Background: A deubiquitinating enzyme OTUB1 inhibits Lys-63-linked ubiquitination by binding to a ubiquitin-conjugating enzyme UBC13.
Results: A mechanism of human OTUB1-UBC13 interaction was revealed by human OTUB1-UBC13-MMS2 complex structure and structure-based mutagenesis.
Conclusion: The atomic-level interactions presented by the OTUB1-UBC13-MMS2 complex structure are critical for Lys-63-linked ubiquitination inhibition.
Significance: Learning how ubiquitination is regulated by the OTUB1-UBC13 interaction is crucial for understanding DNA damage response in biology.
Keywords: DNA Damage Response, Protein Structure, Surface Plasmon Resonance (SPR), Ubiquitin, Ubiquitin Conjugating Enzyme (Ubc), Ubiquitin-dependent Protease, X-ray Crystallography
Abstract
UBC13 is the only known E2 ubiquitin (Ub)-conjugating enzyme that produces Lys-63-linked Ub chain with its cofactor E2 variant UEV1a or MMS2. Lys-63-linked ubiquitination is crucial for recruitment of DNA repair and damage response molecules to sites of DNA double-strand breaks (DSBs). A deubiquitinating enzyme OTUB1 suppresses Lys-63-linked ubiquitination of chromatin surrounding DSBs by binding UBC13 to inhibit its E2 activity independently of the isopeptidase activity. OTUB1 strongly suppresses UBC13-dependent Lys-63-linked tri-Ub production, whereas it allows di-Ub production in vitro. The mechanism of this non-canonical OTUB1-mediated inhibition of ubiquitination remains to be elucidated. Furthermore, the atomic level information of the interaction between human OTUB1 and UBC13 has not been reported. Here, we determined the crystal structure of human OTUB1 in complex with human UBC13 and MMS2 at 3.15 Å resolution. The presented atomic-level interactions were confirmed by surface-plasmon resonance spectroscopy with structure-based mutagenesis. The designed OTUB1 mutants cannot inhibit Lys-63-linked Ub chain formation in vitro and histone ubiquitination and 53BP1 assembly around DSB sites in vivo. Finally, we propose a model for how capping of di-Ub by the OTUB1-UBC13-MMS2/UEV1a complex efficiently inhibits Lys-63-linked tri-Ub formation.
Introduction
Ubiquitin (Ub)3 is a highly conserved 76-residue protein that can be covalently attached to substrate proteins to control a wide variety of biological processes (1, 2). The enzymatic cascade of ubiquitination includes three distinct classes of enzymes: (i) a Ub-activating enzyme E1 for attaching Ub to the active site Cys via a thioester bond in an ATP-dependent manner; (ii) a Ub-conjugating enzyme E2 for receiving Ub from E1 to the active site Cys; (iii) a Ub ligase E3 for aiding transfer of Ub from E2 to specific lysine residues of substrate proteins. E3s are classified into two groups; the HECT domain-containing E3s have a catalytic Cys and transfer Ub from E2 to substrates via Ub-charged E3 intermediates (E3∼Ub), whereas the RING domain-containing E3s bridge E2 and substrates to enable the Ub transfer between them. E1, E2, and E3 enzymes work together to conjugate the terminal carboxyl group of Ub with the ϵ-amino group of lysine residues of substrate proteins including Ub itself. Ub has seven lysine residues and one terminal amino group, all of which can be attached to additional Ub molecules, producing eight types of poly-Ub chains that play important roles for their specific cellular processes. For example, Lys-48-linked polyUb chains (Lys-48 chains) are the most abundant in vivo and served as the canonical signal for degradation by the proteasome (1). On the other hand, Lys-63-linked polyUb chains (Lys-63 chains) function primarily in contexts outside of targeting substrates to the proteasome (2). Lys-63 chain synthesis is solely dependent on an E2 enzyme UBC13 and its cofactor UEV1a or MMS2 (3–5).
In the course of DNA damage response induced by DNA double-strand break (DSB), Lys-63 chains signal sites of DNA damage to recruit a RAP80-BRCA1 complex and 53BP1, which play a critical role for DNA repair, cell cycle checkpoint control, and maintenance of genome stability (6–18). DNA damage-dependent Lys-63-linked polyubiquitination is the downstream event after ATM-dependent phosphorylation of histone H2AX and MDC1 and initiated by the RNF8-dependent ubiquitination on damaged chromatin. Histone ubiquitination promotes the accumulation of a RING E3 enzyme RNF168 at DSB sites and elongates Lys-63 chains on chromatin-bound substrates that likely include histones H2A and H2AX in concert with UBC13 and UEV1a/MMS2. Defective DSB repair associated with mutations on RNF168 causes RIDDLE (radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties) syndrome because of the loss of the RNF168-dependent chromatin ubiquitination (13, 19). RNF8 and RNF168 are served as positive regulators in the context of a ubiquitination-dependent process of DSB signaling and repair (11–13, 16–18).
Typical negative regulators for ubiquitination-dependent signaling pathways are deubiquitinating enzymes (DUBs) that can remove Ub and/or Ub chains from substrates by hydrolysis (20). DNA damage-dependent ubiquitination by RNF168 and UBC13 has been found to be negatively regulated by a DUB OTUB1 (21). Surprisingly, this negative regulation, however, occurs independently of its DUB activity. Rather, OTUB1 can directly bind UBC13 and inhibit RNF168/UBC13-dependent ubiquitination in a non-canonical fashion. Furthermore, in our ubiquitination inhibition assay, we found that Lys-63-linked tri-Ub formation is strongly suppressed, whereas di-Ub formation slowly but steadily occurs. Although mechanisms for preventing di-Ub production have been recently illustrated by the crystal structures of worm OTUB1-human UBC13∼Ub and human OTUB1-UBCH5b complexes (22, 23), a mechanism for suppressing tri-Ub production remains elusive. In addition, no atomic level information of the interaction between human OTUB1 and UBC13 is available because of the lack of a complex structure comprising human OTUB1 and UBC13.
In this study we determined the crystal structure of human OTUB1 in complex with human UBC13 and MMS2 at 3.15 Å resolution. The observed atomic level interactions between human OTUB1 and UBC13 were confirmed by structure-based mutagenic analyses using a surface-plasmon spectrometry, an in vitro ubiquitination assay, and an immunofluorescence assay of DNA damage-dependent responses in vivo. Finally, we propose two additional mechanisms to elucidate the inhibition of Lys-63 chains longer than di-Ub by OTUB1 based on our docking analysis of the OTUB1-UBC13-MMS2 complex and Ub chains.
EXPERIMENTAL PROCEDURES
Preparation of the OTUB1-ΔN-UBC13-MMS2 Complex
The genes encoding human OTUB1-ΔN (residues 45–271) and UBC13 (residues 3–150) were amplified from the cDNAs obtained from Open Biosystems. The gene encoding human MMS2 (residues 6–143) was PCR-amplified from a human cDNA library. The gene encoding yeast SUMO was PCR-amplified from a Saccharomyces cerevisiae cDNA library. The gene encoding SUMO was attached to the N terminus of the genes encoding OTUB1-ΔN, UBC13, and MMS2. The amplified genes were cloned into the pET26b expression vector with NdeI and XhoI sites to produce N-terminal His6-tagged SUMO fusion proteins and confirmed by DNA sequencing. Escherichia coli strain RosettaTM (DE3) cells (Invitrogen) were transformed with the expression vector and cultured in LB containing 50 mg/liter kanamycin at 37 °C. The expression was induced by the addition of 0.1 mm isopropyl-β-d-thiogalactopyranoside at A600 of ∼0.5, and then the E. coli cells were incubated overnight at 15 °C. The cells were collected by centrifugation at 8000 × g for 15 min and disrupted by sonication in the sonication buffer (50 mm Tris-HCl buffer (pH 8.0) containing 150 mm NaCl, 20 mm imidazole, and 0.5% Triton X-100). The lysates were centrifuged at 30,000 × g for 60 min, and the supernatants were then loaded onto a nickel-nitrilotriacetic acid (Qiagen) column that had been pre-equilibrated with the sonication buffer. The column was washed with the sonication buffer and then with 50 mm Tris-HCl buffer (pH 8.0) containing 150 mm NaCl and 20 mm imidazole. The His6-tagged SUMO fusion proteins were eluted with 50 mm Tris-HCl buffer (pH 8.0) containing 150 mm NaCl and 200 mm imidazole. The His6-tagged SUMO was cleaved by Ulp1 protease, and the samples were dialyzed against 50 mm Tris-HCl buffer (pH 8.0) containing 1 mm DTT. The proteins were loaded onto a ResourceQ anion exchange column (GE Healthcare) pre-equilibrated with 50 mm Tris-HCl buffer (pH 8.0) containing 1 mm DTT and were eluted with a linear gradient of 0–1 m NaCl. To prepare the OTUB1-ΔN-UBC13-MMS2 complex, a 1.5-fold molar excess of UBC13 was incubated at 4 °C overnight with OTUB1-ΔN and MMS2. OTUB1-ΔN-UBC13-MMS2 complex was loaded onto a HiLoad Superdex 75 size-exclusion column (GE Healthcare) pre-equilibrated with 10 mm Tris-HCl buffer (pH 7.2), 50 mm NaCl, and 5 mm β-mercaptoethanol to remove unbound monomers.
Crystallography
The purified OTUB1-ΔN-UBC13-MMS2 complex was concentrated to 10 g/liter by using an Amicon Ultra-15 10,000 molecular weight cutoff filter (Millipore). Initial crystallization screening was performed using the sitting drop vapor diffusion method at 20 °C, with a MosquitoR liquid-handling robot (TTP Lab Tech). We tested about 600 conditions using crystallization reagent kits supplied by Hampton Research and Qiagen, and initial hits were further optimized. The best crystals of the OTUB1-ΔN-UBC13-MMS2 complex were obtained at 20 °C with the sitting drop vapor diffusion method by mixing 0.5 μl of protein solution with an equal amount of precipitant solution containing 90 mm Tris-HCl buffer (pH 8.5), 19% PEG3350, and 10 mm EDTA sodium salt and equilibrating against 500 μl of reservoir solution containing 100 mm Tris-HCl buffer (pH 8.5) and 22% PEG3350. For data collection, the crystals were transferred to a cryostabilizing solution (50 mm Tris-Cl (pH 8.5), 24% PEG3350, and 20% ethylene glycol) and flash-frozen in a 100 K nitrogen stream. The diffraction data set was collected at the beamline BL41XU in SPring-8 (Hyogo, Japan) and was processed with the program HKL2000 (24) and the CCP4 program suite (25). The complex structure was determined by molecular replacement using the program MolRep (26). The crystal structures of the human OTUB1 (PDB code 2ZFY) and the human UBC13-MMS2 complex (PDB code 1J7D) were used as search models. Three OTUB1-ΔN molecules and nine UBC13-MMS2 complexes were found in the asymmetric unit. At first, the crystal apparently belonged to the space group P212121, with unit cell dimensions of a = 102.1 Å, b = 137.3 Å, c = 257.2 Å. The model was built and refined using the programs COOT (27) and Refmac5 against the data processed with P212121. However, the Rfree value was not improved (around 35%), and the resultant 2Fo − Fc map was poor. We, therefore, suspected a pseudosymmetry and processed the data in the space group P21 with unit cell dimensions of a = 102.1 Å, b = 137.3 Å, c = 257.1 Å, and β = 90.03°. This improved the Rfree values and the electron density map, and the final model consists of six OTUB1 molecules and 18 UBC13-MMS2 complexes. Refinement was carried out by using Refmac5, including temperature factor, positional, and TLS refinement, treating each protein molecule as one TLS group. The final models have excellent stereochemistry and Rfree values of 28.1% at 3.15 Å resolution. Data collection, phasing, and refinement statistics are shown in Table 1. All molecular graphics were prepared with the program PyMOL.
TABLE 1.
Data collection and refinement statistics
The numbers in parentheses are for the highest resolution shell. r.m.s.d., root mean square deviation.
| Human OTUB1-ΔN·UBC13·MMS2 | |
|---|---|
| Data collection | |
| Space group | P21 |
| Unit cell parameter | a = 102.1 Å, b = 137.3 Å, c = 257.1 Å, β = 90.03° |
| Wavelength (Å) | 1.00000 |
| Resolution (Å) | 50.0-3.15 (3.20-3.15) |
| Unique reflections | 121,981 |
| Total reflections | 341,500 |
| Completeness (%) | 96.9 (92.6) |
| I/σ(I) | 16.1 (3.08) |
| Rsyma | 0.083 (0.322) |
| Refinement | |
| Number of atoms | 51,450 |
| Rmsd bond length (Å) | 0.007 |
| Rmsd bond angle (°) | 1.009 |
| Average B factor | 80.7 |
| Residues in core region (%) | 90.0 |
| Residues in additionally allowed region (%) | 9.5 |
| Residues in generously allowed region (%) | 0.4 |
| Residues in disallowed region (%) | 0.0 |
| Rwork,b Rfreec | 0.223, 0.281 |
a Rsym = Σ|Iavg − Ii|/ΣIi.
b Rwork = Σ|Fo − Fc|/ΣFo for reflections of working set.
c Rfree = Σ|Fo − Fc|/ΣFo for reflections of test set (5% of total unique reflections).
Surface Plasmon Resonance Spectrometry
GST-fused OTUB1, OTUB2, and UBC13 were purified by glutathione-Sepharose FF and ResourceQ anion exchange columns (GE Healthcare). Experiments were carried out on a Biacore T200 instrument (GE Healthcare) equilibrated at 25 °C in HBS-P buffer including 10 mm HEPES (pH 7.4), 150 mm NaCl, and 0.05% surfactant P-20 using a Sensor Chip CM5 (GE Healthcare). GST antibody (GE Healthcare) was covalently immobilized on the sensor chip at a density of about 15,000 resonance units (RUs), and GST-fused OTUB1, OTUB2 and UBC13 were captured on the sensor chip at a density of 500–1000 RUs. OTUB1, UBC13, and UBC13-MMS2 were then injected for 60 s at a flow rate of 10 ml/min. Dissociation constants (Kd) were calculated by fitting to a 1:1 interaction model using Biacore T200 evaluation software (GE Healthcare).
Cell Culture and Plasmid Transfection
U2OS cells and 293T cells were grown in McCoy's 5A medium and Dulbecco's modified Eagle's medium with 10% FBS, respectively. Plasmid transfections were performed using Effectene (Qiagen) following the manufacturer's protocol with minor modifications. pcDNA3-FLAG OTUB1, pcDNA3-HA RNF168, and pCMV-MYC ubiquitin expression vectors were described previously (21). Point mutations and deletions were generated by using PCR.
Antibodies
We used the following antibodies: 53BP1 (NB100-304, Novus Biologicals), histone H3 (ab1791, Abcam), Ub (Clone VU-1, VU101, LifeSensors), UBC13 (#4919, Cell Signaling), α-tubulin (T6074, Sigma), FLAG (M2, F3165, Sigma), HA (clone HA-7, H9658, Sigma), MYC (clone 9E10, sc-40, Santa Cruz).
Western Blotting
Cells were lysed in SDS sample buffer. Whole cell lysate were separated on SDS-PAGE (5–20% or 15% e-PAGEL, ATTO) and then transferred to nitrocellulose membranes by iBlot Gel Transfer System (Invitrogen). For Ub immunodetection, nitrocellulose membranes were incubated with 0.5% glutaraldehyde in PBS before blocking. Membranes were blocked with 5% Difco skim milk (BD Biosciences) in TBST and stained with the indicated primary antibodies followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. Western Lightning ECL Pro (PerkinElmer Life Sciences) was used for chemiluminescent horseradish peroxidase-conjugated antibody detection. Chemiluminescent signals were detected with LAS4000 (Fuji Film).
In Vitro Ubiquitination Assay
Assays were set up in a total volume of 25 μl in 50 mm HEPES-KOH (pH 7.5), 100 mm NaCl, 5 mm MgCl2, 2 mm ATP, 0.5 mm DTT. UBE1 (0.025 μm; Boston Biochem), 0.1 μm E2 (UBC13 and MMS2 or UBC13/UEV1a (Boston Biochem)), and Lys-63-only ubiquitin (20 μm Ub (Boston Biochem)) were incubated with 10 μm of recombinant OTUB1 proteins. The indicated proteins were assembled on ice for 30 min, and then the reaction was initiated by the addition of E1 and ATP. Reaction mixtures were incubated at 37 °C for 1, 2, 4, 6, 10, or 30 min or 2 h (UBC13/UEV1a) or 16 h (UBC13/MMS2).
Immunoprecipitation and Acid Extraction of Histones
10-cm dishes of 293T cells were grown, and each dish was transfected with 0.5 μg of pcDNA3-OTUB1, 4 μg of pcDNA3-HA RNF168. and 1 μg of pCMV-MYC ubiquitin expression vectors using Effectene (Qiagen). After 24 h of post-transfection, cells were harvested and divided in two. One sample was lysed in ice-cold lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% digitonin supplemented with EDTA-free protease and phosphatase inhibitor cocktails (Nacalai Tesque)). Cell lysates were sonicated and then clarified by centrifugation at 4 °C for 30 min at 26,000 × g. Clarified cell lysates were precleared using protein G-agarose (Sigma) and then subjected to immunoprecipitation with 20 μl of anti-FLAG (M2) affinity gel (Sigma). Immunoprecipitations were carried out for 1 h at 4 °C. Immunoprecipitates were washed three times with the lysis buffer followed by two washes with ice-cold PBS. The beads were then boiled in 80 μl of 2× SDS sample buffer. The other sample was lysed in nuclear preparation buffer (10 mm Tris-HCl (pH 7.6), 150 mm NaCl, 1.5 mm MgCl2, 0.65% Nonidet P-40, EDTA-free protease, and phosphatase inhibitor cocktails). Nuclei were collected by centrifugation (5 min, 920 × g, 4 °C). The nuclear pellet was extracted into 0.4 n H2SO4 for 1 h at 4 °C. The acid soluble fraction was collected after centrifugation (5 min, 7500 × g, 4 °C). Trichloroacetic acid was added to the supernatant to a final concentration of 20% and then incubated on ice for 1 h. Precipitates were collected by centrifugation (10 min, 15,000 × g, 4 °C). Precipitates were washed once with 0.1% HCl-acetone and twice with acetone and then resuspended in distilled H2O.
Immunofluorescence Microscopy
U2OS cells were grown on no. 1 glass coverslips (Fisher), and 0.1 μg of pcDNA3 FLAG-OTUB1 expression vectors were transfected at 24-well format using Effectene (Qiagen). Cells were irradiated to 2-gray x-ray 1 day post-transfection and subjected to immunofluorescence 1 h post-IR. Cells were fixed using 3% paraformaldehyde and 2% sucrose in PBS for 15 min at room temperature and permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. After fixation, cells were washed with PBS 3 times and blocked with 2% BSA in PBS for 1 h. Cells were then stained with anti-FLAG and anti-53BP1 antibodies, which were diluted in 2% BSA in PBS. Cells were washed with PBS twice and then stained with Alexa Fluor 488 goat anti-mouse IgG or Alexa Fluor 555 goat anti-rabbit IgG (Invitrogen). Cells were washed with 2% BSA in PBS and PBS alone 2 times each. DNA was counterstained with DAPI (0.2 μg/ml) in PBS, and samples were mounted with Prolong Gold mounting agent (Invitrogen). Confocal images were captured using an inverted microscope (TCS SP5, Leica) equipped with a 63× oil immersion lens. For quantification of the immunostaining data, a minimum of 300 cells were analyzed per experiment. Images were acquired in LAS AF (Leica) format.
In Vitro Deubiquitination Assay
Assays were set up in a total volume of 20 μl in 50 mm HEPES-KOH (pH 7.5), 25 mm NaCl, 5 mm MgCl2, 0.5 mm DTT. 1 μg of Lys-48-linked tetra-Ub (Boston Biochem) was incubated with 0.9 μg of recombinant OTUB1 proteins. Reaction mixtures were incubated at 37 °C for 150 min.
RESULTS
Structure Determination of Human OTUB1-UBC13-MMS2 Ternary Complex
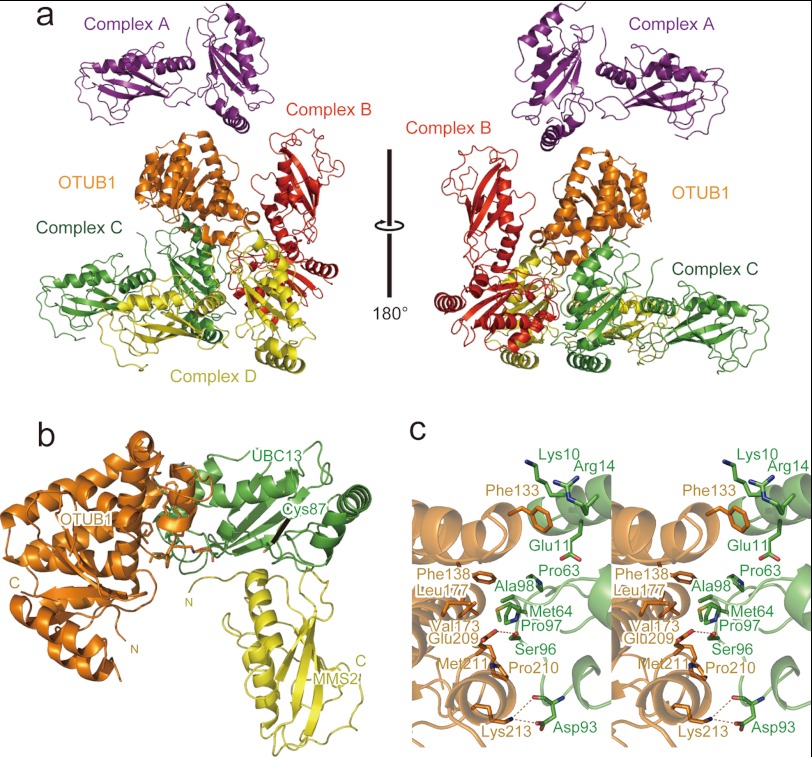
To reveal a mechanism of the interaction between human OTUB1 and UBC13 for the DUB activity-independent, non-canonical inhibition of ubiquitination, we tried crystallization of the full-length or N-terminal-truncated human OTUB1 (i.e. residues 45–271; referred to hereafter as OTUB1-ΔN) in complex with human UBC13 or UBC13(C87S/K92R)∼Ub in the presence or absence of human MMS2 or UEV1a. C87S/K92R double mutation was introduced to UBC13 for preparing a stable oxyester-linked analog of the UBC13∼Ub intermediate without producing a ubiquitinated UBC13 via Lys-92. Most of the crystallization trials failed, but a trial using the N-terminal truncated OTUB1, UBC13, and MMS2 was successful. The crystal comprising the ternary complex belongs to the space group P21 with unit cell dimensions of a = 102.1 Å, b = 137.3 Å, c = 257.1 Å, β = 90.03°. An asymmetric unit contains 18 complexes of UBC13-MMS2 and -6 molecules of OTUB1-ΔN. Three complexes of UBC13-MMS2 embrace one molecule of OTUB1-ΔN and constitute a structural unit OTUB1-ΔN-(UBC13-MMS2)3 (supplemental S1). Furthermore, three OTUB1-ΔN-(UBC13-MMS2)3 units are aligned in parallel and assembled into a cluster [OTUB1-ΔN-(UBC13-MMS2)3]3. The asymmetric unit contains two [OTUB1-ΔN-(UBC13-MMS2)3]3 clusters that are related with each other by a pseudo 21-fold symmetry along x and z axes of the unit cell. Therefore, the crystal of the ternary complex appears to belong to the space group P212121.
In this complicated packing of OTUB1-ΔN and the UBC13-MMS2 complex in the crystal, OTUB1-ΔN contacts four distinct UBC13-MMS2 complexes (Fig. 1a). For convenience, the UBC13-MMS2 complexes contacting α4, α8:α9, α5:α7:α9 and the loop connecting α8 and α9 of OTUB1-ΔN are referred to as Complexes A, B, C, and D, respectively (Fig. 1a). To identify a biologically functional OTUB1-UBC13-MMS2 ternary complex, we first calculated a buried surface area at the interface between OTUB1-ΔN and UBC13 in Complex A, B, C, or D. The combination of OTUB1-ΔN and Complex C (721 Å2) buries a sufficiently larger contact surface than other combinations (114, 129. and 202 Å2 for Complex A, B, and D, respectively). Furthermore, the contact of Complex C with OTUB1-ΔN is mediated exclusively by UBC13, consistent with the fact that neither MMS2 nor UEV1a is involved in the interaction between OTUB1 and UBC13 (Fig. 1, a and b). These structural features suggest that the interactions between OTUB1-ΔN and UBC13 in Complex C should be physiologically functional (Fig. 1 and supplemental S1).
FIGURE 1.
Crystal structure of the OTUB1-ΔN-UBC13-MMS2 ternary complex. a, four UBC13-MMS2 complexes contacting one OTUB1 molecule is shown. OTUB1 is colored orange. Complex A, B, C, and D of the UBC13-MMS2 are colored purple, red, green, and yellow, respectively. b, overall structure of the OTUB1-ΔN-UBC13-MMS2 ternary complex is shown. OTUB1, UBC13, and MMS2 are colored orange, green, and yellow, respectively. c, shown is a stereo view of the interface between OTUB1-ΔN and UBC13.
Interactions between OTUB1 and UBC13
Interactions between OTUB1-ΔN and UBC13 in Complex C are mainly hydrophobic. The side chains of Phe-138, Val-173, Leu-177, and Met-211 in OTUB1 form a hydrophobic surface that interacts with the side chains of Pro-63, Met-64, Pro-97, and Ala-98 in UBC13 (Fig. 1c and supplemental S2). Furthermore, the side chain of Phe-133 in OTUB1 is accommodated in a pocket formed by the aliphatic portions of the Lys-10, Glu-11, and Arg-14 side chains in UBC13. In addition, the Nζ atom of Lys-213 in OTUB1 hydrogen bonds with the Oδ atom and main-chain CO of Asp-93 in UBC13. The Oϵ atom of Glu-209 in OTUB1 hydrogen bonds with the Oγ atom of Ser-96 in UBC13. The Ser-96–Pro97–Ala98 tripeptide sequence is conserved among OTUB1 binding E2s (i.e. UBC13 and UBE2D and UBE2E families) (28, 29) but not among other E2s (supplemental S3), suggesting that UBC13, UBE2D, and UBE2E families share a common strategy for OTUB1 binding.
To test whether these interactions actually contribute to the specific binding between OTUB1 and UBC13 in solution, we replaced Phe-133, Phe-138, Met-211, and Lys-213 by Ala in the full-length OTUB1 and measured the affinities by using surface plasmon resonance spectrometry (supplemental S4 and Table 2). As expected, these mutations reduced the affinities 33–50-fold, clearly indicating that the abovementioned interactions between OTUB1 and UBC13 occur in solution and contribute to the binding between them. Met-211 is conserved in both OTUB1 and OTUB2, and Phe-138 is perfectly conserved in OTUB1 and mostly in OTUB2 (supplemental S5). On the other hand, Phe-133 is conserved in OTUB1 but is replaced by His in OTUB2, and Lys-213 is conserved in OTUB1 but not in OTUB2. Therefore, Phe-133 and Lys-213 of OTUB1 may contribute to the specific binding between OTUB1 and UBC13. Our surface plasmon resonance analyses show that OTUB1 binds UBC13 with a 40-fold higher affinity than OTUB2.
TABLE 2.
Dissociation constants determined by surface plasmon resonance analyses
Results are the means ± S.D. n = 3.
|
Kd (μm) |
||
|---|---|---|
| UBC13 | UBC13-MMS2 | |
| GST-OTUB1 | ||
| WT | 8.98 ± 1.75 | 17.5 ± 6.2 |
| C91S | 2.85 ± 0.14 | 5.68 ± 0.41 |
| F133A | 372 ± 4 | |
| F138A | 296 ± 6 | |
| M211A | 445 ± 40 | |
| M211A/F133A/F138A | ND | |
| K213A | 449 ± 21 | |
| GST-OTUB2 | ||
| WT | 364 ± 25 | |
|
OTUB1 |
||
| 1–271 | 45–271 | |
| GST-UBC13 | 19.2 ± 1.8 | 23.2 ± 1.1 |
Structure-based Mutational Analysis of the Inhibition of Lys-63 Chain Synthesis by OTUB1 in Vitro
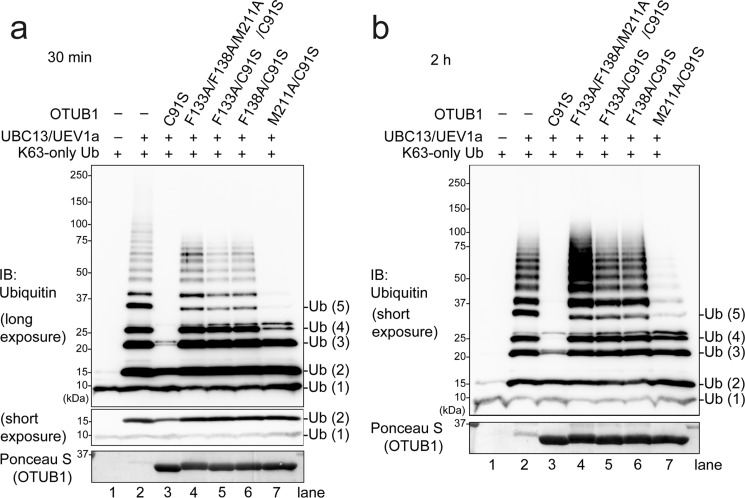
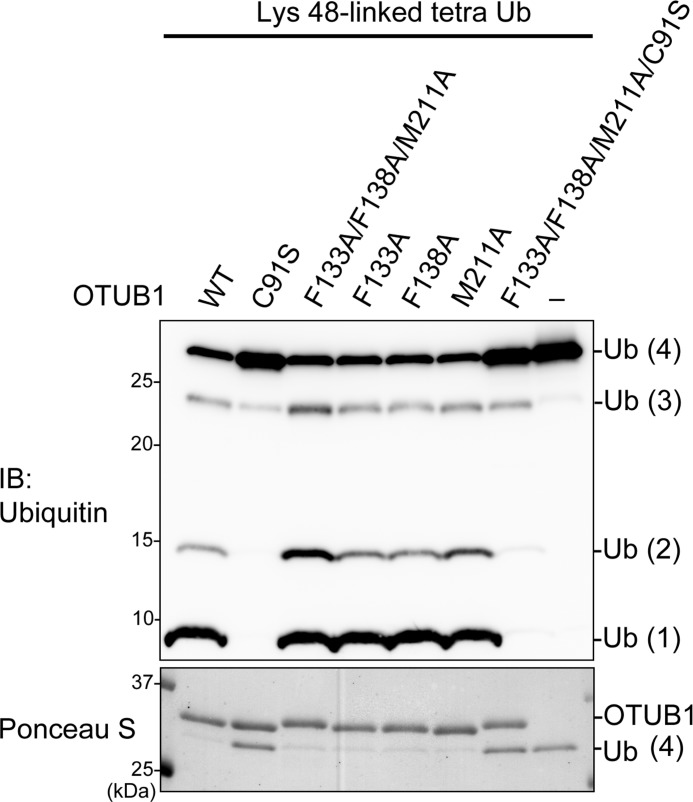
We next asked if these atomic-level interactions for the binding between OTUB1 and UBC13 are critical for the inhibition of Lys-63 chain synthesis in vitro. Lys-63 chain formation was reconstituted with E1, Lys-63-only Ub (i.e. K6R/K11R/K27R/K29R/K33R/K48R), and UBC13/MMS2 or UBC13/UEV1a in the presence or absence of the wild type or mutant OTUB1, and then products were analyzed by Western blotting for Ub. In this experiment, DUB-inactive mutation C91S was additionally introduced to OTUB1 to avoid a nonspecific DUB activity affecting the result. In the presence of C91S mutant of OTUB1, UBC13/UEV1a- or UBC13/MMS2-dependent Lys-63 chain formation was inhibited (Fig. 2, a and b, and supplemental S6, compare lanes 2 and 3). In striking contrast, C91S/F133A, C91S/F138A, and C91S/F133A/F138A/M211A mutants of OTUB1 showed no inhibitory effect (Fig. 2, a and b, and supplemental S6, lanes 4–6), whereas C91S/M211A mutant had a marginal inhibitory effect (Fig. 2, a and b, and supplemental S6, lane 7). Note that proper folding of F133A, F138A, M211A, and F133A/F138A/M211A mutants of OTUB1 was confirmed by their retaining full DUB activities (Fig. 3). These results indicate that the observed atomic-level interactions between OTUB1 and UBC13 are critical for the inhibition.
FIGURE 2.
Phe-133, Phe-138, and Met-211 of OTUB1 are crucial for the inhibition of UBC13-dependent Lys-63 chain formation. In vitro Lys-63 chain synthesis by UBC13-UEV1a was performed in the presence or absence of the indicated recombinant OTUB1. Reaction products were analyzed by Western blotting (IB) for Ub. Reactions after 30 min and 2 h are shown in a and b, respectively.
FIGURE 3.
OTUB1 mutants retain full DUB activities. Lys-48-Ub4 cleavage by the OTUB1 mutants was analyzed by SDS-PAGE. The reaction products were stained with Ponceau S and immunoblotted (IB) with anti-Ub antibodies.
Structure-based Mutational Analysis of the Inhibition of DNA Damage Response by OTUB1 in Vivo
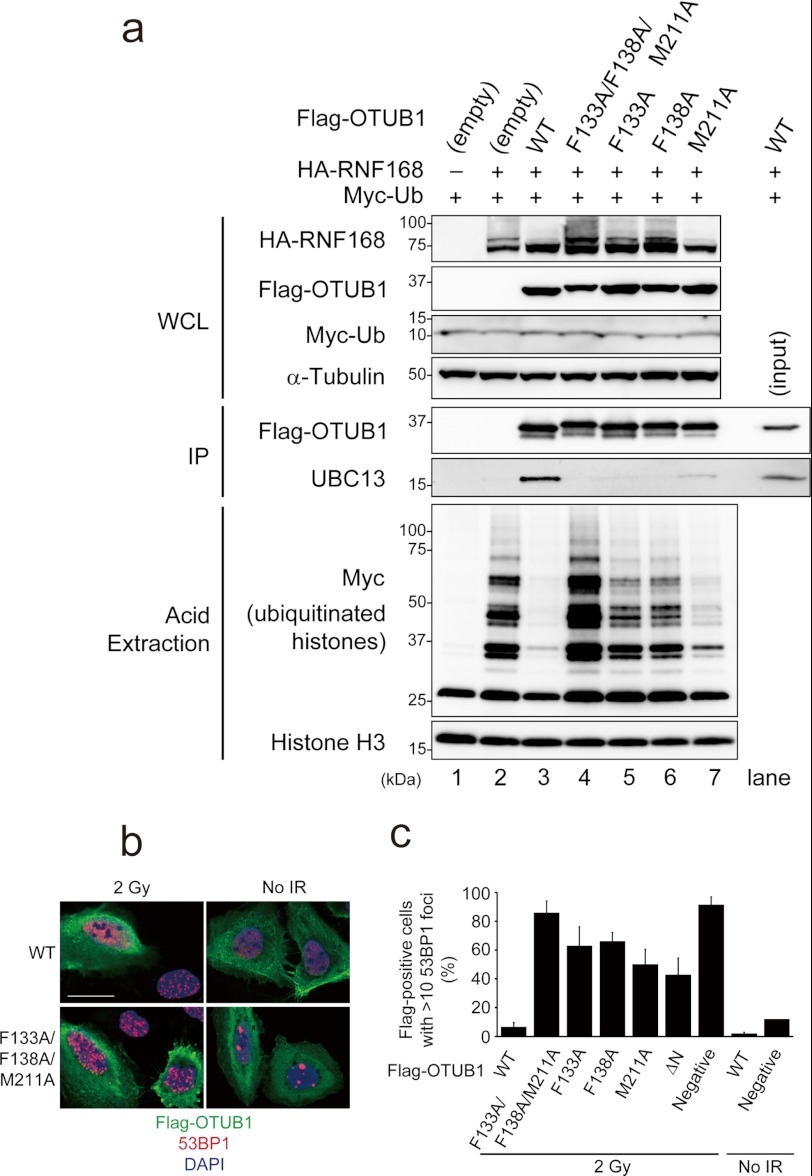
All the above data prompted us to confirm the significance of the OTUB1-UBC13 interaction through Phe-133, Phe-138, and Met-211 of OTUB1 at the cellular level. Binding between UBC13 and the wild type or mutant OTUB1 in cells was assessed by a co-immunoprecipitation experiment. As shown in Fig. 4a, the wild type FLAG-OTUB1 can be efficiently co-precipitated with UBC13. In contrast, F133A, F138A, and F133A/F138A/M211A mutants of FLAG-OTUB1 cannot be co-precipitated with UBC13, whereas M211A mutant can be fairly weakly co-precipitated. These results are consistent with the abovementioned Lys-63 ubiquitination inhibition analyses. We next analyzed the impact of F133A, F138A, and M211A mutations of OTUB1 on RNF168-dependent histone ubiquitination, which can be detected from acid-extracted histone from 293T cells transfected with plasmids expressing HA-RNF168 and MYC-Ub (Fig. 4a, lane 2, lower panels). When these plasmids were co-transfected with FLAG-OTUB1-expressing plasmids, histone ubiquitination was completely suppressed (Fig. 4a, lane 3, lower panels). In contrast, F133A, F138A, and F133A/F138A/M211A mutants of OTUB1 showed no inhibitory effect (Fig. 4a, lanes 4–6, lower panels). M211A, which weakly binds UBC13, mildly inhibited histone ubiquitination (Fig. 4a, lane 7, lower panels).
FIGURE 4.
Phe-133, Phe-138, and Met-211 of OTUB1 are required for the inhibition of UBC13/RNF168-mediated DNA damage response. a, 293T cells cotransfected with hemagglutinin (HA)-RNF168, MYC-ubiquitin, and/or FLAG-OTUB1 were subjected to FLAG immunoprecipitation (IP) or acid extraction of chromatin. WCL, whole cell lysate. b and c, U2OS cells transfected with the indicated FLAG-OTUB1 expression vectors were irradiated with 2 gray (Gy) and processed for 53BP1 and FLAG immunofluorescence 1 h post-ionizing radiation. Scale bar, 25 μm. c, shown is quantification of the data shown in b (mean ± S.D., n = 3). Gy, gray; WCL, whole cell lysate; IP, immunoprecipitation.
RNF168/UBC13-dependent histone ubiquitination is prerequisite for localization of 53BP1 to sites of DSBs, which is visualized as irradiation induced foci by immunofluorescence (11, 13). Overexpression of OTUB1 inhibits Lys-63 chain and 53BP1 irradiation induced foci (21). Therefore, we finally tested whether F133A, F138A, and/or M211A mutations abrogated the OTUB1-dependent suppressive regulation of DNA damage response. U2OS cells overexpressing OTUB1 showed defective 53BP1 irradiation induced foci, whereas overexpression of any OTUB1 mutants did not inhibit 53BP1 irradiation induced foci efficiently. Especially, F133A/F138A/M211A mutant of OTUB1 completely lost its inhibitory effect on DNA damage response (Fig. 4, b and c).
Altogether, we concluded that OTUB1 inhibits E2 activity of UBC13 and the downstream UBC13/RNF168-mediated DNA damage response depending on the affinity between OTUB1 and UBC13 through specific interactions observed in the present OTUB1-ΔN-UBC13-MMS2 ternary complex structure.
OTUB1 Delays di-Ub Production and Completely Blocks Tri-Ub Production in Vitro
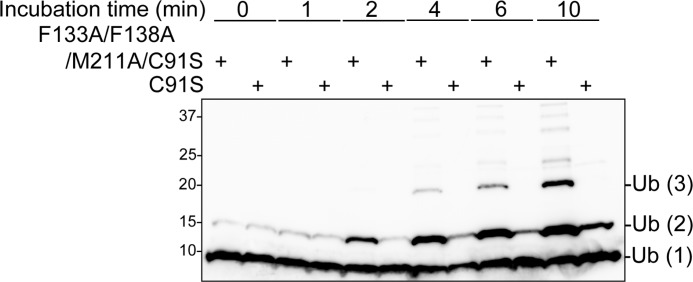
Our in vitro ubiquitination inhibition assay shows that Lys-63-linked di-Ub formation is inhibited in the presence of OTUB1 in a short time scale (i.e. ∼30 min) (Fig. 2b). Furthermore, more detailed time course analysis with time points of 1, 2, 4, 6, and 10 min clearly demonstrates that di-Ub formation is sufficiently delayed in the presence of OTUB1 (Fig. 5).
FIGURE 5.
OTUB1 mildly inhibits UBC13-dependent di-Ub formation. In vitro Lys-63 chain synthesis by UBC13-UEV1a was performed in the presence of C91S or F133A/F138A/M211A/C91S OTUB1. Reaction products were analyzed by Western blotting for Ub after incubation for the indicated time at 37 °C.
Recently, the crystal structures of the Ub∼UBCH5b-OTUB1-Ub and Ub∼UBC13-OTUB1∼Ub complexes have been reported (22, 23). The N-terminal conserved region of OTUB1 (i.e. residues 22–46), which is required for the E2 inhibition, folds into an α-helix and interacts with Ile-44-centered patch of the donor Ub charged to UBCH5b and UBC13. For the inhibition of UBCH5b (and possibly, other OTUB1-interacting E2s), the edge of the helix partly shields a thioester linkage between the active site Cys of E2 and the terminal Gly of the donor Ub, thus likely interfering with a transesterification reaction to form an isopeptide linkage between the donor and acceptor Ub molecules. For UBC13 inhibition, it is likely that the N-terminal helix of OTUB1 sterically interferes with the binding between UBC13 and UEV1a/MMS2, which provides a binding site for the acceptor Ub and thus is required for Lys-63 chain formation. Interfering with the transesterification reaction and/or UEV1a/MMS2 binding by OTUB1 can explain how OTUB1 delays Lys-63-linked di-Ub production.
On the other hand, in the detailed time course analysis, we found that the amount of di-Ub gradually increased, whereas no tri-Ub could be detected. Even in a longer time scale (i.e. ∼2 h), tri-Ub formation is completely blocked in the presence of OTUB1 despite di-Ub being sufficiently produced (Fig. 2b). These results indicate that OTUB1 suppresses Lys-63-linked tri-Ub formation more strongly than Lys-63-linked di-Ub formation, which postulates another additional mechanism for the inhibition of Lys-63 chain synthesis for the complete blocking of tri-Ub production, as discussed below.
DISCUSSION
A molecular mechanism of Lys-63 chain synthesis by the UBC13-MMS2/UEV1a complex has been proposed on the basis of the crystal structure of yeast UBC13∼Ub-MMS2 binary complex. The UBC13-MMS2 complex forms two adjacent Ub binding sites termed “acceptor site” and “donor site.” The donor Ub charged to the active site Cys by a thioester bond binds to the donor site, and then the acceptor Ub is recruited to the acceptor site. This places Lys-63 of the acceptor Ub in the active site, near the thioester bond between the donor Ub and UBC13. The Nζ atom of Lys-63 of the acceptor Ub can then attack the thioester and form an isopeptide bond between the terminal carboxyl group of the donor Ub and Lys-63 of the acceptor Ub. After that, the donor Ub becomes the acceptor Ub for the next round of Lys-63-linked isopeptide formation, although it is not clear whether the donor Ub moves to the acceptor site on the same UBC13-MMS2/UEV1a complex or binds to the acceptor site on another complex. In either case, binding of the distal Ub moiety in the nascent di-Ub to the acceptor site of the UBC13-MMS2/UEV1a complex for tri-Ub chain formation should be a step to be potentially inhibited by OTUB1.
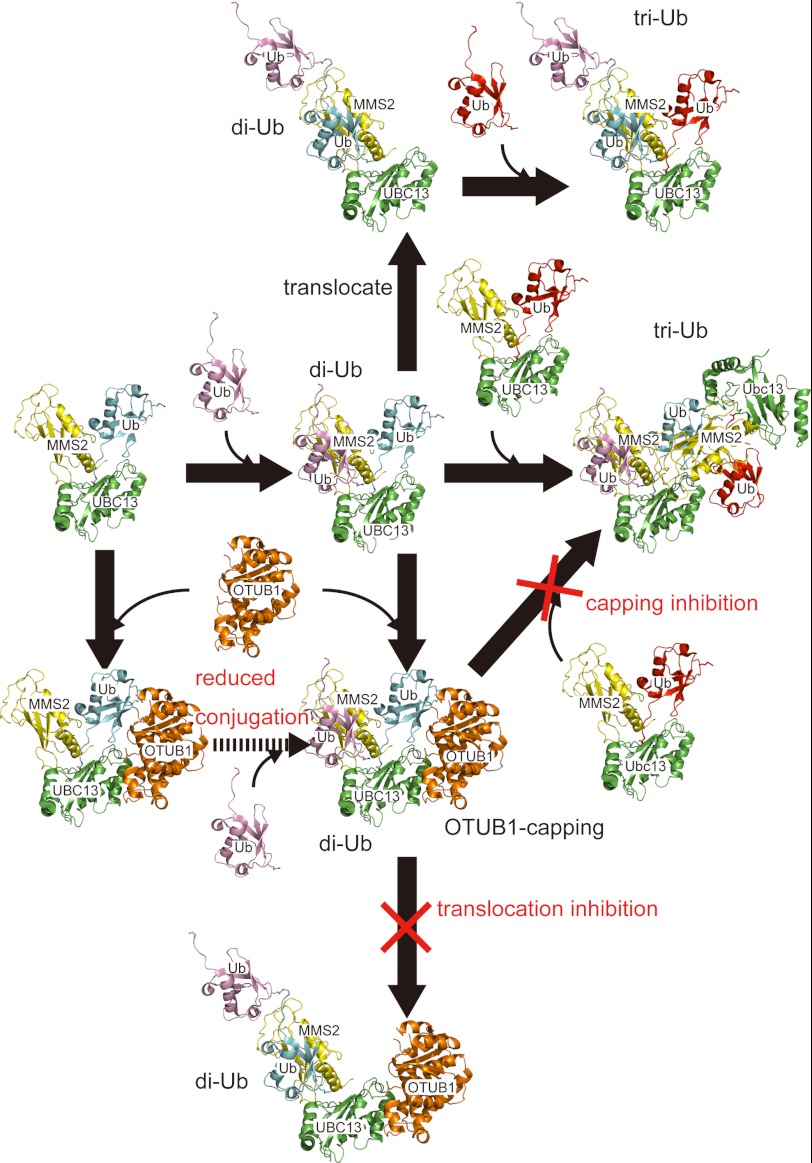
A docking model of the OTUB1-UBC13∼Ub(donor)-MMS2-Ub(acceptor) quaternary complex, which was built by superposing the present OTUB1-ΔN-UBC13-MMS2 complex on the reported UBC13∼Ub-MMS2 and Ub∼UBCH5b-OTUB1-Ub complexes (Cα root mean square deviation values of 2.65 and 2.60 Å over 280 and 362 residues, respectively) enabled us to propose the following models for the inhibition of Lys-63 chain synthesis by OTUB1 (Fig. 6). The OTUB1-UBC13-MMS2 ternary complex remains on the nascent di-Ub in the elongation cycle of Lys-63 chain synthesis, likely due to the enhanced affinity through the interaction with the charged Ub and behaves as a capping protein for the nascent di-Ub. This capping can sterically block the binding of the acceptor Ub at the distal terminus to the acceptor site of a new UBC13∼Ub-MMS2 complex. Additionally, the modeled N-terminal helix of OTUB1 intervenes between the distal and proximal Ub moieties of the nascent di-Ub so as to prevent the translocation of the di-Ub chain on the same UBC13-MMS2/UEV1a complex, thus further blocking the next Ub transfer from E1 to UBC13, for the next round of the isopeptide formation. These capping and translocation inhibition models postulate that the concentration of OTUB1 should be comparable to that of the Ub-charged UBC13, which has been shown by Durocher and co-workers (23). OTUB1 is more abundant than UBC13 in U2OS and 293T cells.
FIGURE 6.
Capping and translocation inhibitions of Lys-63 chain synthesis by OTUB1. Drawing schemes are the same as in Fig. 1. Ub are colored cyan, pink, and red, respectively.
In OTUB1-overexpressing cells, monoubiquitination of histones during DNA damage response could be detected, but diubiquitination could not, possibly due to an intrinsic high DUB activity in cells. On the other hand, OTUB1-E2 complex structures illustrate that OTUB1 binding surfaces of E2s overlap with E3 RING binding surfaces, implying multivalent inhibition strategies for regulating DNA damage responses. Further discussion awaits future studies on E2-E3 relationships and roles of specific types and/or lengths of Ub chains during DNA damage responses, which still remain controversial.
Supplementary Material
This work was supported by KAKENHI from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) Japan (to S. F., Y. S., and S. N.), the Promotion of Environmental Improvement for Independence of Young Researchers “Kanrinmaru Project” from MEXT (to S. N.), and grants from Takeda Science Foundation (to S. N.), Kato Memorial Bioscience Foundation (to S. N.), and The Nakajima Foundation (to S. F.).

This article contains supplemental S1–S6.
The atomic coordinates and structure factors (code 3VON) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Ub
- ubiquitin
- DSB
- double-strand break
- DUB
- deubiquitinating enzymes.
REFERENCES
- 1. Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway. Destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 2. Grabbe C., Husnjak K., Dikic I. (2011) The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 12, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hofmann R. M., Pickart C. M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 4. VanDemark A. P., Hofmann R. M., Tsui C., Pickart C. M., Wolberger C. (2001) Molecular insights into polyubiquitin chain assembly. Crystal structure of the Mms2/Ubc13 heterodimer. Cell 105, 711–720 [DOI] [PubMed] [Google Scholar]
- 5. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 6. Kim H., Chen J., Yu X. (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 7. Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kinner A., Wu W., Staudt C., Iliakis G. (2008) Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36, 5678–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan J., Jetten A. M. (2008) RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett. 271, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 12. Panier S., Durocher D. (2009) Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair 8, 436–443 [DOI] [PubMed] [Google Scholar]
- 13. Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 14. Sims J. J., Cohen R. E. (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin binding preference of rap80. Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sato Y., Yoshikawa A., Mimura H., Yamashita M., Yamagata A., Fukai S. (2009) Structural basis for specific recognition of Lys-63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 18. Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Orchestration of the DNA damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stewart G. S., Stankovic T., Byrd P. J., Wechsler T., Miller E. S., Huissoon A., Drayson M. T., West S. C., Elledge S. J., Taylor A. M. (2007) RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 16910–16915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains. Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 21. Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y. C., O'Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A. C., Natsume T., Suda T., Durocher D. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 [DOI] [PubMed] [Google Scholar]
- 22. Wiener R., Zhang X., Wang T., Wolberger C. (2012) The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Juang Y. C., Landry M. C., Sanches M., Vittal V., Leung C. C., Ceccarelli D. F., Mateo A. R., Pruneda J. N., Mao D. Y., Szilard R. K., Orlicky S., Munro M., Brzovic P. S., Klevit R. E., Sicheri F., Durocher D. (2012) OTUB1 co-opts Lys-48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 26. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr D Biol. Crystallogr 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Natsume T., Yamauchi Y., Nakayama H., Shinkawa T., Yanagida M., Takahashi N., Isobe T. (2002) A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 74, 4725–4733 [DOI] [PubMed] [Google Scholar]
- 29. Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I., Aebersold R., Raught B., Gingras A. C. (2009) A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics 8, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.