a specific siRNA (Fig. 6B). Transient transfection of the siRNA caused a significant decrease in the levels of the a-SMN mRNA, which was accompanied by a reduction in the amounts of the transcripts encoding CCL2, CCL7, and IGF1.
FIGURE 6.
Regulation of target genes by a-SMN, a-SMN-dependent induction and subcellular localization of the IGF1 protein. A, NSC34 cells were transiently transfected with a-SMN or FL-SMN cDNAs or with the void vector. After 48 h, cells were harvested, and the total RNA was extracted. The levels of CCL2, CCL7, and IGF1 transcripts were measured by RT/real time PCR. The data are expressed in fold change relative to the control (vector) taken as 1. The results represent triplicate cultures and are expressed as the means ± S.D. B, a-SMN6 cells were transfected with siRNAs specific for human a-SMN or with scrambled control siRNAs (ns siRNA). After 72 h, total RNAs were extracted, and the levels of a-SMN, CCL2, CCL7, and c-Myc transcripts were measured. The data are expressed in fold change relative to the control (ns siRNA) taken as 1. The results represent triplicate cultures and are expressed as the means ± S.D. C, the indicated a-SMN-expressing clones as well as the parental cell line, TR4, were grown in DMEM supplemented with 5% serum to confluency. Medium was changed, and 24 h later, the levels of the IGF1 protein in cell extracts and conditioned medium were measured with specific ELISAs. D, NSC34, a-SMN22, and a-SMN81 cells were grown and subjected to immunofluorescence staining with an anti-IGF1 antibody. **, statistically significant according to Student's t test (p < 0,01); *, statistically significant according to Student's t test (p < 0.05).
We determined whether induction of the IGF1 and CCL2 mRNAs resulted in increased intracellular accumulation and secretion of the corresponding proteins. In basal conditions, a-SMN22, a-SMN81, and a-SMN16 cells were characterized by intracellular levels of IGF1 protein well above those observed in the TR4 control cell line (Fig. 6C, left panel). This was accompanied by augmented secretion of the growth factor (Fig. 6C, right panel). Addition of TET to the medium did not alter secretion of IGF1 (data not shown). IF studies demonstrated that a-SMN expression did not alter the intracellular distribution of IGF1, which localized predominantly to the cell body and axons (Fig. 6D).
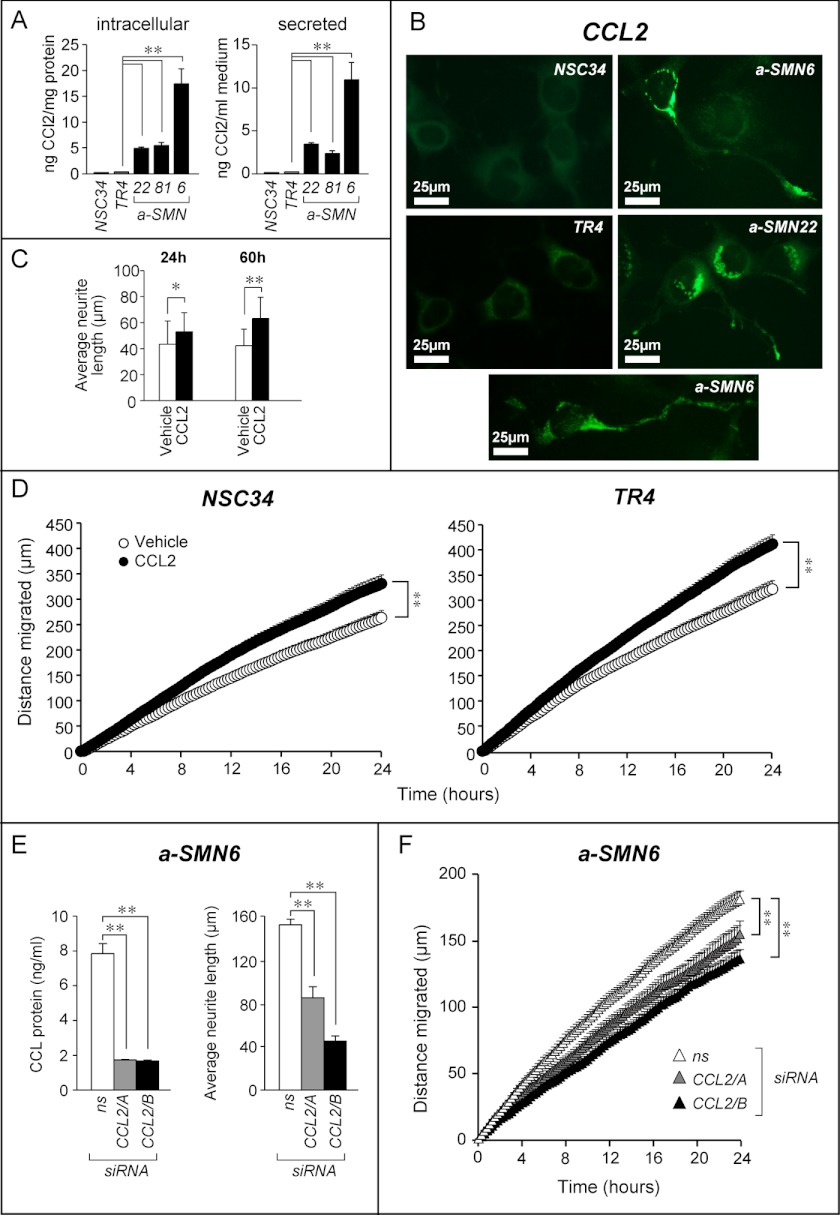
Similar to IGF1, a-SMN22, a-SMN81, and a-SMN6 cells grown in basal conditions synthesized and secreted much more CCL2 than the two control NSC34 and TR4 cell lines (Fig. 7A). Significantly, the levels of CCL2 found in the medium are higher than the range of chemokine concentrations observed in human serum (30–150 pg/ml) (27). Even in this case, treatment of cells with TET had no significant effect on the amounts of CCL2 protein secreted by the various clones (data not shown). IF experiments confirmed augmented intracellular CCL2 in a-SMN22 and a-SMN6 cells. The majority of intracellular CCL2 was distributed in the form of granular structures in the perinuclear area of the cell body, consistent with localization in the secretory machinery (Fig. 7B).
FIGURE 7.
a-SMN-dependent induction and subcellular localization of the CCL2 protein, control of axon growth and cell motility by CCL2. A, indicates that a-SMN-expressing clones as well as the parental cell lines, NSC34 and TR4, were grown to confluency in DMEM supplemented with 5% serum. Medium was changed, and 24 h later, the levels of the CCL2 protein in cell extracts and conditioned medium were measured with specific ELISAs. B, NSC34, TR4, a-SMN6, and a-SMN22 cells were grown and subjected to immunofluorescence staining with an anti-CCL2 antibody. C, NSC34 cells were treated in the presence/absence of CCL2 for the indicated amount of time. The average neurite length was determined. D, NSC34 and TR4 cells were seeded at a density of 3000 cells/ml in the presence/absence of CCL2. Random cell motility was measured after the indicated amount of time. E, cells (a-SMN6) were transfected with two different CCL2 targeting siRNAs (CCL2/A and CCL2/B) and a relative control siRNA (ns, 80 nm) in the presence of a GFP-expressing plasmid (pEGFPN1, 10 nm). Left panel, CCL2 knockdown. Six hours after transfection, the culture medium was changed. Twenty four hours later, the amount of secreted CCL2 was measured in the conditioned medium of transfected cells. Each value is the mean ± S.D. of three replicate cultures. Right panel, axon growth. Forty eight hours after transfection with the siRNAs, cells were re-seeded for time-lapse analysis of axon growth. After 24 h, the average neurite length of GFP-positive cells was measured. Each value represents the mean ± S.D. of at least 20 cells. F, cell motility. The random cell motility of GFP-positive transfected cells was measured starting 24 h after re-seeding of transfected cells. **, statistically significant according to two-way ANOVA (p < 0.01) (p < 0,01); *, statistically significant according to two-way ANOVA (p < 0,05).
CCL2 Is Involved in the Control of Cell Motility and Axonogenesis
We studied the functional links between a-SMN expression and induction of CCL2. We evaluated whether the protein exerted any effect on axon growth and cell motility, two processes controlled by a-SMN expression. To this purpose, parental NSC34 cells were challenged with CCL2 and subjected to time-lapse microscopy. CCL2 caused a measurable and significant augmentation in the average length of the axons, which was evident after 24 h and increased with time (Fig. 7C). In addition, the chemokine produced a statistically significant and reproducible increase in the motility of NSC34 and TR4 cells relative to vehicle-treated controls (Fig. 7D). Thus, our data are consistent with a role of CCL2 secretion in cell motility and axon deposition by a-SMN.
To evaluate the involvement of CCL2 in a-SMN-induced motility and axon growth, we knocked down CCL2 in a-SMN6 cells with two siRNAs (Fig. 7E, left panel). Forty eight hours after transfection with the two siRNAs, cells were re-seeded for time-lapse analysis of axon growth and cell motility. Within 24 h, we observed a significant decrease in the average length of axons in CCL2 silenced cells relative to cells transfected with a control siRNA (Fig. 7E, right panel). In the same experimental conditions, we observed a significant reduction in cell motility after treatment with the two CCL2 targeting siRNAs. We confirmed the results in the a-SMN22 clone (data not shown). These data further demonstrate that CCL2 contributes to the motility and axon growth responses triggered by a-SMN.
DISCUSSION
a-SMN is a recently identified and motor neuron-specific product of SMN1, the disease-gene of SMA (6). Synthesis of a-SMN in the mouse CNS and spinal cord is limited to the last phases of fetal development and the first few days of neonatal life. This represents the time window characterized by maximal development of the CNS with establishment of the majority of synaptic circuits. To better define the function of a-SMN, we developed a cellular model for the expression of a-SMN. The model was instrumental in providing new insights into the biological activity of a-SMN.
The cellular paradigm described herein allowed regulated expression of human a-SMN in NSC34 cells, which is considered a proxy of the motoneuron (28–30). Our model was characterized by basal expression of a-SMN in untreated cells and by time-dependent induction of the protein upon challenge with TET. Basal expression of the protein was an intrinsic characteristic of the cellular system, and it highlighted an interesting aspect of a-SMN biological activity, i.e. threshold effects. Altogether the data were consistent with the idea that maximal a-SMN biological activity was already evident when the intracellular amounts of the protein exceeded a threshold level. This may account for the tight quantitative and time-dependent regulation of a-SMN expression and the low levels of protein observed in the developing CNS and spinal cord.
In our cellular model, a-SMN was shown to play a role in biological processes of pivotal importance for motoneuron homeostasis. We documented that a-SMN controls cell motility and axon outgrowth, stimulating both processes. Stimulation of axon outgrowth was accompanied by an a-SMN-dependent increase in the surface area of growth cones (see Fig. 3A). The observation is interesting, as it has been reported that the area occupied by growth cones is reduced in motoneurons from a SMA mouse model (31–33). An increase in cell motility and axon outgowth effects was accompanied by a cytostatic action on the motoneuron. a-SMN-dependent growth inhibition and axon outgrowth may be coupled processes and the result of a differentiating action along the neuronal pathway. In fact, terminal cellular differentiation is invariably associated with growth arrest in various cell types (14), including neurons (34–37). In addition, some of the morphological features associated with a-SMN expression in NSC34 cells were reminiscent of those induced by treatment of motoneurons with the prototypic differentiating agent, all-trans-retinoic acid (38–41). It is likely that a-SMN induced cellular motility and axonogenesis are also coupled processes. In fact, coupling of cell motility and axon growth has been demonstrated in other cellular models of neuronal differentiation (42–44). Furthermore, establishment of neuronal circuits is the result of axon and neuronal cell movement to their correct destinations by guidance cues in the developing brain (45). Therefore, it is possible that the primary function of a-SMN in neurons, and particularly in motoneurons, is to control differentiation and motility ultimately resulting in the modulation of directional axon growth. Loss of a-SMN and consequent defects in motoneuron motility may contribute to the pathogenesis of SMA. Indeed, the position of spinal motoneurons in type I SMA patients suggests abnormal cell migration, as these cells are aberrantly located along the ventral roots outside the ventral horn (46). A role in neuronal migration is also suggested by the specific SMN localization in the region of the germinative neuroepithelium where young post-mitotic neurons start migration during human brain development (47).
Our transcriptome analysis provided important clues as to some of the molecules potentially involved in a-SMN-induced cell motility and axonogenesis. We identified the growth factor, IGF1, as well as the chemokines, CCL2 and CCL7, as possible mediators of a-SMN biological activity. The transcripts encoding the three factors were up-regulated in all the a-SMN-expressing clones analyzed, and induction was subject to the same threshold effects observed in the case of cell motility and axon growth. Experiments involving transient transfection or silencing of a-SMN in NSC34 cells support a cause-effect relationship between a-SMN expression and induction of CCL2/CCL7 and IGF1. Studies involving addition of CCL2 to the growth medium indicated that the cytokine increased the motility of NSC34 and TR4 cells. Complementary studies performed in the a-SMN-expressing clone a-SMN6 demonstrated that selective silencing of the CCL2 gene was associated not only with a decrease in random motility but also with a reduction in axon growth. Collectively, the evidence gathered suggests that the chemokine mediates part of the effects afforded by a-SMN on the two cellular processes. It is possible that CCL2 contributes to the control of motility and axon growth via modulation of the cytoskeletal dynamics (48, 49). In fact, a-SMN seems to affect tubulin and actin reorganization (see Fig. 2B).
Regulation of CCL2 and CCL7 has far-reaching implications as to the potential relevance of a-SMN functional inactivation for SMA etiopathogenesis. In fact, CCL2 and CCL7 recognize the same CCR2 receptor, which, along with CCR5, is considered to be an important player in myoblast proliferation after skeletal muscle injury (50). Thus, loss of a-SMN activity and degeneration of the motoneuron axons in SMA may result in CCL2 and CCL7 deficits with consequent effects on muscular tropism. With respect to this, there is recent evidence indicating that CCL2 is involved not only in peripheral and CNS inflammatory responses but also in controlling the normal activity of the neuron. CCL2 and its cognate receptor are constitutively expressed in several brain regions (50) and enhance neuronal excitability as well as synaptic transmission via pre-synaptic mechanisms (51). Our results provide the first demonstration that synthesis and secretion of CCL2 (52, 53) contribute to neuronal axonogenesis and motility in a cell autonomous manner. In our model, the effect of CCL2 on cell motility and axon growth is neither directional nor gradient-dependent, two characteristics that are typical of the chemoattractive action exerted by the chemokine on other cell types. The demonstration that CCL2 exerts functions other than chemotactic activity on neuronal cells adds to the emerging evidence that chemokines are involved in neuronal plasticity (54–56).
Up-regulation of IGF1 has relevance for the physiological activity of a-SMN and its involvement in SMA. In fact, a-SMN stimulates synthesis, secretion, and axonal transport of IGF1. The protein possesses neurotrophic properties (57) mediated by activation of the PI3K/AKT pathways (58, 59), and it is involved in myoblast proliferation as well as induction of myogenic differentiation. Through IGF1, a-SMN could exert an action in modulating the maturation of the neuromuscular unit during development. In addition, a recent report demonstrated that the levels of IGF1 were very low in the serum of an SMA mouse model (60), and the amounts of the circulating growth factor were restored to normal levels upon re-expression of the full-length form of SMN2. Finally, increased expression of IGF1 extended the survival of animals with spinal and bulbar muscular atrophy and increased median survival in an SMA (Δ7) mouse model (61–63).
Further studies are needed to establish whether a-SMN deficiency and consequent impairment of the cellular processes controlled by the protein play a role in the etiopathogenesis of SMA.
Acknowledgments
We thank James Neil Fisher for critical reading of the manuscript. We also acknowledge the help of Felice Deceglie and Alessandro Soave with the artwork.
This work was supported by Telethon-Italy Foundation Grant GGP07223, Fondazione Cariplo Grant 2008_2307, Italian Ministry of Health Grant RF-INN-2007-644440, the Fondazione Italo Monzino, Girotondo/Onlus and Smarathon/ONLUS, the Negri-Weizmann Foundation, and in part by Ministero della Salute.
D. Locatelli, unpublished observations.
- SMA
- spinal muscular atrophy
- a-SMN
- axonal survival of motor neuron
- IF
- immunofluorescence
- ANOVA
- analysis of variance
- TET
- tetracycline
- FL-SMN
- full-length survival of motor neuron.
REFERENCES
- 1. Lunn M. R., Wang C. H. (2008) Spinal muscular atrophy. Lancet 371, 2120–2133 [DOI] [PubMed] [Google Scholar]
- 2. Burghes A. H., Beattie C. E. (2009) Spinal muscular atrophy. Why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 10, 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burnett B. G., Muñoz E., Tandon A., Kwon D. Y., Sumner C. J., Fischbeck K. H. (2009) Regulation of SMN protein stability. Mol. Cell. Biol. 29, 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ogawa C., Usui K., Ito F., Itoh M., Hayashizaki Y., Suzuki H. (2009) Role of survival motor neuron complex components in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 284, 14609–14617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meister G., Bühler D., Pillai R., Lottspeich F., Fischer U. (2001) A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3, 945–949 [DOI] [PubMed] [Google Scholar]
- 6. Setola V., Terao M., Locatelli D., Bassanini S., Garattini E., Battaglia G. (2007) Axonal SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gunadi, Sasongko T. H., Yusoff S., Lee M. J., Nishioka E., Matsuo M., Nishio H. (2008) Hypomutability at the polyadenine tract in SMN intron 3 shows the invariability of the a-SMN protein structure. Ann. Hum. Genet. 72, 288–291 [DOI] [PubMed] [Google Scholar]
- 8. Locatelli D., d'Errico P., Capra S., Finardi A., Colciaghi F., Setola V., Terao M., Garattini E., Battaglia G. (2012) Spinal muscular atrophy pathogenic mutations impair the axonogenic properties of axonal-survival of motor neuron. J. Neurochem. 121, 465–474 [DOI] [PubMed] [Google Scholar]
- 9. Cashman N. R., Durham H. D., Blusztajn J. K., Oda K., Tabira T., Shaw I. T., Dahrouge S., Antel J. P. (1992) Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 194, 209–221 [DOI] [PubMed] [Google Scholar]
- 10. Yu W., Ling C., Baas P. W. (2001) Microtubule reconfiguration during axogenesis. J. Neurocytol. 30, 861–875 [DOI] [PubMed] [Google Scholar]
- 11. Elemer G. S., Edgington T. S. (1994) Microfilament reorganization is associated with functional activation of αMβ2 on monocytic cells. J. Biol. Chem. 269, 3159–3166 [PubMed] [Google Scholar]
- 12. Hirano M., Hashimoto S., Yonemura S., Sabe H., Aizawa S. (2008) EPB41L5 functions to post-transcriptionally regulate cadherin and integrin during epithelial-mesenchymal transition. J. Cell Biol. 182, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Terao M., Fratelli M., Kurosaki M., Zanetti A., Guarnaccia V., Paroni G., Tsykin A., Lupi M., Gianni M., Goodall G. J., Garattini E. (2011) Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells. Biological correlates and molecular targets. J. Biol. Chem. 286, 4027–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gianni M., Ponzanelli I., Mologni L., Reichert U., Rambaldi A., Terao M., Garattini E. (2000) Retinoid-dependent growth inhibition, differentiation, and apoptosis in acute promyelocytic leukemia cells. Expression and activation of caspases. Cell Death Differ. 7, 447–460 [DOI] [PubMed] [Google Scholar]
- 15. Dajas-Bailador F., Jones E. V., Whitmarsh A. J. (2008) The JIP-1 scaffold protein regulates axonal development in cortical neurons. Curr. Biol. 18, 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwasato T., Katoh H., Nishimaru H., Ishikawa Y., Inoue H., Saito Y. M., Ando R., Iwama M., Takahashi R., Negishi M., Itohara S. (2007) Rac-GAP α-chimerin regulates motor-circuit formation as a key mediator of EphrinB3/EphA4 forward signaling. Cell 130, 742–753 [DOI] [PubMed] [Google Scholar]
- 17. Kim M. D., Kolodziej P., Chiba A. (2002) Growth cone pathfinding and filopodial dynamics are mediated separately by Cdc42 activation. J. Neurosci. 22, 1794–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vogt D. L., Gray C. D., Young W. S., 3rd, Orellana S. A., Malouf A. T. (2007) ARHGAP4 is a novel RhoGAP that mediates inhibition of cell motility and axon outgrowth. Mol. Cell. Neurosci. 36, 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gomez T. M., Robles E. (2004) The great escape; phosphorylation of Ena/VASP by PKA promotes filopodial formation. Neuron 42, 1–3 [DOI] [PubMed] [Google Scholar]
- 20. Tursun B., Schlüter A., Peters M. A., Viehweger B., Ostendorff H. P., Soosairajah J., Drung A., Bossenz M., Johnsen S. A., Schweizer M., Bernard O., Bach I. (2005) The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 19, 2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Banisor I., Leist T. P., Kalman B. (2005) Involvement of β-chemokines in the development of inflammatory demyelination. J. Neuroinflammation 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edman L. C., Mira H., Arenas E. (2008) The β-chemokines CCL2 and CCL7 are two novel differentiation factors for midbrain dopaminergic precursors and neurons. Exp. Cell Res. 314, 2123–2130 [DOI] [PubMed] [Google Scholar]
- 23. Mirabelli-Badenier M., Braunersreuther V., Viviani G. L., Dallegri F., Quercioli A., Veneselli E., Mach F., Montecucco F. (2011) CC and CXC chemokines are pivotal mediators of cerebral injury in ischaemic stroke. Thromb. Haemost. 105, 409–420 [DOI] [PubMed] [Google Scholar]
- 24. Cheng C. M., Mervis R. F., Niu S. L., Salem N., Jr., Witters L. A., Tseng V., Reinhardt R., Bondy C. A. (2003) Insulin-like growth factor 1 is essential for normal dendritic growth. J. Neurosci. Res. 73, 1–9 [DOI] [PubMed] [Google Scholar]
- 25. Llorens-Martín M., Torres-Alemán I., Trejo J. L. (2009) Mechanisms mediating brain plasticity. IGF1 and adult hippocampal neurogenesis. Neuroscientist 15, 134–148 [DOI] [PubMed] [Google Scholar]
- 26. Prochownik E. V. (2008) c-Myc. Linking transformation and genomic instability. Curr. Mol. Med. 8, 446–458 [DOI] [PubMed] [Google Scholar]
- 27. Comini-Frota E. R., Teixeira A. L., Angelo J. P., Andrade M. V., Brum D. G., Kaimen-Maciel D. R., Foss N. T., Donadi E. A. (2011) Evaluation of serum levels of chemokines during interferon-β treatment in multiple sclerosis patients. A 1-year observational cohort study. CNS Drugs 25, 971–981 [DOI] [PubMed] [Google Scholar]
- 28. Chestnut B. A., Chang Q., Price A., Lesuisse C., Wong M., Martin L. J. (2011) Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 31, 16619–16636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirby J., Halligan E., Baptista M. J., Allen S., Heath P. R., Holden H., Barber S. C., Loynes C. A., Wood-Allum C. A., Lunec J., Shaw P. J. (2005) Mutant SOD1 alters the motor neuronal transcriptome. Implications for familial ALS. Brain 128, 1686–1706 [DOI] [PubMed] [Google Scholar]
- 30. Simeoni S., Mancini M. A., Stenoien D. L., Marcelli M., Weigel N. L., Zanisi M., Martini L., Poletti A. (2000) Motoneuronal cell death is not correlated with aggregate formation of androgen receptors containing an elongated polyglutamine tract. Hum. Mol. Genet. 9, 133–144 [DOI] [PubMed] [Google Scholar]
- 31. Rossoll W., Jablonka S., Andreassi C., Kröning A. K., Karle K., Monani U. R., Sendtner M. (2003) Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jablonka S., Beck M., Lechner B. D., Mayer C., Sendtner M. (2007) Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J. Cell Biol. 179, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sendtner M. (2010) Therapy development in spinal muscular atrophy. Nat. Neurosci. 13, 795–799 [DOI] [PubMed] [Google Scholar]
- 34. Mao L., Ding J., Zha Y., Yang L., McCarthy B. A., King W., Cui H., Ding H. F. (2011) HOXC9 links cell cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. Cancer Res. 71, 4314–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marzinke M. A., Clagett-Dame M. (2012) The all-trans-retinoic acid (atRA)-regulated gene Calmin (Clmn) regulates cell cycle exit and neurite outgrowth in murine neuroblastoma (Neuro2a) cells. Exp. Cell Res. 318, 85–93 [DOI] [PubMed] [Google Scholar]
- 36. Murakami M., Ito H., Hagiwara K., Yoshida K., Sobue S., Ichihara M., Takagi A., Kojima T., Tanaka K., Tamiya-Koizumi K., Kyogashima M., Suzuki M., Banno Y., Nozawa Y., Murate T. (2010) ATRA inhibits ceramide kinase transcription in a human neuroblastoma cell line, SH-SY5Y cells. The role of COUP-TFI. J. Neurochem. 112, 511–520 [DOI] [PubMed] [Google Scholar]
- 37. Howard M. K., Burke L. C., Mailhos C., Pizzey A., Gilbert C. S., Lawson W. D., Collins M. K., Thomas N. S., Latchman D. S. (1993) Cell cycle arrest of proliferating neuronal cells by serum deprivation can result in either apoptosis or differentiation. J. Neurochem. 60, 1783–1791 [DOI] [PubMed] [Google Scholar]
- 38. Farrar N. R., Dmetrichuk J. M., Carlone R. L., Spencer G. E. (2009) A novel nongenomic mechanism underlies retinoic acid-induced growth cone turning. J. Neurosci. 29, 14136–14142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maden M. (2007) Retinoic acid in the development, regeneration, and maintenance of the nervous system. Nat. Rev. Neurosci. 8, 755–765 [DOI] [PubMed] [Google Scholar]
- 40. Novitch B. G., Wichterle H., Jessell T. M., Sockanathan S. (2003) A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron 40, 81–95 [DOI] [PubMed] [Google Scholar]
- 41. Reimer M. M., Kuscha V., Wyatt C., Sörensen I., Frank R. E., Knüwer M., Becker T., Becker C. G. (2009) Sonic hedgehog is a polarized signal for motor neuron regeneration in adult zebrafish. J. Neurosci. 29, 15073–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hou S. T., Jiang S. X., Smith R. A. (2008) Permissive and repulsive cues and signaling pathways of axonal outgrowth and regeneration. Int. Rev. Cell Mol. Biol. 267, 125–181 [DOI] [PubMed] [Google Scholar]
- 43. Kapfhammer J. P., Schwab M. E. (1992) Modulators of neuronal migration and neurite growth. Curr. Opin. Cell Biol. 4, 863–868 [DOI] [PubMed] [Google Scholar]
- 44. Killeen M. T., Sybingco S. S. (2008) Netrin, Slit, and Wnt receptors allow axons to choose the axis of migration. Dev. Biol. 323, 143–151 [DOI] [PubMed] [Google Scholar]
- 45. Sanes J. R., Lichtman J. W. (1999) Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22, 389–442 [DOI] [PubMed] [Google Scholar]
- 46. Simic G., Mladinov M., Seso Simic D., Jovanov Milosevic N., Islam A., Pajtak A., Barisic N., Sertic J., Lucassen P. J., Hof P. R., Kruslin B. (2008) Abnormal motoneuron migration, differentiation, and axon outgrowth in spinal muscular atrophy. Acta Neuropathol. 115, 313–326 [DOI] [PubMed] [Google Scholar]
- 47. Giavazzi A., Setola V., Simonati A., Battaglia G. (2006) Neuronal specific roles of the survival motor neuron protein. Evidence from survival motor neuron expression patterns in the developing human central nervous system. J. Neuropathol. Exp. Neurol. 65, 267–277 [DOI] [PubMed] [Google Scholar]
- 48. Cross A. K., Woodroofe M. N. (1999) Chemokines induce migration and changes in actin polymerization in adult rat brain microglia and a human fetal microglial cell line in vitro. J. Neurosci. Res. 55, 17–23 [DOI] [PubMed] [Google Scholar]
- 49. Hall A. A., Herrera Y., Ajmo C. T., Jr., Cuevas J., Pennypacker K. R. (2009) σ receptors suppress multiple aspects of microglial activation. Glia 57, 744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yahiaoui L., Gvozdic D., Danialou G., Mack M., Petrof B. J. (2008) CC family chemokines directly regulate myoblast responses to skeletal muscle injury. J. Physiol. 586, 3991–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Y., Tang H., Liu J., Dong J., Xiong H. (2011) Chemokine CCL2 modulation of neuronal excitability and synaptic transmission in rat hippocampal slices. J. Neurochem. 116, 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Steenwinckel J., Reaux-Le Goazigo A., Pommier B., Mauborgne A., Dansereau M. A., Kitabgi P., Sarret P., Pohl M., Mélik Parsadaniantz S. (2011) CCL2 released from neuronal synaptic vesicles in the spinal cord is a major mediator of local inflammation and pain after peripheral nerve injury. J. Neurosci. 31, 5865–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dansereau M. A., Gosselin R. D., Pohl M., Pommier B., Mechighel P., Mauborgne A., Rostene W., Kitabgi P., Beaudet N., Sarret P., Melik-Parsadaniantz S. (2008) Spinal CCL2 pronociceptive action is no longer effective in CCR2 receptor antagonist-treated rats. J. Neurochem. 106, 757–769 [DOI] [PubMed] [Google Scholar]
- 54. Gordon R. J., Mehrabi N. F., Maucksch C., Connor B. (2012) Chemokines influence the migration and fate of neural precursor cells from the young adult and middle-aged rat subventricular zone. Exp. Neurol. 233, 587–594 [DOI] [PubMed] [Google Scholar]
- 55. Turbic A., Leong S. Y., Turnley A. M. (2011) Chemokines and inflammatory mediators interact to regulate adult murine neural precursor cell proliferation, survival, and differentiation. PLoS One 6, e25406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Valerio A., Ferrario M., Martinez F. O., Locati M., Ghisi V., Bresciani L. G., Mantovani A., Spano P. (2004) Gene expression profile activated by the chemokine CCL5/RANTES in human neuronal cells. J. Neurosci. Res. 78, 371–382 [DOI] [PubMed] [Google Scholar]
- 57. Caroni P., Grandes P. (1990) Nerve sprouting in innervated adult skeletal muscle induced by exposure to elevated levels of insulin-like growth factors. J. Cell Biol. 110, 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Johnson-Farley N. N., Travkina T., Cowen D. S. (2006) Cumulative activation of akt and consequent inhibition of glycogen synthase kinase-3 by brain-derived neurotrophic factor and insulin-like growth factor-1 in cultured hippocampal neurons. J. Pharmacol. Exp. Ther. 316, 1062–1069 [DOI] [PubMed] [Google Scholar]
- 59. Serra C., Palacios D., Mozzetta C., Forcales S. V., Morantte I., Ripani M., Jones D. R., Du K., Jhala U. S., Simone C., Puri P. L. (2007) Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell 28, 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C. F., Krainer A. R. (2011) Peripheral SMN restoration is essential for long term rescue of a severe spinal muscular atrophy mouse model. Nature 478, 123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bosch-Marcé M., Wee C. D., Martinez T. L., Lipkes C. E., Choe D. W., Kong L., Van Meerbeke J. P., Musarò A., Sumner C. J. (2011) Increased IGF-1 in muscle modulates the phenotype of severe SMA mice. Hum. Mol. Genet. 20, 1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dobrowolny G., Giacinti C., Pelosi L., Nicoletti C., Winn N., Barberi L., Molinaro M., Rosenthal N., Musarò A. (2005) Muscle expression of a local Igf-1 isoform protects motor neurons in an ALS mouse model. J. Cell Biol. 168, 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaspar B. K., Lladó J., Sherkat N., Rothstein J. D., Gage F. H. (2003) Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science 301, 839–842 [DOI] [PubMed] [Google Scholar]